Abstract
Huntington’s disease (HD) is caused by a CAG triplet repeat expansion in exon 1 of the Huntingtin (Htt) gene, encoding an abnormal expanded polyglutamine (polyQ) tract that confers toxicity to the mutant Htt (mHtt) protein. Recent data suggest that posttranslational modifications of mHtt modulate its cytotoxicity. To further understand the cytotoxic mechanisms of mHtt, we have generated HEK293 cell models stably expressing Strep- and FLAG-tagged Htt containing either 19Q (wild type Htt), 55Q (mHtt), or 94Q (mHtt) repeats. Following tandem affinity purification, the tagged Htt and associated proteins were subjected to tandem mass spectrometry or 2D-nano-LC tandem mass spectrometry and several novel modification sites of mHtt containing 55Q or 94Q were identified. These were phosphorylation sites located at Ser 431 and Ser 432, and ubiquitination site located at Lys 444. The two phosphorylation sites were confirmed by Western blot analysis using phosphorylation site-specific antibodies. In addition, prevention of phosphorylation at the two serine sites altered mHtt toxicity and accumulation. These modifications of mHtt may provide novel therapeutic targets for effective treatment of the disorder.
Keywords: tandem affinity purification, mass spectrometry, post-translational modification, phosphorylation, ubiquitination
Huntington’s disease (HD) is an incurable autosomal dominant, late onset neurodegenerative disorder caused by a polyglutamine (polyQ) repeat expansion in the N-terminus of Huntingtin protein (Htt)[1]. HD develops when this polyQ expansion exceeds 35 glutamine residues[2]. The polyQ number is correlated directly with the severity of HD and is inversely correlated with the age of onset[3].
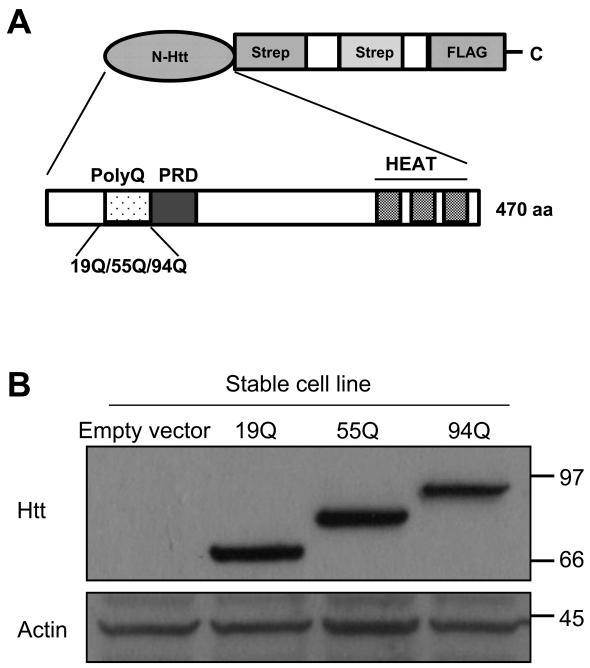
The molecular pathogenesis of expanded polyQ-induced neuronal cell death still remains poorly understood. However, recent studies have revealed that covalent posttranslational modifications of Htt play important roles in the pathogenesis of HD. For example, SUMOylation of mutant Htt (mHtt) increases its soluble form and elicits its cytotoxicity[4]. Phosphorylation of mHtt at Ser 13 and Ser 16 abolishes neurodegeneration and prevent the accumulation of mHtt aggregates[5-7]. Similarly, phosphorylation of mHtt on Ser 421 enhances its function in axonal transport and reduces its toxicity [8]. Palmitoylation of Htt at cysteine 214 is essential for its trafficking and function. Mutant Htt, with expanded polyQ, shows decreased palmitoylation, which may contribute to the accumulation of the mHtt aggregates and neurotoxicity[9]. Acetylation of mHtt at K444 facilitates trafficking of mHtt into autophagosomes for degradation and reverses the toxic effects of mHtt [10]. Thus, different modifications of mHtt protein may have distinct effects on the properties and function of the protein. To identify additional posttranslational modification sites of mHtt, we employed a novel strategy that combines tandem affinity purification (TAP) with 2D-nano-LC-MS/MS[11]. Three novel modification sites of mHtt were identified: two phosphorylation sites at Ser431and Ser432 and one ubiquitination site at Lys444. The TAP strategy was originally used in the analysis of protein-protein interaction in yeast. This technique allows to efficient identification of interacting proteins under near-physiological conditions[12]. In this study, we employed a modified TAP approach, consisting of a tandem Strep tag II and a FLAG tag (SF-TAP) to generate the Htt fusion protein constructs [11]. Since the N-terminal fragment of Htt containing the expanded polyQ is sufficient to cause neurotoxicity and disease in both animal and cell models[13, 14], we elected to introduce the N-terminal Htt fragment (470 amino acids) containing either 19Q (wild type, WT), 55Q or 94Q (mutant) into pDEST/C-SF-TAP vector having the Strep/FLAG (SF) tandem tags at its C-terminus as described previously[15] (Fig. 1A). To obtain a sufficient level of expression of the SF-Htt fusion proteins for the TAP assay, we generated stable cell lines expressing the fusion proteins. We transfected HEK293 cells with the SF-Htt constructs containing either 19Q, 55Q, or 94Q and then selected individual positively transfected colonies with G418[11]. After passaging the cells for at least two months, the cell lines that showed high level expression of the SF-Htt fusion proteins (Fig. 1B) were used in the TAP experiments.
Figure 1.
Purification of Htt protein by TAP
(A)Schematic illustration of SF-Htt construct. The N-terminal fragment of Htt including the first 470 amino acids containing either 19Q (wtHtt), 55Q, or 94Q (mHtt) was introduced into pDEST/C-SF-TAP vector containing two tandem Strep II tags and one FLAG tag at the C-terminus. PRD, proline rich domain.
(B) Western blot showing HEK293 cell lines stably overexpressing SF-Htt fusion proteins. Top panel: stable cell lines expressing SF-Htt fusion protein (anti-FLAG tag); bottom panel: loading control (anti-actin). 19Q, 55Q, and 94Q represent stable cell lines expressing SF-Htt19Q, SF-Htt55Q, and SF-Htt94Q respectively.
We next determined the efficiency of the TAP purification by incubating the supernatants from the cell lysates derived from the SF-Htt expressing cells with both Strep and FLAG superflow resin columns. The very low level of SF-tagged Htt protein in the flow-through of either the first Strep column or the second FLAG column (Supplemental Figures, S-1A, upper panel) and the presence of the fusion proteins in the TAP eluates (Supplemental Figures, S-1A, lower panel) indicate a highly efficient capture of SF-Htt fusion protein by the columns. The proteins in the final TAP eluates (19Q, 55Q, and 94Q) were separated by SDS-PAGE and the protein gel was stained with SYPRO Ruby, which allows us to visualize numerous proteins associated with the Htt containing different polyQ repeats (Supplemental Figures, S-1B). To determine the identities of these proteins, the final TAP eluates were subjected to 2D-nano-LC-MS/MS (see Supplemental Data, Tables 3a, 3b, 4a, 4b, 5a and 5b).
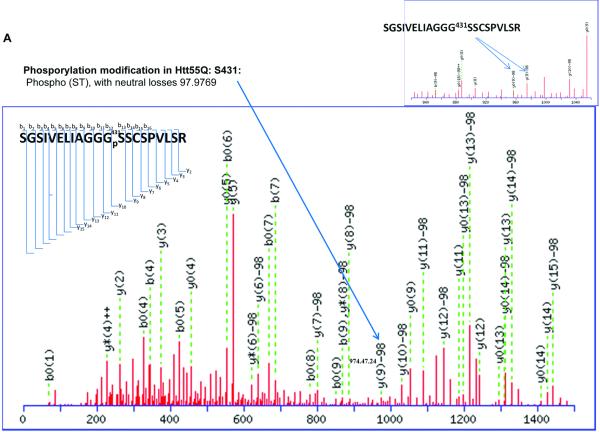
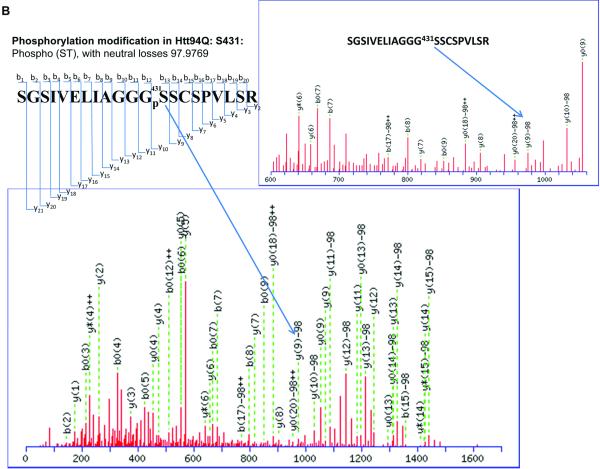
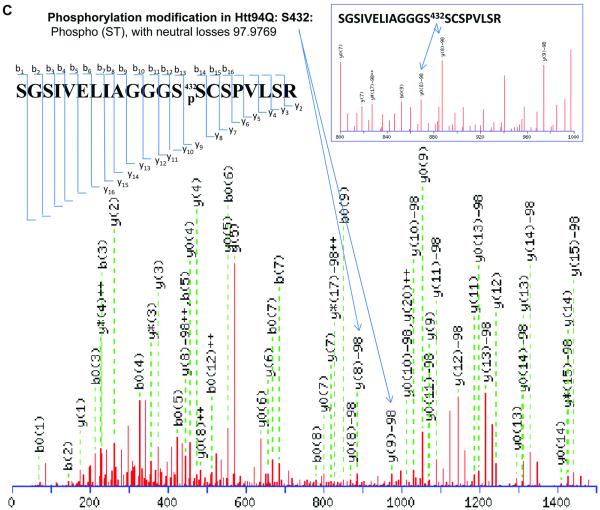
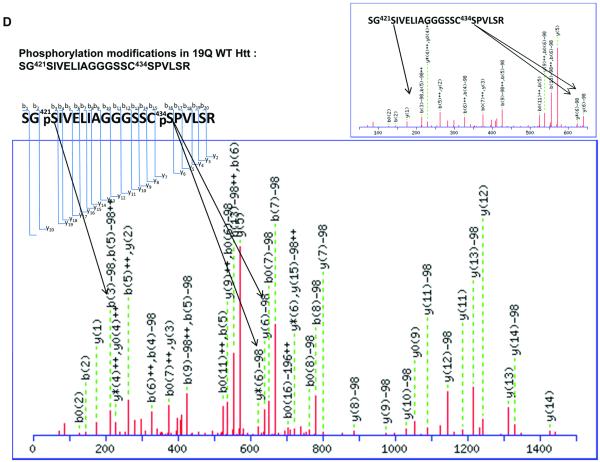
It has been reported that human Htt can be phosphorylated at Ser13, Ser 16[5-7], and Ser 421[8]. Phosphorylation at all of these sites suppresses the pathogenesis and reduces the toxicity caused by mHtt. Other previously identified phosphorylation sites of Htt include Ser 536, Ser 1181, Ser 1201, Ser 2076, Ser 2653 and Ser 2657. Phosphorylation at Ser 536 inhibits calpain cleavage and reduces mHtt toxicity[16]. Our results show two novel phosphorylation sites in Htt proteins. Phosphorylation at Ser 431 is only detected in the mHtt protein, in both Htt55Q (Fig. 2A) and Htt 94Q (Fig. 2B), whereas phosphorylation at Ser 432 is only found in Htt94Q (Fig. 2C). The MS/MS maps exhibited a clear neutral loss of 98 Da, which corresponds to the cleavage of phosphoric acid from the phosphorylated peptide ion (Figure 2A, 2B, 2C, and Supplementary Table 7, 8 and 9). However, phosphorylation at either Ser431 or Ser432 was not found in Htt19Q protein (Fig. 2D). A previous study indicated that CDK5 phosphorylates Htt at Ser434 in the presence of p35, which reduces aggregate formation and toxicity of mHtt[17]. However, thus far, there has been no report of the phosphorylation of mHtt protein at Ser 431 and Ser 432.
Figure 2.
2D-nano-LC-MS/MS identification of novel posttranslational modification sites in mHtt.
(A)2D-nano-LC-MS/MS analysis of tryptic Htt55Q protein shows phosphorylation of mHtt occurs at Ser 431.
(B) 2D-nano-LC-MS/MS analysis of tryptic Htt94Q protein shows phosphorylation of mHtt occurs at Ser 431.
(C)) 2D-nano-LC-MS/MS analysis of tryptic Htt94Q protein shows another phosphorylation site at Ser 432.
(D)2D-nano-LC-MS/MS analysis of tryptic Htt19Q protein shows no phosphorylation occurs at Ser 431 and Ser432.
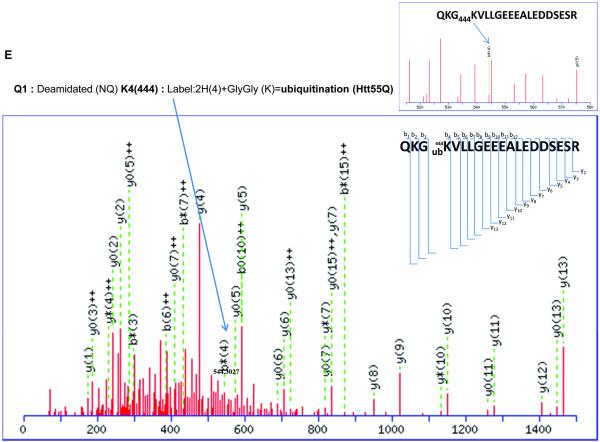
(E) 2D-nano-LC-MS/MS analysis of tryptic Htt55Q protein shows that ubiquitination occurs at Lys 444 residue.
Our results also indicate a novel ubiquitination modification (labeled 2H (4) + GlyGly) at Lys 444 in Htt55Q as our MS/MS peptide fragmentation map clearly showed an addition of 118.06 Da to Lysine 444 (Fig. 2E and Supplementary Table 6). Ubiquitination is a reversible posttranslational modification that plays important roles in the regulation of several cellular processes, including protein quality control, protein trafficking, cell cycle and transcriptional regulation[18]. Interestingly, Jeong et al recently reported that Lys444 of N-terminal mHtt containing 480 amino acids and 68Q was modified by acetylation in the presence of histone deacetylase (HDAC) inhibitors or in the condition of overexpressing a histone acetyltransferase (HAT) [10]. This difference from our observation is probably due to the fact that our experiments were performed under a normal physiological condition while most of their experiments were carried out in the presence of HDAC inhibitors or under overexpression of a HAT.
To further confirm the presence of the two phosphorylation sites, Ser431 and Ser432, we generated two phosphorylation site-specific antibodies. As shown in Fig. 3A, specificity of the antibodies was verified by dot blot. Using the antibodies, we performed Western blot analysis of proteins from the cells stably expressing SF-Htt19Q or SF-Htt94Q. As shown in Fig. 3B and 3C, Western blot results were consistent with the observations obtained from mass spectrometry, confirming that phosphorylation of Ser431 and Ser432 were present in SF-Htt94Q but not in SFHtt19Q.
Figure 3.
Verification of the two novel phosphorylation sites
(A)Verification of the specificity of anti-phosphorylation Ser431 and -Ser432 antibodies by dot blot. Increasing amount of phospho-Ser431 or phospho-Ser432 peptide and unmodified peptide were spotted onto a nitrocellulose membrane and probed with the antibody indicated on the right of each panel.
(B) & (C) Confirmation of phosphorylation at Ser431 (B) and Ser432 (C) in mHtt by Western blot using phosphorylation site-specific antibodies. The primary antibodies used were indicated at the right side of each panel.
To further examine the role of phosphorylation at Ser431 and Ser432, we generated mutant constructs by mutating serine either to phosphomimetic aspartate (SD) or to phosphoresistant alanine (SA). These mutants were transiently transfected into HEK293 cell line and then, cell viability and Htt aggregation was analyzed in the two cell lines, respectively. Compared to the cells transfected with phosphomimetic mutant, Ser431SD, the cells transfected with phosphoresistant Ser431SA plasmid showed a remarkable increase in cell viability (Supplementary Figures, S-2) and reduction of Htt accumulation (Supplemental Figures, S-3A, S-3B and S-3C). In contrast, expression of SF-Htt94 mutant containing Ser432SA mutation reduced cell viability (Supplemental Figures, S-2) but enhanced accumulation of the protein (Supplemental Figures, S3A, 3B and 3C). These data indicate that phosphorylation at Ser431 and Ser432 not only modulates mHtt toxicity but also influences its degradation.
In summary, we have identified three novel posttranslational modification sites in mHtt, phosphorylation at Ser431 and Ser 432, and ubiquitination at K444, using a TAP strategy combined with 2D-Nano-LC-MS/MS. The two novel phosphorylation sites were confirmed by Western blot using phosphorylation site-specific antibodies. Functional studies indicated that prevention of phosphorylation at the Ser431 and Ser432 altered mHtt toxicity and aggregation. Since these posttranslational modification sites are located in the same short region that is involved in Htt cleavage, aggregate formation and trafficking[19], they may act as novel molecular switches to regulate mHtt toxicity.
Supplementary Material
Acknowledgments
We would like to thank Dr. Robin Miskimins for critical reading of the manuscript and Drs.Marian DiFiglia, Shihua Li and Xiao-Jiang Li for providing huntingtin constructs. This work was supported by Start-up Funds from the University of South Dakota (HW), a Competitive Research Grant Award of the South Dakota Board of Regents (HW), and a New Faculty Development Award of the University of South Dakota (HW). The Proteomics Core Facility at the University of South Dakota was supported by NIH Grant Number 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources.
Abbreviations
- HD
Huntington’s disease
- CAG
cytosine, adenine, guanine
- Htt
Huntingtin
- PolyQ
polyglutamine
- mHtt
mutant Huntingtin
- K
lysine
- SUMO
small ubiquitin-like modifier
- TAP
tandem affinity purification
- 2D-nano-LC-MS/MS
two-dimensional-nano-liquid chromatography tandem mass spectrometry
- Ser
serine
- Lys
lysine
- SF
Strep/Flag
- Gly
glycine
- HDAC
histone deacetylase
- HAT
histone acetyltransferase
- SD
serine aspartate
- SA
serine alanine
Footnotes
Conflict of interest statement The authors have declared no conflict of interest.
References
- [1].A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- [2].Lombardi MS, Jaspers L, Spronkmans C, Gellera C, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- [3].Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- [4].Steffan JS, Agrawal N, Pallos J, Rockabrand E, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- [5].Gu X, Greiner ER, Mishra R, Kodali R, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aiken CT, Steffan JS, Guerrero CM, Khashwji H, et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zala D, Colin E, Rangone H, Liot G, et al. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Human molecular genetics. 2008;17:3837–3846. doi: 10.1093/hmg/ddn281. [DOI] [PubMed] [Google Scholar]
- [9].Yanai A, Huang K, Kang R, Singaraja RR, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jeong H, Then F, Melia TJ, Jr., Mazzulli JR, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gloeckner CJ, Boldt K, Schumacher A, Ueffing M. Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Methods Mol Biol. 2009;564:359–372. doi: 10.1007/978-1-60761-157-8_21. [DOI] [PubMed] [Google Scholar]
- [12].Rigaut G, Shevchenko A, Rutz B, Wilm M, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- [13].Mangiarini L, Sathasivam K, Seller M, Cozens B, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- [14].Ratovitski T, Gucek M, Jiang H, Chighladze E, et al. Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J Biol Chem. 2009;284:10855–10867. doi: 10.1074/jbc.M804813200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dong G, Callegari EA, Gloeckner CJ, Ueffing M, Wang H. Prothymosin-alpha Interacts with Mutant Huntingtin and Suppresses Its Cytotoxicity in Cell Culture. J Biol Chem. 2012;287:1279–1289. doi: 10.1074/jbc.M111.294280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schilling B, Gafni J, Torcassi C, Cong X, et al. Huntingtin phosphorylation sites mapped by mass spectrometry. Modulation of cleavage and toxicity. J Biol Chem. 2006;281:23686–23697. doi: 10.1074/jbc.M513507200. [DOI] [PubMed] [Google Scholar]
- [17].Luo S, Vacher C, Davies JE, Rubinsztein DC. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J Cell Biol. 2005;169:647–656. doi: 10.1083/jcb.200412071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kimura Y, Yashiroda H, Kudo T, Koitabashi S, et al. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell. 2009;137:549–559. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- [19].Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.