Abstract
Tropomyosin is a key factor in the molecular mechanisms that regulate the binding of myosin motors to actin filaments in most eukaryotic cells. This regulation is achieved by the azimuthal repositioning of tropomyosin along the actin:tropomyosin:troponin thin filament to block or expose myosin binding sites on actin. In striated muscle, including involuntary cardiac muscle, tropomyosin regulates muscle contraction by coupling Ca2+ binding to troponin with myosin binding to the thin filament. In smooth muscle, the switch is the post-translational modification of the myosin. Depending on the activation state of troponin and the binding state of myosin, tropomyosin can occupy the blocked, closed, or open position on actin. Using native cryogenic 3DEM, we have directly resolved and visualized cardiac and gizzard muscle tropomyosin on filamentous actin in the position that corresponds to the closed state. From the 8-Å resolution structure of the reconstituted Ac:Tm filament formed with gizzard-derived Tm we discuss two possible mechanisms for the transition from closed to open state and describe the role Tm plays in blocking myosin tight binding in the closed state position.
Keywords: Actin, Tropomyosin, cryogenic 3DEM, electron microscopy, thin filament, cytoskeleton
Introduction
Striated and smooth Actin:Tropomyosin
Tropomyosin (Tm) is an important component in most actin filament (F-Actin) structures and is therefore essential to cell motility, morphology, organization, and structure (reviewed by 1). Different Tm isoforms are required for survival of organisms from yeast to humans: they play a critical role in muscle contraction as part of muscle thin filament and support F-Actin’s diverse cellular function outside the context of muscle contraction (reviewed by 2).
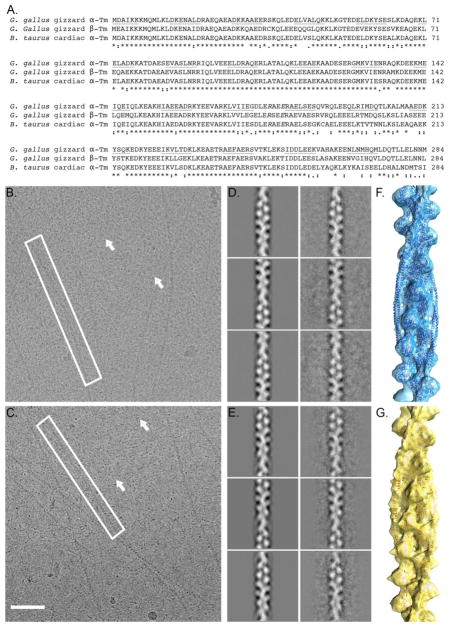
High molecular weight (HMW) Tm is a 40 nm-long, dimeric coiled coil that cooperatively polymerizes end-to-end along the surface of F-Actin 3; 4; 5; 6; 7. Striated and smooth muscle contain HMW Tm, where its coiled coil wraps continuously around F-Actin to form part of the thin filament that helps elicit voluntary and involuntary muscle contraction 8. Additionally, both cardiac and skeletal thin filaments from striated muscle contain troponin (Tn), a regulator that links muscle activation to cellular Ca2+ concentration 9. Smooth muscle Ac:Tm, such as that found in the gizzard, does not use Tn to control activity but relies on post-translational modification of the myosin with which it interacts 10. HMW Tm is 284 amino acids long and is strongly conserved between species, despite being expressed from multiple genes, multiple promoters, and as multiple splice isoforms (Figure 1) 1; 2; 11.
Figure 1. Imaging Ac:Tm reconstituted with Tm derived from cardiac tissue and Tm derived from gizzard tissue.
A. Sequence alignment of the G. gallus (chicken) gizzard α and β Tm with B. taurus (cow) α Tm. Identity is marked with an asterisk (*), homology is marked with a colon (:), and similarity is marked with a period (.). The heptad repeat is marked with alternating underlines. B. Representative raw image of cardiac Ac:Tm filaments show long, flat filaments (white box) as well as short, punctate molecules that correspond to unbound cardiac Tm molecules (white arrows). C. Representative raw image of gizzard Ac:Tm filaments show similar filaments (white box) and free Tm (white arrows). D. Representative projection and corresponding class average of the cardiac Tm-containing Ac:Tm filaments. E. Representative projection and corresponding class average of the gizzard Tm-containing Ac:Tm filaments. F. Reconstruction of the cardiac Tm-containing Ac:Tm filament. G. Reconstruction of the gizzard Tm-containing Ac:Tm filament.
Tm binds F-Actin and regulates muscle contraction
The sequence of amino acids in cardiac and gizzard Tm (and other HMW Tms) forms a pattern that determines how Tm binds F-Actin (Figure 1A). 41 sequential heptad repeats comprise the Tm α-helix that dimerizes as a coiled coil and then binds in a repeating pattern along the length of the F-Actin molecule 12. The protein products from the α and β isoforms, the predominant forms found in cardiac (αα) or gizzard (αβ) tissue, are 72% identical, with the least conserved segment lying in the C-terminus of the protein (Figure 1A). Those heptad repeats translate into seven pseudo-repeating units per dimer, marked by the coiled coil crossover, that conform to the path of seven successive Ac monomers within one half of the two-start F-Actin helix 13; 14; 15. Strategically located alanines interspersed along the heptad repeats allow smooth, anisotropic bending of the long coiled coil, suggesting Tm is preformed into a shape that is complementary to F-Actin’s twist 12; 16; 17; 18; 19. Further, this innate superhelical twist suggests that movement of Tm along one part of the Ac:Tm filament can propagate along the Tm polymer, promoting the cooperative nature of F-Actin regulation by Tm 20; 21.
In muscles, the thin filament works with myosin-containing thick filaments to regulate muscle contraction 22; 23; 24. Tm plays an integral role in this regulation by cooperatively regulating access of other Ac binding proteins onto the F-Actin surface. Specifically, striated muscle Tm and Tn alternatively inhibit, allow, or enhance myosin binding to F-Actin to control the myosin crossbridge cycling in response to Ca2+ concentration, and consequently relax or promote muscle contraction 25. In this model, known as the three-state steric blocking model of muscle regulation, Tm can occupy three states that are defined biochemically and structurally: blocked, closed, and open 26; 27. Smooth muscle is proposed to work in a similar fashion, but post-translational modification of myosin by myosin light chain kinase, regulated by the Ca2+-binding calmodulin, performs a similar role to Tn/Ca2+ control of Tm position 28; 29.
Blocked, closed, and open states are defined biochemically and structurally
In the blocked state (“B”), myosin binding to F-Actin is inhibited by Tm:Tn – Ca2+ (Tm:Tn minus Ca2+ will be abbreviated Tm:Tn – Ca2+ throughout the manuscript). According to the structural interpretation of the B state inferred from negatively stained EM images of Ac:Tm and Ac:Tm:Tn, Tm is pinned in place by Ca2+-free Tn such that Tm covers the myosin binding site on Ac 30. In solution, the B state is occupied only in Ca2+-free Tn-containing filaments and never in the Tn-free Ac:Tm filament or in the presence of Tn and Ca2+ 26. Depending on the isoform, however, the Tm position that corresponds to the B-state can be visualized in Tn-free Ac:Tm filaments in some negative stain reconstructions, including those reconstituted with Tm derived from both cardiac and gizzard muscle 31.
In the closed state (“C”), myosin binding to Ac:Tm(:Tn) is weak, but cooperative 32; 33. According to the structural interpretation of the C state, also inferred from negatively stained filaments, Tm shifts from its B-state position across the surface of F-Actin through a 25° rotation around the F-Actin axis to partially expose the previously buried myosin binding site 22; 30. In solution, this state is 80% occupied both in Ac:Tm filaments and Ac:Tm:Tn filaments with Ca2+26.
In the open state (“M”), myosin binding to Ac:Tm(:Tn) is strong and cooperative. According to the structural interpretation of the M state, this position is marked by further azimuthal movement of Tm from its C-state position, through a rotation of 10° around the F-Actin axis and across the F-Actin surface, coupled to an axial shift along the F-Actin helical axis to fully expose myosin binding sites on F-Actin 22. From the B-state to the M-state, this represents a 23 Å shift up and across with a 31° rotation around the F-Actin axis 34. In solution, the M state is 20% occupied both in Ac:Tm filaments and Ac:Tm:Tn filaments with Ca2+, even without any myosin binding 26.
The mechanism for how the Tm filament cooperatively moves across F-Actin has remained an intriguing question ever since this 3-state model and its physical interpretation, inferred from negatively-stained 3DEM structures, was proposed. Possible mechanisms have contradicting experimental support that are not always resolved by the lower resolution negative stain structures. Do the Tm filaments roll or shift 35; 36? Does the Tm filament act as a stiff rod or as a continuously flexible strand 17; 37? Clarification of the mechanism for Tm movement on F-Actin provides a context for understanding regulation of cardiac and smooth muscle contraction, and the linkage between polymorphisms of Tm and F-Actin and disease.
Here, we show the cryo-EM structures of Ac:Tm filaments reconstituted with either bovine cardiac muscle- or chicken gizzard muscle-derived Tm that corresponds to the C-state position. By comparing the two datasets, we show that the Tn-dependent cardiac Tm reconstitutes with F-Actin into a less ordered filament than the Tn-independent gizzard Tm. From the 8-Å resolution gizzard Tm-containing Ac:Tm structure, we propose a novel, rocking mechanism that transitions Tm from the C- to M-state position and discuss the details about how the C-state position blocks myosin binding.
Results
Cryogenic 3DEM Imaging of Ac:Tm
Ac:Tm filaments reconstituted either from bovine cardiac striated muscle-derived Tm or chicken gizzard smooth muscle-derived Tm, both assembled on F-Actin derived from rabbit skeletal muscle, were visualized under native, cryogenic conditions (Figure 1). Incubating a 10-fold molar excess of Tm over F-Actin allowed stable binding during the vitrification process (Figures 1B and C). This ratio of Tm to Ac results in full binding in solution 38; 39 and is similar to the Ac:Tm ratio used in imaging the recently-determined high resolution structure of Ac:Tm bound to the S1 fragment of myosin (the Behrmann model of Ac:Tm:S1) 34.
Heterogeneity
Iterative helical real space reconstruction (IHRSR) was used to align, classify, and reconstruct structures of both isoforms. The resulting initial reconstructions, at about 20 Å-resolution, reveal the Tm to be in a similar position to the C- and M-state positions, which are quite close to one another and thus difficult to distinguish at moderate resolution (Figure 1F and G) 22. Further, biochemical data suggests that Tm is in equilibrium between the C- and M-state positions in solution, so one possibility that could explain the moderate resolution is that there is underlying structural heterogeneity in the sample 26.
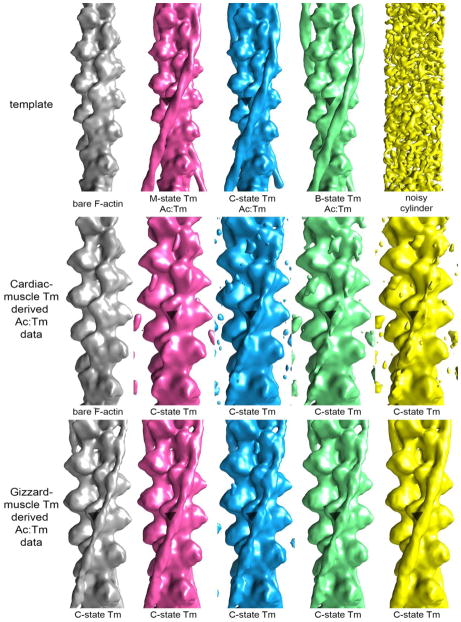
To test the hypothesis that our structure represents a mixture of Tm positions on F-Actin, we used competitive alignment against a mixed set of references for assessing heterogeneity in each data set. Both datasets were aligned against projections of 5 different models: bare F-Actin; the M-state Ac:Tm generated by superimposing a 14 actin-long F-Actin and two full length Tms from the B-state position40 onto the shorter Ac:Tm:S1 model (PDB code 4A7F34); C-state Ac:Tm, generated by docking a 14 actin-long F-Actin and two full length Tms from the B-state position40 into the gizzard-Tm containing Ac:Tm structure; the B-state from the atomic coordinates determined from the negative stain structure 40; and a noisy cylinder of diameter 7 nm (Figure 2, row 1). Segments segregated into each model at about the same frequency, but favoring the spatially similar M- and C-state positions (Table 1 breaks down the number of segments and the corresponding percentage of data that sort with each of the 5 models for both datasets).
Figure 2. Multi-reference alignment reveals that the cardiac-derived Tm containing Ac:Tm filament is less occupied than the gizzard-derived Tm containing Ac:Tm filament.
Top row: template used for projection matching alignment. Each column represents the structure that resulted by aligning the subset of data that segregated with each model against a bare actin filament reference. Middle row: Alignment of cardiac-derived Tm containing Ac:Tm images that segregate with the various models refine to show that the filaments are either bare (first and last column), in the C-state and well ordered (second column), or in the C-state and not well ordered (third and fourth column). Bottom row: Alignment of gizzard-derived Tm containing Ac:Tm images that segregate with the various models refine to show that the majority of the filaments are occupied with C-state Tm. The arrow heads demark similar positions on the actin to help visualize the transition of the Tm.
Table 1.
Mutireference Alignment against diverse Ac:Tm filament models divides the single particle segments into almost evenly distributed subsets of data.
| Cardiac | Gizzard | |
|---|---|---|
|
|
||
| F-Actin | 23,795 (16%) | 73,773 (15%) |
|
| ||
| M-state | 31,718 (22%) | 108,849 (23%) |
|
| ||
| C-state | 30,424 (21%) | 106,326 (22%) |
|
| ||
| B-state | 34,876 (24%) | 114,587 (24%) |
|
| ||
| cylinder | 24,139 (17%) | 76,982 (16%) |
Next, each subset of data was aligned against a bare F-Actin filament and reconstructed. In the case of the cardiac muscle-derived Ac:Tm data (Figure 2, row 2), the 16% of filament segments that segregated with the bare F-Actin refined to produce filaments with no bound Tm. The remaining data, when aligned against the bare F-Actin model, resulted in Tm-decorated filaments where the Tm is in the C-state position, even if poorly defined for some subsets. In the case of the gizzard muscle-derived Tm data (Figure 2, row 3), each of the subsets refine to the C- state position seen in the original refinements, visible in the raw maps.
Cardiac-derived Tm
The data from the cardiac Tm-containing Ac:Tm filaments that had bound Tm was separated from the rest and further refined, resulting in a structure whose Tm is clearly shifted axially around the F-Actin, away from the B-state position defined by negative stain 3DEM (Figure 3). The relatively small dataset results in a moderate resolution structure, limiting the precision with which the Tm can be fit into the density axially (Figure 3 and S1). Regardless, the azimuthal Tm position is clear. Further, there is sufficient detail to see Tm in single filament reconstructions and to ascertain the polarity of the filaments (Figure S2), showing that the azimuthal shift that is apparent in the refined structure accurately represents the position of the Tm in the portion of the data with bound, well-ordered Tm.
Figure 3. Cardiac-derived Tm containing Ac:Tm.
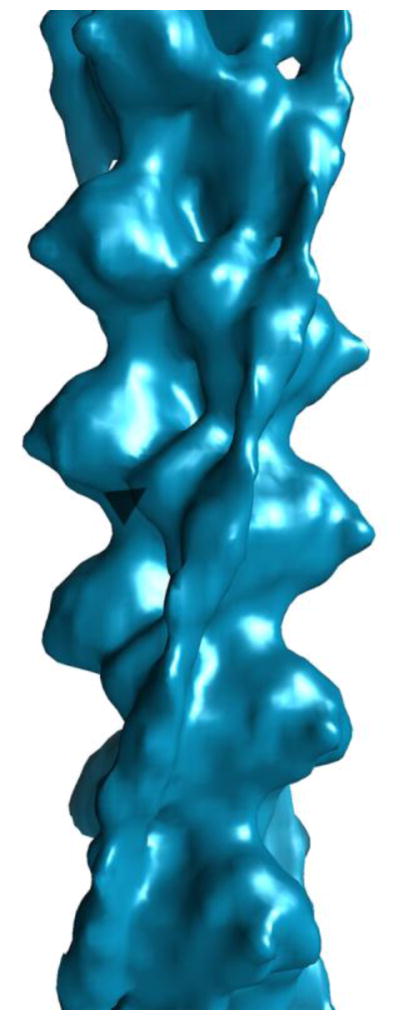
Refinement of the occupied filaments results in a moderate resolution structure of the C-state Ac:Tm filament.
Gizzard-derived Tm
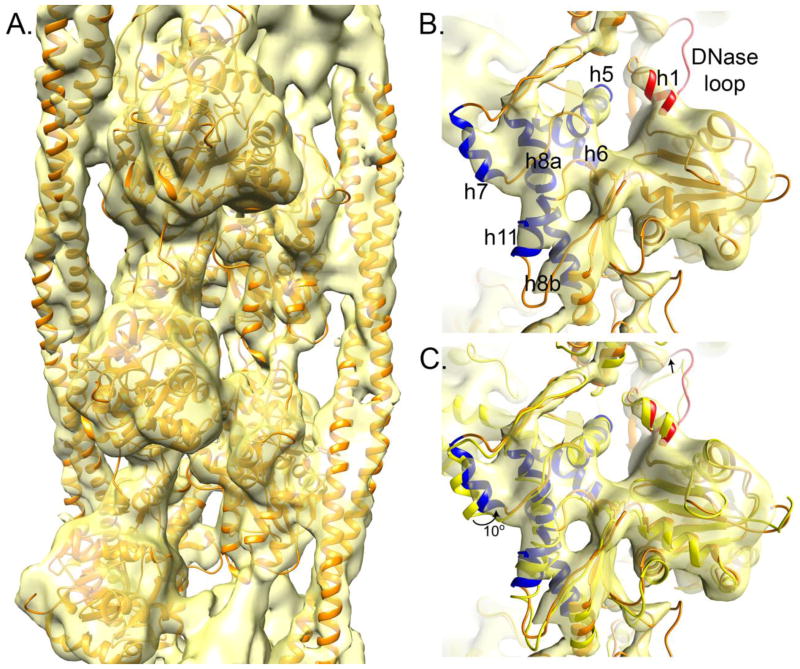
Data from the gizzard Tm-containing Ac:Tm filaments showed that, unlike the cardiac-derived Tm-containing Ac:Tm, all of the filaments had bound Tm that appeared to be in the C-state position (Figure 2). Correspondence analysis of the members of each reference-based class average, followed by subclassification and averaging, was independently performed to reduce the effects of misalignment in the final reconstruction. After classification and averaging, only the subclasses with the highest correlation to the initial reprojection were used in the final reconstruction. In this way, the amount of data was reduced from 483,139 segments (used for the initial reconstruction and MRA) to 224,337 segments. Reconstruction of the classes not used for the final structure did not show heterogeneity, as expected from the MRA analysis (Figure S3). Reconstruction of the best data improved the FSC resolution by about 5 Å and resolved structural details in both the F-Actin and Tm such that the Tm can be positioned both axially and azimuthally into the contours of the twisting ribbon of density that corresponds to the Tm (Figure 4A). Only slight adjustment to the F-Actin secondary structural elements were required to accommodate the density (Figure 4B).
Figure 4. Gizzard-derived Tm containing Ac:Tm.
A. At 8 Å resolution, separation of the α-helices in the Ac and Tm are evident, allowing the independent fitting of F-Actin and Tm from Ac:Tm:S1 (PDB code 4A7F)34. B. At high contour, helices corresponding to the internal features of F-Actin are evident, showing a slight mismatch of helix 7. Helices corresponding to the inner domain (subdomains 3 and 4) are blue and those corresponding to the outer domain (subdomains 1 and 2) are red. C. Flexible fitting rotates helix 7 by about 10° from where it sits when the myosin S1 fragment is bound (yellow).
pH
Next, we tested the effect of pH similar to that of uranyl formate or acetate stains to judge the potential for artifacts in the negative stain structures to which these cryogenic structures would be compared 31. Both G-Actin and Tm are acidic proteins whose interaction is controlled primarily by electrostatics 40. The pI of G-Actin is about 5.4 41 whereas the pI of Tm is about 4.0 42. Their assembly is, of course, sensitive to salt and pH 31. To test the effect of pH on the structure of the Ac:Tm filament, we attempted to determine the cryogenic structure of a low pH, vitrified filament. Immediately before blotting and plunging pre-formed Ac:Tm into ethane to preserve the filaments for cryogenic data collection, the pH of the solution was dropped by addition of a small amount of NaH2PO4. Rather than finding nicely spaced, individual filaments, as with the pH=7 specimen (Figure S4A), the filaments were bundled (Figure S4B) such that they were unsuitable for single particle helical analysis.
Discussion
Structural details of the gizzard-derived Tm containing Ac:Tm filament
A model of a 5-subunit long F-Actin and two dimeric Tm coiled-coils (PDB code 4A7F)34, with each element docked separately, fit into the major structural elements of the F-Actin well and line up with the clear twist of the Tm (Figure 4A). Delineated features correspond to the inner domain helices 5, 6, 7, 8a/b and 11, which all orient to the center of the helix, as well as the outer domain DNase I-binding loop and adjacent helix 1, which bridge between subunits along the helix (Figure 4B). The bulk of these elements are in the same orientation as they are when the myosin S1 domain binds (Figure 4C). One inner domain feature, helix 7, pivots by 10°. Likewise, the outer domain features are slightly mismatched – the DNase I-binding loop and helix 1 are slightly out of the density, as if they should be drawn over toward the Tm. Moving those elements into the density requires only slight adjustments to the shape of the loop and orientation of the helix; these elements of the structure are variable, even when no Tm 43; 44 is bound or Tm and myosin are bound 34.
The width and contour of the density corresponding to Tm agrees well with the expected oblong shape and unambiguously shows the azimuthal position of the coiled-coil (Figure 4A). The C-state position here is about 15 Å away from the M-state position, measured by the tmArg187 Cα–Cα distance between the Tm docked into the Ac:Tm or Ac:Tm:S1, where the adjacent F-Actins are superimposed. Arg187 was chosen as a good point of measurement because it is one of two amino acids identified in the Ac:Tm:S1 structure as a possible contact point with F-Actin, with acAsp311 (tmGlu181 is the other, interacting with acLys315)34.
C-state position
The thin filament transmits stress in muscle fibers when myosin binds, hydrolyzes ATP, and changes conformation to elicit the power stroke 45. This process is regulated by elements in the cell that sense and respond to cellular Ca2+ concentration to activate muscle contraction. Biochemically, three states of the thin filament serve as a framework for understanding how the proteins work together. According to this three-state steric blocking model of thin filament regulation, Tm is in equilibrium between different states that correspond to its three physical positions on F-Actin. In the absence of other regulators or in the presence of Tn and Ca2+, the equilibrium is between the C and M states, in which myosin binds weakly and strongly, respectively 26.
Negative-stain 3DEM has provided a physical framework for understanding the Tm position corresponding to each of the three biochemical states. According to these structures, Tm moves azimuthally across the F-Actin surface from a position where it blocks myosin binding to where the myosin site is fully exposed 22. The fully blocked position is only populated when Tn (in the absence of Ca2+) pins it in place 26. Within the confines of the three-state model, as defined by the negative stain Ac:Tm structures, one would expect to find Tm either in the C- or M-state position according to the equilibrium constant, which predicts they would be in a 4:1 ratio (80% closed, 20% open) 26. The structures presented here, determined from cryogenically preserved Ac:Tm filaments, contribute structural details to this model and two competing mechanisms for the transition from C- to M-state.
Differences between Tm isoforms
Tm’s density is sufficiently well resolved to unambiguously identify the rotation around the F-Actin helical axis in both the cardiac Tm- and gizzard Tm-containing Ac:Tm filaments, which places the Tm helical coiled coil essentially in the same location in both isoforms, when Tm is bound (Figure 1). Analysis of the individual segments that went into each reconstruction showed that about 15% of the cardiac-derived Tm-containing Ac:Tm filaments is undecorated and in many of the subclasses the Tm is poorly resolved, limiting the resolution of the resulting structure (Figures 2 and 3 and S1, Table 1). In contrast, the majority, if not all, of the gizzard-derived Tm-containing filaments were bound with Tm that wraps around F-Actin in the same, well-resolved C-state position (Figure 2 and Table 1). Given that the filaments were prepared in the same way, the difference in filament composition is likely due to the different roles these proteins play in the cell.
Cardiac and gizzard muscle work differently in the cell, so it is not surprising that they behave differently biochemically and structurally. Tm is the gatekeeper of myosin activation in both muscle types regardless of Tn dependence, but despite strong sequence conservation Ac:Tm works with different protein partners to effect change (Figure 1A) 9; 10. It is possible that the Ac:Tm filament formed from cardiac-derived Tm and skeletal-derived F-Actin is less well-ordered than that formed from gizzard-derived Tm and skeletal-derived F-Actin because it is lacking Tn, which pins it in the B-state position in the absence of Ca2+. Smooth muscle, in contrast, does not need Tn so it is held in the C-state position only by interactions between F-Actin and Tm. The C-terminal regions of the sequence show the highest sequence variation between the two isoforms, pointing to the last three heptad repeats as the region of the structure that could modulate this response (Figure 1A); the amino terminal end of the TnT subunit also binds along the C-terminal end of Tm 46, supporting the idea that this region of the highly repetitive Tm structure is responsible for its differential regulation between isoforms.
Different behavior of Tm isoforms prepared on skeletal F-Actin has been observed 31, but has not been characterized in terms of Tm occupancy. Here, we show that Ac:Tm filaments reconstituted from gizzard (i.e. smooth) muscle-derived Tm is more occupied by Tm than that from cardiac (i.e. striated), suggesting differential binding affinities between isoforms that correlates with Tn dependency. The Tn-independent isoform more fully occupied by Tm and the images can be better aligned, leading to a more detailed structure, even when normalized for particle number (Figure 3, 4 and S1). Further, if the ability to align particles is a proxy for structural rigidity, this result would suggest that Ac:Tm filaments formed from components that do not rely on Tn, like the gizzard-derived smooth muscle Tm, are more ordered than those that rely on Tn, like the cardiac-derived striated muscle Tm, when Tn is not present.
Alternative mechanisms could explain the movement of Tm
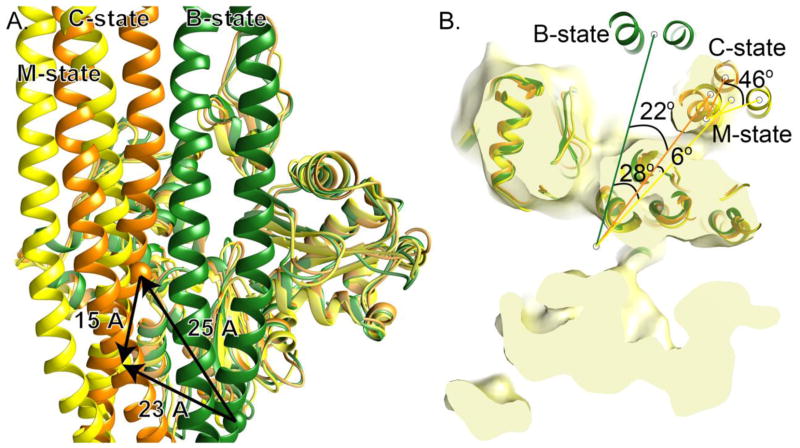
Tm is surprisingly agile in the positions it can accommodate on the surface of F-Actin. In the C state reported here, Tm sits over 25 Å away from where it sits in the B-state position reported from negative-stain structures 40, measured by the Cα-Cα distance between the two equivalent tmArg178 when the adjacent Actin monomer is superimposed (Figure 5A). This represents a shift across the face of the F-Actin of about 25 Å with a rotation of 22° around the helical axis (Figure 6B). By this same measurement, from the B-state to the M-state, Tm moves about 21 Å, shifting across the face of the F-Actin with a rotation of 28° around the helical axis measured in the same way as for the C-state position (the reported values are 23 Å shift and 31° rotation 34). If these three positions represent a transition of Tm during myosin binding during which the same face of Tm remains in contact with F-Actin at all times, then Tm moves from the B-state position, across and around the face of the F-Actin to the C-state position, and then settles back into the M-state position with another shift of about 15 Å coupled to a 6° rotation when myosin binds (Figure 5). Measurements involving the B-state position assume an accurately determined B-state position. Persistence length measurements define Tm as a semi-rigid rod that would more easily slide than roll, supporting this mechanism for Tm movement across the face of F-Actin 16; 17; 18. On the other hand, florescence measurement shows some amino acids believed to form the F-Actin:Tm interface are differentially solvated in the Tn - Ca2+ state (presumably the B-state) compared with the Tn + Ca2+ state (presumably the C-state), suggesting a change in the Tm environment upon repositioning that could arise if the Tm rolls by 90° around the filament 35. Both sets of experiments deal primarily with the larger transition from B- to C-state position, implying, but not directly addressing, the smaller change from C- to M- state positions.
Figure 5. B to C to M-state transition.
A. Tm in the C-state position (orange) is about 25 Å across the face of F-Actin from where it is in the B-state position (yellow), measured from cross over to cross over. It is about 15 Å away from where it is in the M-state (yellow), also measured from cross over to cross over. M-state Tm is about 23 Å away from B-state Tm. B. Tm rotates 22° around the central axis of F-actin to transition from the B- to C-state position (green and orange, respectively) and then it rotates another 6° to move into the M-state position (yellow). A single Tm helix in the C- and M-state position is superimposable at the F-actin interface. To move from the C- to M-state position the other helix rotates about 46° around that common position.
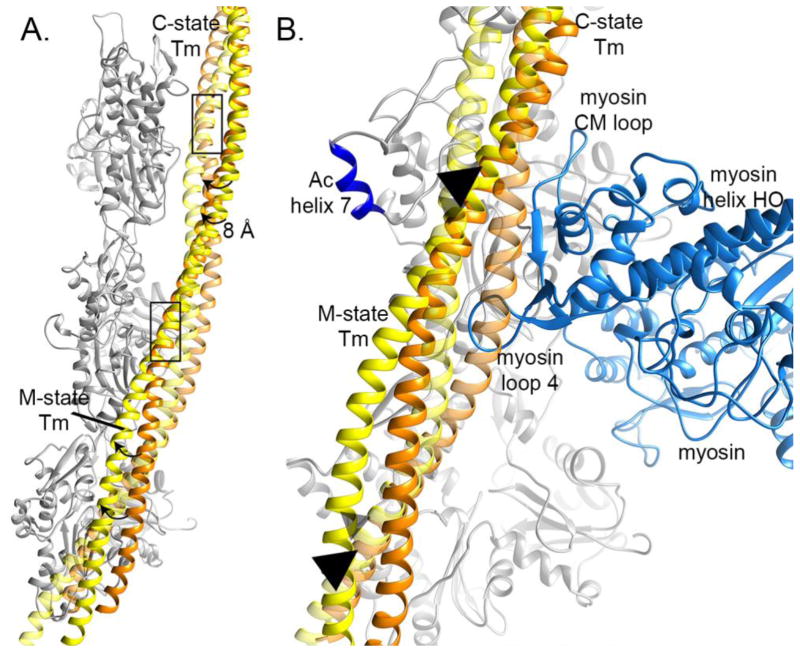
Figure 6. Tm could rotate around a fixed position to move from C- to M-state.
A. When C-state Tm (orange) and M-state Tm (orange) are independently docked into superimposed maps, a stretch of F-actin bound helix aligns in register, marked by a rectangle, suggesting Tm could transition from the C- to M-state through a rotation around this common element. B. In the C-state (orange), steric conflict between myosin loop 4 (light blue) and the Tm helix that is not bound to F-actin would prevent tight myosin binding. Triangles mark the overlapping helix that could be the anchor point for a rotation that allows Tm to rock out of the way, buttressed by F-Actin helix 7 (dark blue), as it moves to the M-state (yellow).
The transition from C- to M-state positions, defined by two reconstructions (one described in this manuscript and the other from the Ac:Tm:S1 structure34), each at about 8 Å resolution, shows the possibility of a different path that requires much less movement across the F-Actin surface than a slide of 15 Å through a path almost parallel to the F-Actin axis. In both the Ac:Tm and Ac:Tm:S1 structures, the separation of the helices in the Tm is visible, so the coils can be placed precisely (Figure 4). In both, a short stretch of the Tm, about 5 turns of one α helix that makes up the Tm dimer, interacts with F-Actin (Figure 6A). In the independently docked pseudo atomic structures, that stretch of Tm is virtually superimposed, regardless of the method of superposition (i.e. docking the maps and then the models into the maps or superimposing just the models onto one another) (Figures 5 and 6). This superposition occurs along the length of the Ac:Tm helix, owing to the symmetrized structure, but alternating which helix of the Tm dimer interacts with the F-Actin, echoing the broken Tm symmetry enforced by applying the full symmetry of the F-Actin to the full Tm dimer.
To move from the C-state position to the M-state position, the second Tm α-helix, which is not in contact with F-Actin at that position, must rock over the first by 8 Å to move from one helical position to the other, pivoting by 46° around this short stretch of Tm that remains in place (Figures 5B and 6). In doing so, the rest of Tm follows into its new position, ultimately shifting the position of the Tm by only small amount (Figure 6 and movie). This proposed rocking would be quite different from the rolling that has been suggested as an alternative to the sliding mechanism that connects the B, C, and M-states 35; 36 because it does not involve movement across the face of the F-actin and, as such, requires much less bending of the semi-rigid Tm coiled.
Myosin binding
As docked in the C-state position, the Tm α-helix that is not in contact with the F-Actin is in direct steric conflict with myosin. Specifically, myosin loop 4 clashes with the Tm helix that is not bound to the F-Actin (Figure 6B). Myosin loop 4 sits at the end of helix HO and participates in closure of the 50-kDa cleft upon F-Actin binding, showing how the C-state Tm position partially blocks myosin’s final transition to tightly binding Ac:Tm (Figure 6B). Interestingly, F-Actin helix 7 is positioned just on the opposite side of Tm from the myosin binding site, at the same height along the helix as the 5-helical turn segment of Tm that is common between the C- and M-states. This helix rotates about 10° away from actin subdomain 4 upon myosin binding (Figure 4B), as if the rocking of Tm into place for myosin binding must push it slightly out of the way. Once repositioned, F-Actin helix 7 serves as a buttress, holding Tm in place after it rocks over for participation in myosin binding (Figure 6B).
Conclusions
We present here an 8-Å resolution cryo-EM structure of the gizzard-derived Tm-containing Ac:Tm filament that shows Tm in the C-state position. This Tm position is recapitulated in a lower resolution Ac:Tm reconstituted with Tm-derived from cardiac muscle that shows lower occupancy of Tm, suggesting binding differences between different isoforms that do or do not incorporate Tn. For the smooth muscle, gizzard-derived Tm structure, which does not use Tn, two different mechanisms could explain the trajectory of Tm as it moves between the C- and M-state positions. In one, Tm moves across the F-Actin face from the B-state position, past its binding site in the M-state to the C-state position, and then slides back into the tightly bound configuration when myosin binds. In another, B-state slides to the C-state, where tight myosin binding is blocked by steric interactions between myosin loop 4 and Tm. Upon a slight rotation around a Tm helix that remains bound to F-Actin, Tm rocks over to facilitate myosin binding.
Methods
Ac:Tm Filament Preparation
Rabbit skeletal muscle F-Actin, bovine cardiac Tm, and chicken gizzard Tm were purified as described elsewhere 47. Both samples were mixed at a ratio of 1:10 Tm binding sites on F-Actin to Tm dimers, applied to well-washed QUANTIFOIL perforated carbon grids (EMS, Hatfield, PA) that had been glow discharged for 5 seconds in a Solarus plasma cleaner (Gatan, Warrendale, PA), and plunged at room temperature and 100% humidity using an Vitrobot plunger (FEI, Hillsboro, OR). For low pH experiments, 0.5 μL of 1 M NaH2PO4 was added to the droplet of 1:10 diluted, buffered cardiac-derived Tm containing Ac:Tm immediately before blotting and plunging into liquefied ethane.
Imaging
Images of either cardiac or gizzard Tm-containing Ac:Tm filaments were collected with an FEI Titan Krios operating at 300 kV at 101,555 times magnification that resulted in a pixel size of 1.477 Å/pixel at the specimen level. A dose rate of ~30 electrons/Å2 and defocus values of 1.5–4 μm were used. Images were recorded on a 4096 × 4096 Gatan Ultrascan 4000 CCD using the Leginon automated electron microscopy package 48; 49.
Image Processing
145,673 segments from 728 cardiac Tm-containing Ac:Tm filaments and 483,139 segments from 2,992 gizzard Tm-containing Ac:Tm filaments were selected using ImageJ 50 and Eman’s Boxer 51. The segments were aligned using Matlab (The MathWorks Inc., Natick, MA), SPIDER 52, and EMAN 51. The contrast transfer function was estimated with the Automated CTF Estimation (ACE) program 53 from Appion and corrected using EMAN 54. The iterative helical real space reconstruction (IHRSR) technique was used to align individual segments 55. The classes resulting from this reference-based alignment in EMAN were subsequently subclassified into 2–7 classes based on correspondence analysis performed with the CA S function in SPIDER 52. Only images from the best matching class matched the reference projection were kept for the final reconstruction of the gizzard-derived Tm-containing Ac:Tm data. In the final gizzard Tm-containing reconstructions, 224,337 images were reconstructed with a 28-Å rise and 167° rotation. Only images that had well-defined Tm visible in the raw structure after MRA were used in the refinement of cardiac Tm-containing data; segments included in the final reconstruction were also subject to subclassification. For this structure, 49,318 images were reconstructed with a 28-Å rise and 167° rotation. The structures observed in the resulting reconstructions of the gizzard derived Tm containing Ac:Tm filaments are about 8 Å-resolution, judging by Fourier-shell correlation at the 0.5 criteria and by features that can be visualized in the structures (Figures 4 and S1). A B-factor of −487 Å2 and an 8-Å low pass filter were applied to all maps of the gizzard Tm-containing Ac:Tm structure; all reconstructions were rendered in Chimera 56. Flexible fitting was performed with NAMD 57.
EMDB deposition
All reconstructions were deposited in the EMDB with the following map codes: cardiac - EMD-5752; gizzard - EMD-5751. The coordinates with a poly-alanine Tm model docked into the gizzard Ac:Tm are submitted to the PDB with the codes RCSB160242/PDB 3J4K.
Supplementary Material
Highlights.
We used cryo-3DEM to explore cardiac and gizzard muscle tropomyosin on FActin.
At 8-Å resolution, we define the C-state position of Tm on F-Actin.
A model is proposed that invokes rocking of Tm to make way for myosin binding.
Acknowledgments
The authors kindly thank Ms. Cynthia Stephan-Reyes for her assistance in particle selection and to Dr. P. Bryant Chase for constructive conversations. This work was funded by an American Heart Association award to MES (#10IRG4300065) and National Institutes of Health awards to William Lehman (R37 HL036153), Roger Craig (AR034711), Larry Tobacman (HL0-63774), and SMS (GM086892). WL, RC, and LT provided specimen. This work was initiated while DRS was supported by an NIH training grant (HL007224) to the Whitaker Cardiovascular Institute at Boston University (J.E. Freedman, P.I). Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 2.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 3.Yang YZ, Korn ED, Eisenberg E. Cooperative binding of tropomyosin to muscle and Acanthamoeba actin. J Biol Chem. 1979;254:7137–40. [PubMed] [Google Scholar]
- 4.Stewart M, McLachlan AD. Fourteen actin-binding sites on tropomyosin? Nature. 1975;257:331–3. doi: 10.1038/257331a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A. 2005;102:18878–83. doi: 10.1073/pnas.0509269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Moore PB, Huxley HE, DeRosier DJ. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J Mol Biol. 1970;50:279–95. doi: 10.1016/0022-2836(70)90192-0. [DOI] [PubMed] [Google Scholar]
- 8.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251:191–6. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- 9.Ebashi S, Kodama A. Native tropomyosin-like action of troponin on trypsin-treated myosin B. J Biochem. 1966;60:733–4. doi: 10.1093/oxfordjournals.jbchem.a128504. [DOI] [PubMed] [Google Scholar]
- 10.Kamm KE, Stull JT. The function of myosin and myosin light chain kinase phosphorylation in smooth muscle. Annu Rev Pharmacol Toxicol. 1985;25:593–620. doi: 10.1146/annurev.pa.25.040185.003113. [DOI] [PubMed] [Google Scholar]
- 11.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–41. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–59. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 13.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 14.Parry DA. Analysis of the primary sequence of alpha-tropomyosin from rabbit skeletal muscle. J Mol Biol. 1975;98:519–35. doi: 10.1016/s0022-2836(75)80084-2. [DOI] [PubMed] [Google Scholar]
- 15.Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A. 2001;98:8496–501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XE, Holmes KC, Lehman W, Jung H, Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J Mol Biol. 2010;395:327–39. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 17.Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys J. 2010;99:862–8. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loong CK, Zhou HX, Chase PB. Persistence length of human cardiac alpha-tropomyosin measured by single molecule direct probe microscopy. PLoS One. 2012;7:e39676. doi: 10.1371/journal.pone.0039676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–9. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 20.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–81. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 21.Brown JH. Deriving how far structural information is transmitted through parallel homodimeric coiled coils: A correlation analysis of helical staggers. Proteins. 2012 doi: 10.1002/prot.24218. [DOI] [PubMed] [Google Scholar]
- 22.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–84. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Geeves MA, Lehrer SS. Dynamics of the muscle thin filament regulatory switch: the size of the cooperative unit. Biophys J. 1994;67:273–82. doi: 10.1016/S0006-3495(94)80478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–65. [PubMed] [Google Scholar]
- 25.Ebashi S, Kodama A. Interaction of troponin with F-actin in the presence of tropomyosin. J Biochem. 1966;59:425–6. doi: 10.1093/oxfordjournals.jbchem.a128320. [DOI] [PubMed] [Google Scholar]
- 26.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 28.Walsh MP, Vallet B, Cavadore JC, Demaille JG. Homologous calcium-binding proteins in the activation of skeletal, cardiac, and smooth muscle myosin light chain kinases. J Biol Chem. 1980;255:335–7. [PubMed] [Google Scholar]
- 29.Dabrowska R, Sherry JM, Aromatorio DK, Hartshorne DJ. Modulator protein as a component of the myosin light chain kinase from chicken gizzard. Biochemistry. 1978;17:253–8. doi: 10.1021/bi00595a010. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, Craig R, Tobacman L, Horowitz R, Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys J. 1999;77:985–92. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer SS, Morris EP. Dual effects of tropomyosin and troponin-tropomyosin on actomyosin subfragment 1 ATPase. J Biol Chem. 1982;257:8073–80. [PubMed] [Google Scholar]
- 33.Maytum R, Lehrer SS, Geeves MA. Cooperativity and switching within the three-state model of muscle regulation. Biochemistry. 1999;38:1102–10. doi: 10.1021/bi981603e. [DOI] [PubMed] [Google Scholar]
- 34.Behrmann E, Muller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–38. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holthauzen LM, Correa F, Farah CS. Ca2+-induced rolling of tropomyosin in muscle thin filaments: the alpha- and beta-band hypothesis revisited. J Biol Chem. 2004;279:15204–13. doi: 10.1074/jbc.M308904200. [DOI] [PubMed] [Google Scholar]
- 36.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J Struct Biol. 2010;170:313–8. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehrer SS, Golitsina NL, Geeves MA. Actin-tropomyosin activation of myosin subfragment 1 ATPase and thin filament cooperativity. The role of tropomyosin flexibility and end-to-end interactions. Biochemistry. 1997;36:13449–54. doi: 10.1021/bi971568w. [DOI] [PubMed] [Google Scholar]
- 38.Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269:24310–5. [PubMed] [Google Scholar]
- 39.Moraczewska J, Nicholson-Flynn K, Hitchcock-DeGregori SE. The ends of tropomyosin are major determinants of actin affinity and myosin subfragment 1-induced binding to F-actin in the open state. Biochemistry. 1999;38:15885–92. doi: 10.1021/bi991816j. [DOI] [PubMed] [Google Scholar]
- 40.Li XE, Tobacman LS, Mun JY, Craig R, Fischer S, Lehman W. Tropomyosin position on F-actin revealed by EM reconstruction and computational chemistry. Biophys J. 2011;100:1005–13. doi: 10.1016/j.bpj.2010.12.3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zechel K, Weber K. Actins from mammals, bird, fish and slime mold characterized by isoelectric focusing in polyacrylamide gels. Eur J Biochem. 1978;89:105–12. doi: 10.1111/j.1432-1033.1978.tb20901.x. [DOI] [PubMed] [Google Scholar]
- 42.Giometti CS, Anderson NL. Tropomyosin heterogeneity in human cells. J Biol Chem. 1984;259:14113–20. [PubMed] [Google Scholar]
- 43.Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17:1318–23. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Splettstoesser T, Holmes KC, Noe F, Smith JC. Structural modeling and molecular dynamics simulation of the actin filament. Proteins. 2011;79:2033–43. doi: 10.1002/prot.23017. [DOI] [PubMed] [Google Scholar]
- 45.Geeves MA, Fedorov R, Manstein DJ. Molecular mechanism of actomyosin-based motility. Cell Mol Life Sci. 2005;62:1462–77. doi: 10.1007/s00018-005-5015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White SP, Cohen C, Phillips GN., Jr Structure of co-crystals of tropomyosin and troponin. Nature. 1987;325:826–8. doi: 10.1038/325826a0. [DOI] [PubMed] [Google Scholar]
- 47.Tobacman LS, Adelstein RS. Mechanism of regulation of cardiac actin-myosin subfragment 1 by troponin-tropomyosin. Biochemistry. 1986;25:798–802. doi: 10.1021/bi00352a010. [DOI] [PubMed] [Google Scholar]
- 48.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Shrum DC, Woodruff BW, Stagg SM. Creating an infrastructure for high-throughput high-resolution cryogenic electron microscopy. J Struct Biol. 2012;180:254–8. doi: 10.1016/j.jsb.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 51.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 52.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 53.Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–34. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.