Abstract
Activation of xenobiotic metabolism pathways has been linked to lifespan extension in different models of aging. However, the mechanisms underlying activation of xenobiotic genes remain largely unknown. Here we showed that although FXR mRNA levels do not change significantly, FXR (farnesoid X receptor, Nr1h4) protein levels are elevated in the livers of the long-lived Little mice, leading to increased DNA binding activity of FXR. Hepatic FXR expression is sex-dependent in wild-type mice but not in Little mice, implying that up-regulation of FXR might be dependent on the reduction of growth hormone in Little mice. Growth hormone treatment decreased hepatic expression of FXR and xenobiotic genes Abcb1a, Fmo3 and Gsta2 in both wild-type and Little mice, suggesting an association between FXR and xenobiotic gene expression. We found that Abcb1a is transactivated by FXR via direct binding of FXR/retinoid X receptor α (RXRα) heterodimer to a response element at the proximal promoter. FXR also positively controls Fmo3 and Gsta2 expression through direct interaction with the response elements in these genes. Our study demonstrates that xenobiotic genes are direct transcriptional targets of FXR and suggests that FXR signaling may play a critical role in the lifespan extension observed in Little mice.
Keywords: FXR, xenobiotic detoxification gene, Little mice, growth hormone
1. Introduction
Little mice (Ghrhrlit/lit) are a very useful mammalian model for studying aging because of their retarded aging and extended lifespan. These mice have a mutation in the gene Ghrhr (growth hormone-releasing hormone receptor) and correspondingly have very low levels of circulating growth hormone (GH) and insulin-like growth factor1 (IGF1) (Donahue and Beamer, 1993; Godfrey et al., 1993). The GH/IGF1 pathway has been associated with lifespan extension in several species including C.elegans, Drosophila melanogaster, and Mus muscus and is the focus of several studies to understand the beneficial aspects of this pathway on longevity (Berryman et al., 2008). Our previous studies have proposed that alterations in xenobiotic metabolism and increased xenobiotic resistance may contribute to the longevity in Little mice (Amador-Noguez et al., 2004, 2007). Genetic studies showed that the up-regulation of xenobiotic detoxification genes is likely to be mediated by the nuclear receptor FXR (Amador-Noguez et al., 2007). Levels of primary bile acids, the endogenous ligands for FXR, are elevated in Little mice and treatment of wild-type mice with cholic acid mimics the up-regulation of xenobiotic detoxification genes observed in Little mice (Amador-Noguez et al., 2007). We further found that knockout of FXR in Little mice reverses or decreases the up-regulation of these genes (Amador-Noguez et al., 2007). However, the mechanism(s) by which FXR regulates these genes remained unclear.
FXR is a member of the nuclear receptor superfamily and is expressed in liver, small intestine, kidney, adrenals, adipose tissue and vascular smooth muscle (Calkin and Tontonoz, 2012; Modica et al., 2010; Wang et al., 2008). FXR has been shown to control expression of various genes in bile acid, lipid, and glucose metabolism (Modica et al., 2010). Upon activation by its natural ligands, such as bile acids and their metabolites, or synthetic agonists including GW4064, FXR regulates the expression of its target genes by binding either as a monomer or as a heterodimer with RXRα to FXR response elements (FXREs) (Calkin and Tontonoz, 2012; Modica et al., 2010; Wang et al., 2008). The typical FXRE is an inverted repeat of the AGGTCA half-site spaced by 1 nucleotide (IR1). Other FXREs include direct repeat (DR), everted repeat (ER) and monomeric binding sites (Modica et al., 2010; Wang et al., 2008). In addition to regulation of target genes via binding to FXREs, FXR represses a group of genes indirectly via the FXR/SHP (small heterodimer partner) pathway (Calkin and Tontonoz, 2012; Goodwin et al., 2000; Li et al., 2005; Lu et al., 2000). Recently, several coactivators of FXR, including PGC-1α, SRC-1, Brg-1, CARM1, PRMT1, GPS2, DRIP205 and TRRAP, have been reported to interact with FXR and enhance FXR-mediated transactivation of different target genes (Ananthanarayanan et al., 2004; Kemper, 2011; Miao et al., 2009; Pineda Torra et al., 2004; Rizzo et al.,2005; Sanyal et al., 2007; Unno et al., 2005; Wang et al., 2006; Zhang et al., 2004), while Ku proteins are identified as FXR corepressors (Ohno et al., 2009).
Our previous study has shown that the loss of FXR, rather than the classic xenobiotic receptors Car (Constitutive Androstane receptor) and Pxr (Pregnane X receptor), had a major influence on the up-regulation of xenobiotic detoxification genes in Little mice (Amador-Noguez et al., 2007). The up-regulation of Abcb1a, Aldh1a1, Cyp2b10, Cyp2c38, Cyp4a10, Fmo3, Gsto2, Gstt2, Papp2s, Por, Sult1d1, Temt, and Ugt1a1 was abolished in the Fxr−/−/Little double deficient mice (Amador-Noguez et al., 2007). Knockout of FXR also reduced the magnitude of the up-regulation of Cyp2b13, Cyp2b9, Cyp4a14 and mOat6 (Amador-Noguez et al., 2007). Interestingly, several genes, including Gsta2, Gstm2, Gstm3, Mt1, and Sult1e1, were more strongly up-regulated in the Fxr−/−/Little mice than in Little mice (Amador-Noguez et al., 2007). The majority of these genes were not known targets of FXR and whether they were directly regulated by FXR was not clear. A recent ChIP-seq study analyzed genome-wide FXR binding in mice and revealed potential FXR binding sites in several of these xenobiotic detoxification genes up-regulated in Little mice (Thomas et al., 2010). Therefore, we hypothesize that these genes might be directly regulated by FXR via binding to these FXREs. In this study, we found that FXR protein levels are elevated in the livers of Little mice of different ages. We identified novel FXREs in three xenobiotic detoxification genes belonging to different phases, Fmo3 (phase 1), Gsta2 (phase 2), and Abcb1a (phase 3), and revealed that FXR activates these genes via direct binding to these regulatory elements as a heterodimer with RXRα. EMSA and ChIP experiments showed that FXR binding is increased in Little mice compared to that in wild type mice. On the other hand, growth hormone treatment reduced the expression levels of FXR and several xenobiotic detoxification genes in Little mice. This study elucidated the mechanism by which FXR: RXRα heterodimer regulates xenobiotic detoxification genes in Little mice and suggested that regulation of FXR by the GH/IGF1 pathway may play a critical role in the lifespan extension observed in this mouse strain.
2. Materials and Methods
2.1 Antibodies and reagents
Antibodies to FXR (H-130 and C-20), Lamin A (H-102) and RXRα (D-20) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal FXR antibody (clone A9033A) was purchased from Perseus Proteomics (Tokyo, Japan). Anti-p-Glycoprotein (C219) antibody was purchased from Calbiochem (San Diego, CA). Mouse monoclonal β-actin antibody, chenodeoxycholic acid, GW4064 and LG100268 were purchased from Sigma-Aldrich (St. Louis, MI). Mouse recombinant growth hormone was purchased from Dr. A. F. Parlow, National Hormone and Peptide Program. Charcoal-stripped fetal bovine serum (FBS) was purchased from Invitrogen (Carlsbad, CA).
2.2 Mice and treatments
Little mice are described in our previous papers (Amador-Noguez et al., 2004; 2007). All animal protocols followed the provisions of the Animal Welfare Act and were approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine. For growth hormone treatment experiments in male Little mice, mice were subcutaneously injected with mouse recombinant growth hormone (2mg/kg) once daily for 10 days. For growth hormone treatment of 24-month old female wild-type and Little mice, mouse recombinant growth hormone (2mg/kg) was injected into mice once daily for 7 days.
2.3 Cell culture and treatment with FXR ligands
Hepa1-6 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin at 37 and 5% CO2. Hep3B2 cells were maintained in Eagle's Minimum Essential Medium with 10% (v/v) FBS and 1% penicillin/streptomycin. For treatments with FXR ligands, Hepa1-6 cells were cultured in 65mm dishes. Forty eight hours after seeding, cells were treated with indicated concentrations of compounds in DMEM supplemented with 10% (v/v) charcoal-stripped FBS for indicated periods.
2.4 Preparation of nuclear extracts and whole cell lysates
Nuclear extracts from mouse livers or cell cultures were prepared using Nuclear Extract Kit (Active Motif, Carlsbad, CA) according to the instruction manual. Whole cell lysates were prepared with RIPA buffer. Protein concentrations were determined with BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL).
2.5 qRT-PCR
Total RNA from liver tissues or Hepa1-6 cells was extracted with RNeasy mini kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. cDNA was synthesized using SuperScript III First-strand kit (Invitrogen) and random primer hexamers. Quantitative PCR was performed using Brilliant II SYBR Green qPCR master mix kit (Agilent Technologies, Palo Alto, CA). Sequences of primers used in these studies are presented in Supplementary Table 1.
2.6 Plasmid construction and luciferase reporter assay
Three different fragments containing the potential FXREs in the genes of Abcb1a (736bp, position −500 to +236 relative to the transcription start site), Fmo3 (1183bp, +18149 to +18683) and Gsta2 (1131bp, −1840 to −709) were amplified using mouse genomic DNA as template and inserted onto pGL3-basic luciferase vector respectively. The Abcb1a promoter mutant was generated using a site-directed mutagenesis approach. Sequences of primers used are presented in Supplementary Table 2. These gene reporter constructs were co-transfected with FXR or RXRα expression plasmids as indicated into Hep3B2 cells using FuGENE HD Transfection Reagent (Roche Applied Science, Basel, Switzerland) respectively. 24 h after transfection, cells were treated with different ligands for FXR or RXRα in fresh media containing 10% charcoal-stripped FBS for 24 hours. Luciferase activity was determined with Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and values were normalized by Renilla luciferase activity.
2.7 Electrophoresis mobility shift assay (EMSA)
Double-stranded oligonucleotides were labeled with [ α −32P] dCTP using Klenow enzyme (Roche). Binding reactions containing nuclear extracts isolated from liver tissues and labeled probes were performed for 30 min on ice. For supershift assay, nuclear extracts were incubated with FXR antibody (C-20 or H-130) or normal rabbit IgG for 15 min on ice prior to addition of labeled probes. Protein-DNA complexes were separated on 6% polyacrylamide gels. The sequences of the probes used are in Supplementary Table 3.
2.8 Chromatin immunoprecipitation assay (ChIP)
ChIP assay was performed with ChIP-IT Kit (Motif) according to the instruction manual. Briefly, mouse liver tissues were fixed in formaldehyde before being quenched with glycine. The nuclei were extracted and sonicated with a Bioruptor UCD-200 (Diagenode) to yield 500 base pair (bp) DNA fragments. FXR (H-130) antibody or RXRα (D-20) antibody was used for immunoprecipitation. The captured chromatins were analyzed by semi-quantitative PCR. The sequences of the primers used for PCR amplification are in Supplementary Table 4.
2.9 Statistical analysis
All data are presented as mean ± SD, and the Student’s t test was used to calculate p values. P values less than 0.05 were considered statistically significant.
3. Results
3.1 Levels of FXR protein are elevated in the livers of Little mice and growth hormone treatment reduces the expression levels of FXR and xenobiotic detoxification genes
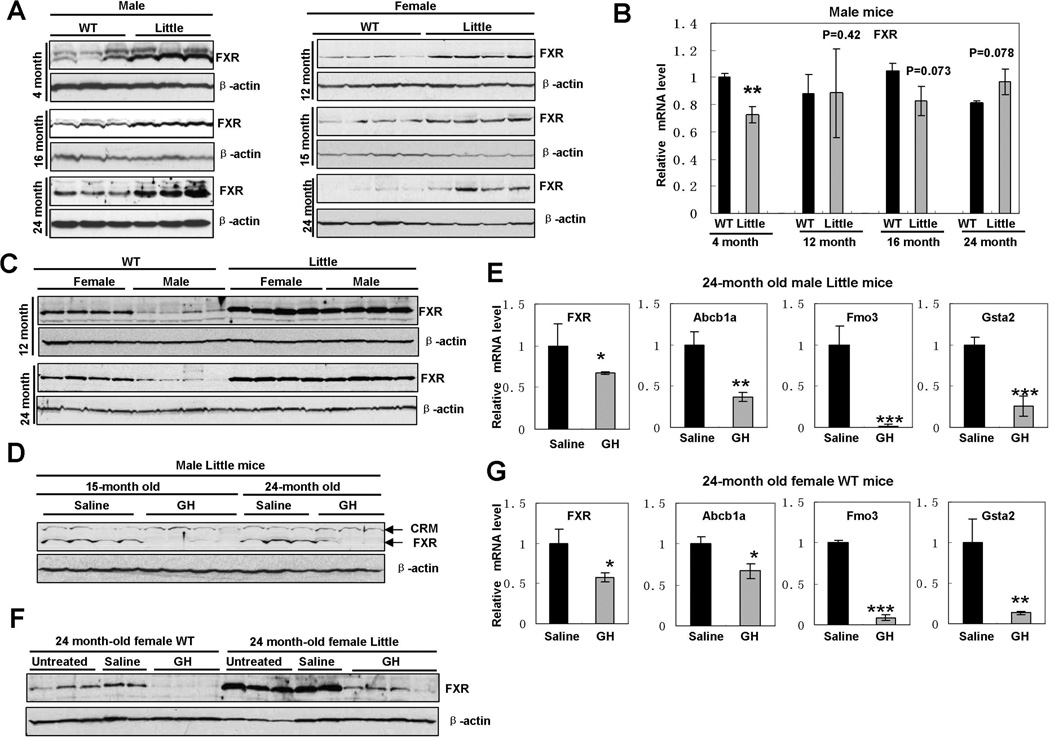
Because crossing Little mice to FXR knockout mice corrected the expression levels of xenobiotic detoxification genes (Amador-Noguez et al., 2007), we have performed a careful analysis of the expression of FXR in Little mice. We found that FXR protein was increased in the livers of both male and female Little mice of different ages (Fig. 1A and Fig. S1A). However, we did not observe a significant change of the levels of FXR mRNA between wild type and Little mice (Fig. 1B), whereas the mRNA levels of Fmo3, a putative downstream target of FXR, were significantly elevated in these Little mice (Fig. S1B). Since gender differences in the expression of hepatic genes have been reported (Waxman and Holloway, 2009), we next examined the effects of gender on hepatic FXR protein levels. As shown in Fig.1C, hepatic FXR is reduced in male wild-type mice compared to that in female counterparts, whereas there is no significant difference in hepatic FXR between male and female Little mice. These results suggested that the elevation of FXR protein in Little mice is dependent on the reduction of growth hormone but independent of gender, since both male and female Little mice have very low levels of circulating growth hormone compared to their wild-type controls. To further determine whether growth hormone regulates FXR expression and signaling, we treated male Little mice with mouse recombinant growth hormone for 10 days and then examined the expression of FXR in the livers. Growth hormone treatment markedly reduced FXR protein levels and nuclear accumulation in the livers of Little mice (Fig. 1D and Fig. S1C). We also observed a small reduction (∼30%) of FXR mRNA levels after growth hormone treatment (Fig 1E), implying that the GH:IGF1 pathway might affect FXR expression either directly or indirectly. As shown in Fig. 1E, growth hormone treatment significantly reduced the mRNA levels of Abcb1a, Fmo3 and Gsta2 in Little mice, indicating that the down-regulation of these xenobiotic detoxification genes by GH:IGF1 pathway is associated with reduction of FXR. We further found that growth hormone treatment also down-regulates hepatic expression of FXR and the xenobiotic detoxification genes in female wild-type mice (Fig. 1F and 1G). Taken together, these results suggested that growth hormone signaling controls the expression and signaling of FXR in Little mice.
Fig. 1. Elevation of FXR in the livers of Little mice and reduction of expression of FXR and xenobiotic detoxification genes by growth hormone treatment.
A. Elevation of FXR protein in the livers of Little mice of different ages. Whole cell protein extracts were isolated from the livers of male or female wild-type and Little mice of different ages and immunoblotted with rabbit polyclonal FXR antibody (H-130).
B. Levels of FXR mRNA in the livers of male wild-type and Little mice of different ages. Total RNA was isolated from the livers of male wild-type (n=3 per group) and Little (n=3 per group) mice of different ages. The mRNA levels for FXR were analyzed using quantitative RT-PCR and normalized to the internal control β-actin. (**p<0.01)
C. Gender effects on hepatic FXR protein levels in wild-type and Little mice. Whole cell protein extracts were isolated from the livers of male or female wild wild-type and Little mice of indicated ages and immunoblotted with rabbit polyclonal FXR antibody (H-130).
D. Growth hormone treatment decreased the levels of FXR protein in the livers of male Little mice. Whole cell protein extracts were isolated from the livers of control (saline-treated) and GH-treated Little mice respectively and immunoblotted with rabbit polyclonal FXR antibody (H-130). CRM = cross reacting material.
E. Analysis of gene expression in the livers of control and GH-treated male Little mice by quantitative RT-PCR. Total RNA was isolated from the livers of 24-month old control (saline-treated) (n=3) and GH-treated (n=3) male Little mice and quantitative RT-PCR was performed to measure the mRNA levels for FXR and three putative FXR target genes: Abcb1a, Fmo3 and Gtsa2. (*p<0.05, **p<0.01, ***p<0.001)
F. Growth hormone treatment decreased the levels of FXR protein in the livers of female wild-type mice. Whole cell protein extracts were isolated from the livers of untreated, control (saline-treated) and GH-treated female wild-type or Little mice respectively and immunoblotted with rabbit polyclonal FXR antibody (H-130).
G. Analysis of gene expression in the livers of control and GH-treated female wild-type mice by quantitative RT-PCR. Total RNA was isolated from the livers of 24-month old control (saline-treated) (n=2) and GH-treated (n=3) female wild-type mice and quantitative RT-PCR was performed to measure the mRNA levels for FXR and three putative FXR target genes: Abcb1a, Fmo3 and Gtsa2. (*p<0.05, **p<0.01, ***p<0.001)
3.2 Activation of FXR by ligands induces Abcb1a expression in vitro
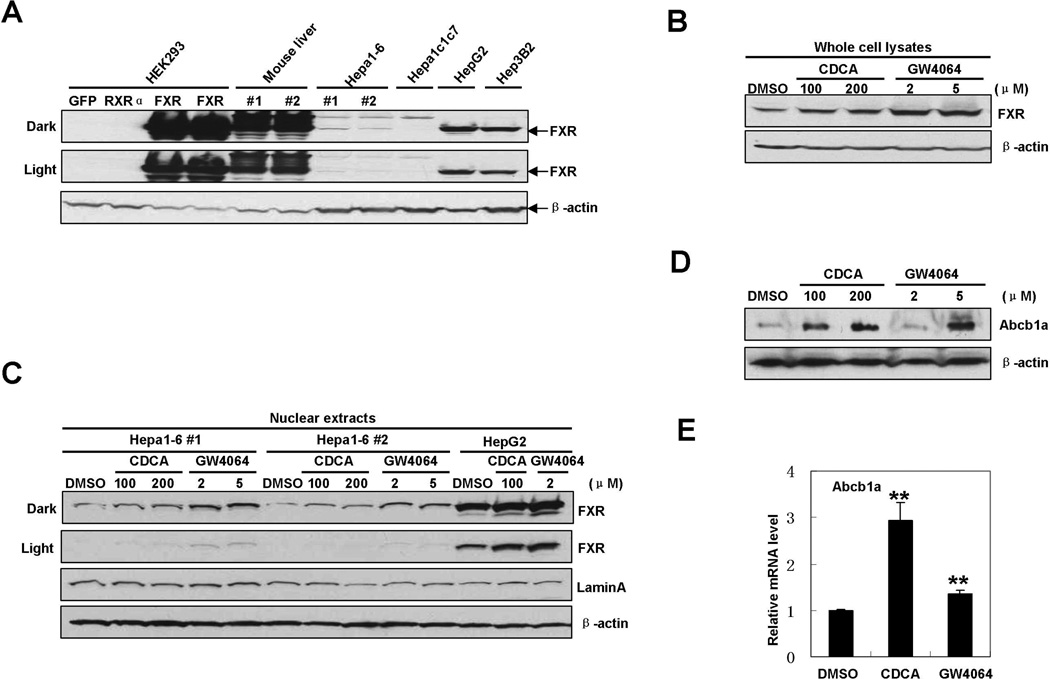
We next sought to determine the direct effects of FXR signaling on the expression of xenobiotic genes using in vitro cell models. Abcb1a, also named MDR1, is one member of a family of ATP-binding proteins that export various xenobiotic and endogenous compounds from the liver (Wu et al., 2009). Our previous study showed that the level of Abcb1a mRNA in Little mice was increased 4.4-fold as compared to that in wild type mice and knockout of FXR in Little mice reduced the level of Abcb1a mRNA to only 0.5-fold of that in wild type mice, indicating that the alteration of Abcb1a expression is dependent on the presence of FXR (Amador-Noguez et al., 2007). Therefore we decided to use Abcb1a as a representative of those FXR-sensitive genes to investigate the mechanisms by which FXR regulates xenobiotic detoxification genes in Little mice. We utilized a mouse hepatoma cell line, Hepa1-6, to evaluate the impact of ligand-induced FXR activation on Abcb1a expression in vitro. We first determined whether FXR protein is expressed in Hepa1-6 cells. Using two different antibodies against FXR, we found that FXR protein was expressed at very low levels in Hepa1-6 cells compared to the levels of FXR in wild type mouse livers or human hepatoma cell lines HepG2 and Hep3B (Fig. 2A and Supplement Fig. S2). Interestingly, FXR was not detectable in a subclone Hepa1c1c7. This result is consistent with a recent report that very low levels of FXR mRNA were detected in Hepa1-6 cells compared with mouse livers (Perez et al., 2011). Treatment with the endogenous FXR ligand, CDCA, or the synthetic agonist, GW4064, enhanced expression and nuclear accumulation of FXR protein in Hepa1-6 cells (Fig. 2B and C), consistent with the previous observations in human hepatoma HepG2 cells, breast cancer MCF-7 cells, rat aortic artery smooth muscle cells and rabbit that FXR ligands increased FXR expression through an auto-regulatory loop (Cui et al., 2003; Journe et al., 2009; Lew et al., 2004; Zhang et al., 2008; Xu et al., 2002). Consequently, Abcb1a protein was increased by treatment of CDCA or GW4064 in a dose-dependent manner (Fig. 2D). Abcb1a mRNA was also increased after a 16-hour treatment with FXR ligands in Hepa1-6 cells (Fig. 2E), indicating that FXR might regulate Abcb1a expression at the transcriptional level. These observations are consistent with our previous data showing that treatment of wild-type mice with cholic acid significantly up-regulated the expression of 23 xenobiotic genes including Abcb1a (Amador-Noguez et al., 2007).
Fig.2. Activation of FXR by ligands increases Abcb1a expression in vitro.
A. Western blot analysis of FXR expression in mouse livers, murine and human cell lines. Whole cell lysates were prepared from mouse livers, murine or human hepatoma cells respectively, and immunoblotted with mouse monoclonal FXR antibody (Clone A9033A). Whole cell lysates from HEK293 cells transfected with plasmids expressing GFP, murine RXRα, or FXR proteins respectively are used as controls.
B. Activation of FXR by ligands increased FXR expression in Hepa1-6 cells. Hepa1-6 cells were incubated with indicated concentrations of CDCA, GW4064 or vehicle (DMSO) for 24 hours. Whole cell lysates were prepared and immunoblotted with mouse monoclonal FXR antibody (Clone A9033A).
C. Activation of FXR by ligands increased nuclear accumulation of FXR protein in Hepa1-6 cells. Two different batches of Hepa1-6 cells were incubated with indicated concentrations of CDCA, GW4064 or vehicle (DMSO) for 24 hours respectively. Nuclear extracts were prepared and immunoblotted with mouse monoclonal FXR antibody (Clone A9033A). Nuclear extracts from ligand-treated HepG2 cells were used as positive control.
D. Activation of FXR by ligands increased the protein level of Abcb1a in Hepa1-6 cells. Hepa1-6 cells were incubated with indicated concentrations of CDCA, GW4064 or vehicle (DMSO) for 24 hours. Whole cell lysates were prepared and immunoblotted with indicated antibodies.
E. Activation of FXR by ligands increased the mRNA level of Abcb1a in Hepa1-6 cells. Hepa1-6 cells were incubated with CDCA (200µM), GW4064 (5µM), or vehicle (DMSO) for 16 hours. Levels of Abcb1a mRNA were measured by a quantitative RT-PCR. β-actin mRNA was used as an internal control. (**p<0.01 )
3.3 Abcb1a is a direct transcriptional target of FXR
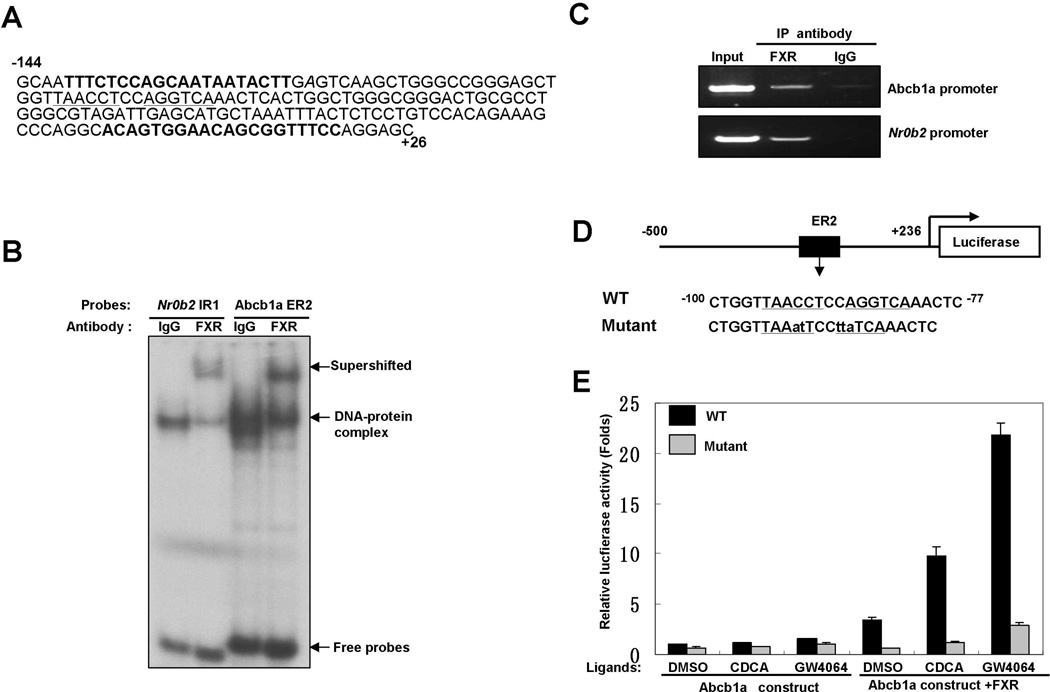
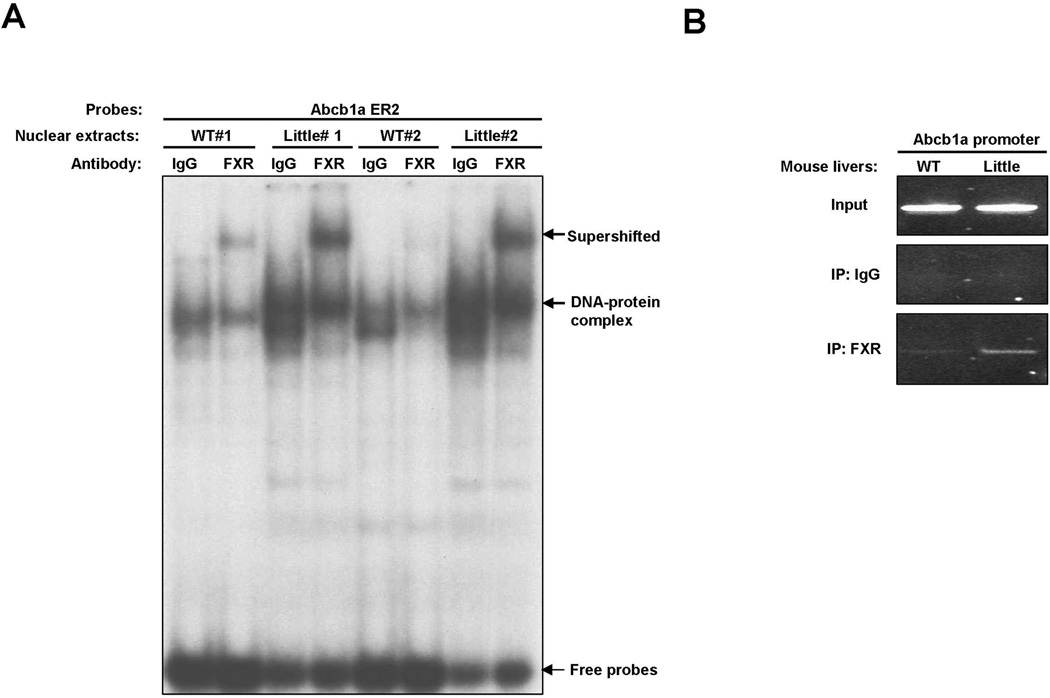
The genome-wide ChIP-seq study for FXR binding in mouse liver and intestine discovered a possible FXR binding site in the 5’-proximal region of Abcb1a gene (Thomas et al., 2010), leading us to hypothesize that FXR might regulate Abcb1a expression through direct binding to Abcb1a promoter. Analysis of the nucleotide sequences in 5’-proximal region of Abcb1a gene with NUBIScan (Podvinec et al., 2002) revealed a potential FXRE, an everted repeat separated by two base pairs (ER2: TAACCTccAGGTCA, score: 0.924571, P=0.000652445), located at base pairs −95 to −82 relative to the transcriptional start site (TSS) (Fig. 3A). As shown in Fig. 3B and 3C, EMSA and ChIP experiments confirmed that FXR directly bound to this site. A known FXRE IR1 in Nr0b2 (−320 to −220 bp upstream of TSS) was used as a positive control in these experiments (Chanda et al., 2008; Goodwin et al., 2000; Thomas et al., 2010). Subsequently, we cloned a 736-bp fragment containing this ER2 into pGL3-basic luciferase vector and generated a mutant construct that abrogates FXR binding (Fig. 3D). As shown in Fig. 3E, either CDCA or GW4064 treatment slightly induced the luciferase activity of Abcb1a promoter construct. Over-expression of exogenous FXR significantly increased the luciferase activity and dramatically amplified ligand-induced activity of the wild type construct. In contrast, both activation of FXR by ligands and over-expression of exogenous FXR had no significant effect on the luciferase activity of the mutant construct. Taken together, these results indicated that FXR regulates the expression of Abcb1a on the transcriptional level through direct binding to an ER2 element in the promoter region.
Fig.3. Abcb1a is a direct transcriptional target of FXR.
A. The nucleotide sequence of the mouse Abcb1a locus from base pair −144 (first base pair upstream of the transcription start site is numbered −1) to +26. The proposed ER2 (underlined) is located at base pairs −95 to −82. The sequences for the primers used in the following ChIP assay are highlighted in bold.
B. FXR bound to Abcb1a promoter in vitro. EMSAs were performed with nuclear extracts from livers of Little mice. FXR antibody (C-20) was used for the supershift assay to confirm binding specificity. A probe containing a known FXRE IR1 found in the promoter of Nr0b2 was used as positive control.
C. FXR bound to Abcb1a promoter in vivo. The chromatin fractions from the liver of a Little mouse and FXR antibody (H-130) were used in ChIP assay. Semi-quantitative PCR was performed with primers covering the putative binding site in the promoter of Abcb1a (the sequences of primers are shown in Figure 3A). Primers covering a known FXRE IR1 found in the promoter of Nr0b2 were used as positive control.
D. Scheme showing the wild type and the mutant luciferase reporter constructs. Underlined letters define the ER2 element. Mutations are indicated in lowercase letters.
E. Activation of FXR induced Abcb1a promoter activity. WT or mutant constructs were co-transfected with an FXR-expressing plasmid or control vector into Hep3B2 cells. Sixteen hours later, the cell cultures were treated with vehicle (DMSO), CDCA (100 µM) or GW4064 (2 µM) for 24 hours and harvested for reporter assay. Luciferase activity was normalized to Renilla luciferase activity. Values are means ± SD.
3.4 Activation of RXRα enhances FXR-mediated transactivation of Abcb1a promoter
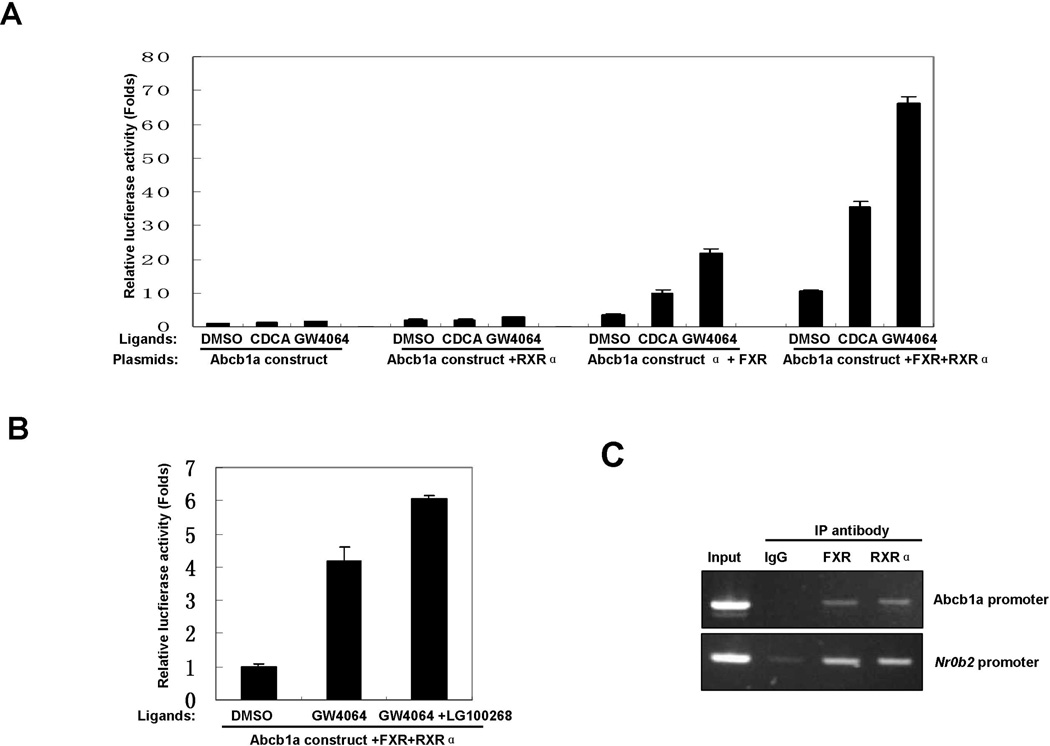
Because FXR binds to DNA as a monomer or a heterodimer with RXRα (Modica et al., 2010; Wang et al., 2008), we sought to determine whether RXRα is involved in FXR-mediated transactivation of Abcb1a promoter. As shown in Fig. 4A, ectopic expression of RXRα efficaciously increased the luciferase activity of Abcb1a promoter construct and amplified ligand-induced Abcb1a promoter activity. Meanwhile, over-expression of both FXR and RXRα significantly increased the luciferase activity and dramatically amplified ligand-induced Abcb1a promoter activity. LG100268, a synthetic ligand for RXRα, further increased GW4064-induced luciferase activity in the presence of exogenous FXR and RXRα (Fig.4B). ChIP assay showed that RXRα bound to the ER2 element in the promoter region of Abcb1a in the liver of Little mice (Fig. 4C). These results strongly suggested that RXRα is involved in FXR-mediated transactivation of Abcb1a promoter.
Fig.4. Activation of RXRα enhances FXR-mediated transactivation of Abcb1a promoter.
A. Over-expression of RXRα enhanced FXR-mediated transactivation of Abcb1a promoter. Abcb1a promoter-reporter construct was co-transfected with FXR or RXRα expression plasmids into Hep3B2 cells. Sixteen hours later, the cell cultures were treated with vehicle (DMSO), CDCA (100 µM) or GW4064 (2 µM) for 24 hours and harvested for luciferase reporter assay.
B. LG100268 increased GW4064-induced Abcb1a promoter activity in the presence of exogenous FXR and RXRα.Abcb1a promoter-reporter constructs were co-transfected with FXR or RXRα expression plasmids into Hep3B2 cells. Sixteen hours later, the cell cultures were treated with GW4064 (1 µM) or both GW4064 (1 µM) and LG100268 (0.5 µM) for 24 hours and harvested for reporter assay.
C. RXRα bound to Abcb1a promoter in vivo. The chromatin fraction from the liver of a Little mouse was used in ChIP assay with indicated antibodies. PCR was performed with primers covering the putative binding site in the promoter of Abcb1a. Primers covering a known IR1 found in the promoter of Nr0b2 were used as positive control.
3.5 DNA binding activity of FXR is higher in the livers of Little mice
Since the level of FXR protein is elevated in the livers of Little mice, we tested whether FXR binding to its target is increased in Little mice. We examined FXR binding to the promoter region of Abcb1a gene in the livers of wild type and Little mice using EMSA and ChIP assay. As shown in Fig. 5A, the binding activity of FXR toward Abcb1 promoter is significantly higher in Little mice compared to that in wild type mice. ChIP assay confirmed this observation and showed that the amounts of FXR on the Abcb1a promoter are significantly increased in the livers of Little mice (Fig. 5B). Thus, these studies demonstrated that the elevation of FXR protein level results in an increase of DNA binding of FXR in Little mice and that the occupation of Abcb1a promoter by FXR is also increased in Little mice.
Fig.5. DNA binding activity of FXR is increased in the livers of Little mice.
A. EMSAs were performed with nuclear extracts from livers of two wild type and two Little mice respectively. FXR antibody (C-20) was used for the supershift assay to confirm binding specificity.
B. Amounts of FXR are increased on the Abcb1a promoter in the liver of Little mice. The chromatin fractions from livers of wild type and Little mice were used in ChIP assay with FXR antibody (H-130) or with control rabbit IgG.
3.6 FXR directly up-regulates the expression of other xenobiotic detoxification genes
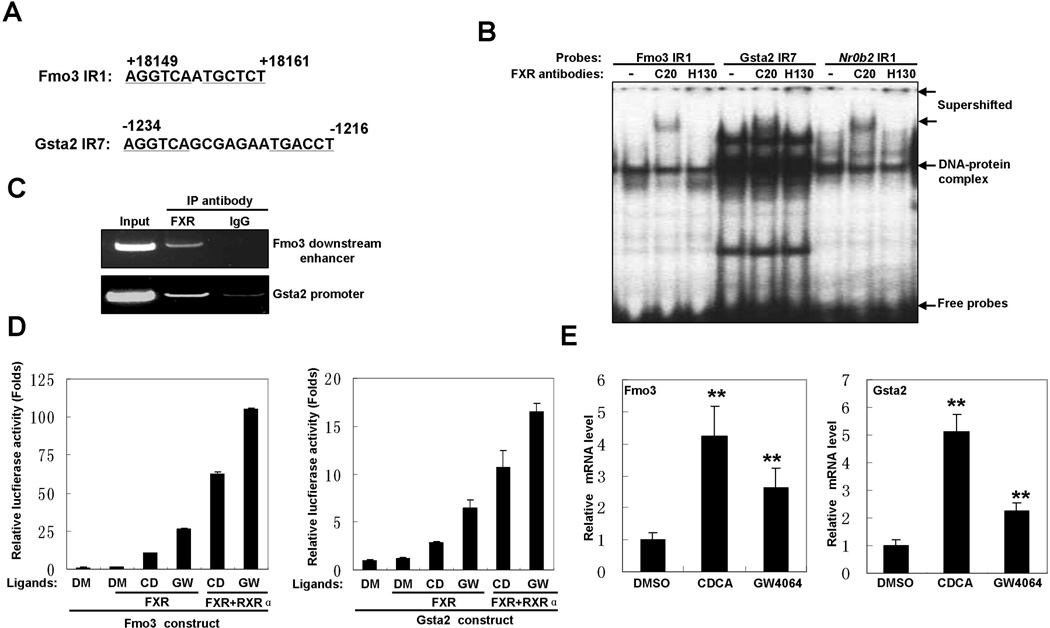
Given the increased binding of FXR to Abcb1a promoter, we asked if FXR might also directly regulate other xenobiotic detoxification genes. Data from Thomas et al (Thomas et al., 2010) showed that FXR might bind to a downstream region of Fmo3 gene and the promoter region of Gsta2 gene in mice. A recent paper also suggested that Fmo3 might be a direct target of activated FXR (Bennett et al., 2013). After analyzing these regions with NUBIScan, we found a putative FXRE IR1 (AGGTCAaTGCTCT) in the downstream region of Fmo3 and an IR7 (AGGTCAgcgagaaTGACCT) in the promoter of Gsta2 (Fig. 6A). Both EMSA and ChIP experiments confirmed that FXR bound to these two sites in vitro and in vivo (Fig. 6B and C). To further investigate regulation of these genes by FXR, we cloned the fragments containing the response elements into pGL3-basic vector and examined the effects of FXR activation on the luciferase activity of these reporter constructs. As shown in Fig. 6D, the luciferase activity was induced by CDCA or GW4064 for both constructs in the presence of exogenous FXR. Co-overexpression of RXRα with FXR further amplified ligand-induced transactivation of these constructs. Finally, treatment of Hepa1-6 cells with CDCA and GW4064 increased the mRNA levels of Fmo3 and Gsta2 (Fig. 6E). Taken together, these results indicated that FXR regulates expression of Fmo3 and Gsta2 through direct binding to the regulatory regions.
Fig.6. FXR directly up-regulates the expression of other xenobiotic detoxification genes.
A. Schemes for IR1 element found in Fmo3 downstream region and IR7 element in Gsta2 promoter.
B. FXR bound to the regulatory regions of Fmo3 and Gsta2 genes in vitro. EMSAs were performed with nuclear extracts from liver of a Little mouse. Two different FXR antibodies (C-20 and H-130) were used for the supershift assay to confirm binding specificity. A probe containing a known FXRE IR1 found in the promoter of Nr0b2 was used as positive control.
C. FXR bound to the regulatory regions of Fmo3 and Gsta2 genes in vivo. The chromatin fraction from the liver of a Little mouse was used in ChIP assay. PCR was performed with primers covering the Fmo3 IR1 element or Gsta2 IR7 element.
D. Activation of FXR induced the promoter activities of Fmo3 and Gsta2 genes. Fmo3 reporter construct or Gsta2 reporter construct was co-transfected with FXR expression plasmid into Hep3B2 cells. Sixteen hours later, the cell cultures were treated with FXR ligands (CD = CDCA, GW = GW4064) or vehicle (DM = DMSO) for 24 hours and harvested for luciferase reporter assay.
E. Activation of FXR by ligands increased the mRNA levels of Fmo3 and Gsta2 genes. Hepa1-6 cells were incubated with CDCA (200µM), GW4064 (5µM) or vehicle (DMSO) for 16 hours. Levels of Fmo3 and Gsta2 mRNAs were measured by quantitative RT-PCR. β-actin mRNA was used as internal control. Values are means ±SD. (**p<0.01 )
4. Discussion
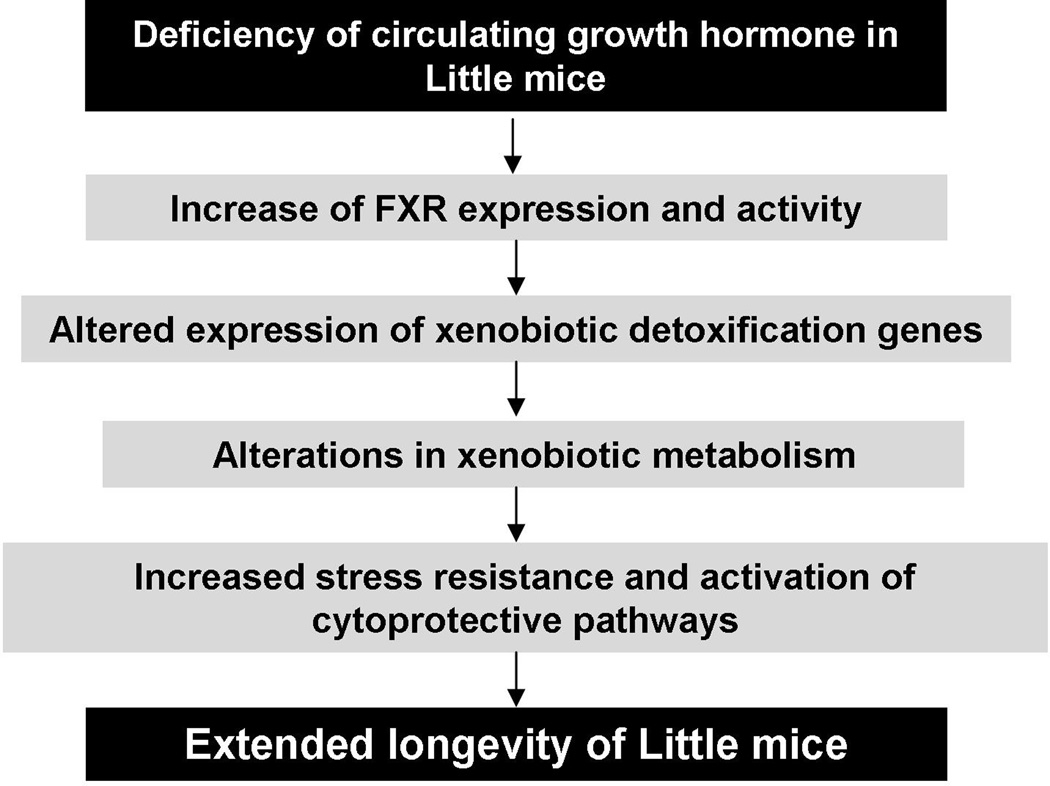
Our previous studies showed a concerted up-regulation of xenobiotic detoxification genes in the long-lived Little mice (Amador-Noguez et al., 2004; 2007). The elevation of xenobiotic metabolism pathways has also been observed in calorically restricted mice, C.elegans and the fruit fly with extended lifespan (McElwee et al., 2004; Pletcher et al., 2002; Swindell, 2007). A recent study reported that rapamycin treatment caused transcriptional activation of xenobiotic genes in mice as well (Steinbaugh et al., 2012). These findings imply that alterations in xenobiotic metabolism might be a common feature in different model organisms in which longevity is modulated by genetic manipulation, dietary restriction or pharmacological treatments. However, how the elevation of xenobiotic metabolism confers increased lifespan is still poorly understood. One hypothesis is that up-regulation of xenobiotic genes increases xenobiotic resistance and activates multiple cytoprotective pathways, which then contribute to lifespan extension (Amador-Noguez et al., 2007; Shore et al., 2012). Meanwhile, although alterations in xenobiotic metabolism have been known for several years as a strong correlate of lifespan extension in different animal models for aging, the molecular mechanism underlying the up-regulation of xenobiotic genes was undefined. Previously we showed that FXR was a major regulator of the elevation of xenobiotic gene expression in Little mice albeit by an unknown mechanism (Amador-Noguez et al., 2007). This manuscript demonstrated that FXR positively regulates the expression of Abcb1a, Fmo3 and Gst2a through direct interactions with the response elements in these genes. Protein levels and DNA binding activity of FXR are increased in Little mice compared to those in wild type mice, suggesting that FXR is a potential regulator of the longevity extension phenotype. Based on our data, we propose a signaling mechanism that mediates extended lifespan in Little mice as depicted in Fig.7. It would be interesting to test whether the signaling mechanism described here is involved in the longevity in other mouse models for aging.
Fig.7. A working model of the signaling mechanism underlying the extended lifespan in the Little mice.
Our current study demonstrates for the first time that FXR expression is regulated by the GH/IGF1 pathway in mice potentially at both transcriptional and post-transcriptional levels. Given the role of FXR as a key regulator in multiple metabolic pathways, further study of molecular mechanisms underlying regulation of FXR expression and signaling by the GH/IGF1 pathway is warranted. Recently two different groups reported that humans with mutations in GH/IGF1 pathway are protected from cancer development (Guevara-Aguirre et al., 2011; Steuerman et al., 2011), which is in agreement with our previous observations that Little mice do not develop liver cancer with age and are resistant to DEN-mediated liver cancer (Jiang et al., 2012). We found that FXR prevents development of liver cancer in Little mice by inhibiting gankyrin, a small subunit of the proteasome (Jiang et al., 2012), which is elevated in many cancers and eliminates tumor suppressor proteins Rb, p53, C/EBPα and HNF4α (Iakova et al., 2011). In addition, a recent report showed that FXR functions as a tumor suppressor in enterohepatic tissues through directly controlling the expression of NDRG2 (N-myc downstream regulated gene 2) (Deuschle et al., 2012). In view of our results and previously published studies we propose that elevation of FXR in these individuals with GH receptor deficiency might be involved in the protection from development of cancer. Since reduction of FXR expression has been linked to the development of several common cancers in human and mice (Koutsounas et al., 2012), our results of regulation of FXR expression by growth hormone indicate that the growth hormone signaling pathway might be a new candidate therapeutic target.
Supplementary Material
Highlights.
Expression and activity of FXR is increased in Little mice.
Growth hormone treatment reversed alteration of FXR expression in Little mice.
Xenobiotic detoxification genes are direct transcriptional targets of FXR.
Acknowledgements
This work was supported by National Institute of Health grants AG028865 (to G.J.D.) and GM551888, CA100070, AG039885, AG028865, CA159942 (to N.A.T.).
Abbrevations
- FXR
farnesoid X receptor
- RXR
retinoid X receptor
- Ghrhr
growth hormone-releasing hormone receptor
- GH
growth hormone
- IGF
insulin-like growth factor
- FXREs
FXR response elements
- IR
inverted repeat
- DR
direct repeat
- ER
everted repeat
- TSS
transcriptional start site
- CDCA
chenodeoxycholic acid
- DMSO
dimethyl sulfoxide
- EMSA
electrophoresis mobility shift assay
- ChIP
chromatin immunoprecipitation assay
- PCR
polymerase-chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J. Biol. Chem. 2004;279:54348–54357. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell. 2004;3(6):423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Dean A, Huang W, Setchell K, Moore D, Darlington G. Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell. 2007;6(4):453–470. doi: 10.1111/j.1474-9726.2007.00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17(1):49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman DE, Christiansen JS, Johannsson G, Thorner MO, Kopchick JJ. Role of the GH/IGF-1 axis in lifespan and healthspan: lessons from animal models. Growth Horm. IGF Res. 2008;18(6):455–471. doi: 10.1016/j.ghir.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012;13(4):213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor Small Heterodimer Partner. Endocr. J. 2008;55(2):253–268. doi: 10.1507/endocrj.k07e-103. [DOI] [PubMed] [Google Scholar]
- Cui J, Huang L, Zhao A, Lew JL, Yu J, Sahoo S, Meinke PT, Royo I, Pelaez F, Wright SD. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J. Biol. Chem. 2003;278(12):10214–10220. doi: 10.1074/jbc.M209323200. [DOI] [PubMed] [Google Scholar]
- Deuschle U, Schüler J, Schulz A, Schlüter T, Kinzel O, Abel U, Kremoser C. FXR controls the tumor suppressor NDRG2 and FXR agonists reduce liver tumor growth and metastasis in an orthotopic mouse xenograft model. PLoS One. 2012;7(10):e43044. doi: 10.1371/journal.pone.0043044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue LR, Beamer WG. Growth hormone deficiency in 'little' mice results in aberrant body composition, reduced insulin-like growth factor-I and insulin-like growth factor-binding protein-3 (IGFBP-3), but does not affect IGFBP-2, −1 or −4. J. Endocrinol. 1993;136(1):91–104. doi: 10.1677/joe.0.1360091. [DOI] [PubMed] [Google Scholar]
- Godfrey P, Rahal JO, Beamer WG, Copeland NG, Jenkins NA, Mayo KE. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat. Genet. 1993;4(3):227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6(3):517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, de Cabo R, Cohen P, Longo VD. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 2011;3(70) doi: 10.1126/scitranslmed.3001845. 70ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Timchenko L, Timchenko NA. Intracellular signaling and hepatocellular carcinoma. Semin. Cancer Biol. 2011;21:28–34. doi: 10.1016/j.semcancer.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Iakova P, Jin J, Sullivan E, Sharin V, Hong I, Anakk S, Mayor A, Darlington G, Finegold M, Moore D, Timchenko TA. FXR inhibits gankyrin in mouse livers and prevents development of liver cancer. Hepatology. 2013;57(3):1098–1106. doi: 10.1002/hep.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journe F, Durbecq V, Chaboteaux C, Rouas G, Laurent G, Nonclercq D, Sotiriou C, Body JJ, Larsimont D. Association between farnesoid X receptor expression and cell proliferation in estrogen receptor-positive luminal-like breast cancer from postmenopausal patients. Breast. Cancer. Res. Treat. 2009;115(3):523–535. doi: 10.1007/s10549-008-0094-2. [DOI] [PubMed] [Google Scholar]
- Kemper JK. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Biochim. Biophys. Acta. 2011;1812(8):842–850. doi: 10.1016/j.bbadis.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsounas I, Giaginis C, Theocharis S. Farnesoid X Receptor (FXR) from normal to malignant state. Histol. Histopathol. 2012;27(7):835–853. doi: 10.14670/HH-27.835. [DOI] [PubMed] [Google Scholar]
- Lew JL, Zhao A, Yu J, Huang L, De Pedro N, Peláez F, Wright SD, Cui J. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 2004;279(10):8856–8861. doi: 10.1074/jbc.M306422200. [DOI] [PubMed] [Google Scholar]
- Li H, Chen F, Shang Q, Pan L, Shneider BL, Chiang JY, Forman BM, Ananthanarayanan M, Tint GS, Salen G, Xu G. FXR-activating ligands inhibit rabbit ASBT expression via FXR-SHP-FTF cascade. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288(1):G60–G66. doi: 10.1152/ajpgi.00170.2004. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6(3):507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J. Biol. Chem. 2004;279(43):44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Miao J, Fang S, Lee J, Comstock C, Knudsen KE, Kemper JK. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol. Cell. Biol. 2009;29:6170–6181. doi: 10.1128/MCB.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl. Recept. Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Kunimoto M, Nishizuka M, Osada S, Imagawa M. Ku proteins function as corepressors to regulate farnesoid X receptor-mediated gene expression. Biochem. Biophys. Res. Commun. 2009;390:738–742. doi: 10.1016/j.bbrc.2009.10.040. [DOI] [PubMed] [Google Scholar]
- Perez MJ, Gonzalez-Sanchez E, Gonzalez-Loyola A, Gonzalez-Buitrago JM, Marin JJ. Mitochondrial genome depletion dysregulates bile acid- and paracetamol-induced expression of the transporters Mdr1, Mrp1 and Mrp4 in liver cells. Br. J. Pharmacol. 2011;162(8):1686–1699. doi: 10.1111/j.1476-5381.2010.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda Torra I, Freedman LP, Garabedian MJ. Identification of DRIP205 as a coactivator for the Farnesoid X receptor. J. Biol. Chem. 2004;279:36184–36191. doi: 10.1074/jbc.M405126200. [DOI] [PubMed] [Google Scholar]
- Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 2002;12(9):712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- Podvinec M, Kaufmann MR, Handschin C, Meyer UA. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol. Endocrinol. 2002;16(6):1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol. Pharmacol. 2005;68:551–558. doi: 10.1124/mol.105.012104. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Bavner A, Haroniti A, Nilsson LM, Lundasen T, Rehnmark S, Witt MR, Einarsson C, Talianidis I, Gustafsson JA, Treuter E. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15665–15670. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore DE, Carr CE, Ruvkun G. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet. 2012;8(7) doi: 10.1371/journal.pgen.1002792. e1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh MJ, Sun LY, Bartke A, Miller RA. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am. J. Physiol. Endocrinol. Metab. 2012;303(4):E488–E495. doi: 10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerman R, Shevah O, Laron Z. Congenital IGF1 deficiency tends to confer protection against post-natal development of malignancies. Eur. J. Endocrinol. 2011;164(4):485–489. doi: 10.1530/EJE-10-0859. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Gene expression profiling of long-lived dwarf mice: longevity-associated genes and relationships with diet, gender and aging. BMC Genomics. 2007;8:353. doi: 10.1186/1471-2164-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51(4):1410–14719. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno A, Takada I, Takezawa S, Oishi H, Baba A, Shimizu T, Tokita A, Yanagisawa J, Kato S. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem. Biophys. Res. Commun. 2005;327:933–938. doi: 10.1016/j.bbrc.2004.12.095. [DOI] [PubMed] [Google Scholar]
- Wang S, Lai K, Moy FJ, Bhat A, Hartman HB, Evans MJ. The nuclear hormone receptor farnesoid X receptor (FXR) is activated by androsterone. Endocrinology. 2006;147(9):4025–4033. doi: 10.1210/en.2005-1485. [DOI] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Moore DD, Huang W. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18(11):1087–1095. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76(2):215–228. doi: 10.1124/mol.109.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Ueno M, Kusaka T, Onodera M, Huang CL, Hosomi N, Kanenishi K, Sakamoto H. Abcb1a and Abcb1b expression in senescence-accelerated mouse (SAM) Neurosci. Lett. 2009;456(1):34–38. doi: 10.1016/j.neulet.2009.03.067. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, He F, Kuruba R, Gao X, Wilson A, Li J, Billiar TR, Pitt BR, Xie W, Li S. FXR-mediated regulation of angiotensin type 2 receptor expression in vascular smooth muscle cells. Cardiovasc. Res. 2008;77(3):560–569. doi: 10.1093/cvr/cvm068. [DOI] [PubMed] [Google Scholar]
- Xu G, Pan LX, Li H, Forman BM, Erickson SK, Shefer S, Bollineni J, Batta AK, Christie J, Wang TH, Michel J, Yang S, Tsai R, Lai L, Shimada K, Tint GS, Salen G. Regulation of the farnesoid X receptor (FXR) by bile acid flux in rabbits. J. Biol. Chem. 2002;277(52):50491–50496. doi: 10.1074/jbc.M209176200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.