Abstract
Skp2 E3 ligase is overexpressed in numerous human cancers and plays a critical role in cell cycle progression, senescence, metabolism, cancer progression and metastasis. In the present study, we identified a specific Skp2 inhibitor using high-throughput in silico screening of large and diverse chemical libraries. This Skp2 inhibitor selectively suppresses Skp2 E3 ligase activity, but not activity of other SCF complexes. It also phenocopies the effects observed upon genetic Skp2 deficiency, such as suppressing survival, Akt-mediated glycolysis as well as triggering p53-independent cellular senescence. Strikingly, we discovered a critical function of Skp2 in positively regulating cancer stem cell populations and self-renewal ability through genetic and pharmacological approaches. Notably, Skp2 inhibitor exhibits potent anti-tumor activities in multiple animal models and cooperates with chemotherapeutic agents to reduce cancer cell survival. Our study thus provides pharmacological evidence that Skp2 is a promising target for restricting cancer stem cell and cancer progression.
Introduction
Cancer is a complex disease characterized by multiple steps of genetic alterations occurring primarily through mutations of tumor suppressors and oncogenes. To date, chemotherapy and radiotherapy represent two major options for cancer treatment through inducing p53-dependent cellular senescence and apoptosis (Vazquez et al., 2008). However, advanced cancers often develop resistance to these treatments as they often lose their p53 response due to frequent mutations on the p53 gene. In this scenario, developing cancer treatment strategies via boosting p53-independent senescence and/or apoptosis responses is a key to the success of advanced cancer treatments. In addition, targeting aerobic glycolysis has recently emerged as a promising strategy for cancer therapies (Vander Heiden et al., 2009). Cancer cells display elevated glycolysis irrespective of the presence or absence of oxygen, which warrants cancer cell proliferation and survival. Aerobic glycolysis is orchestrated by Akt, whose activation is achieved by its membrane translocation and subsequent phosphorylation (Elstrom et al., 2004; Robey and Hay, 2009). As our recent reports uncovered that distinct E3 ligases are utilized to trigger non-proteolytic K63-linked ubiquitination of Akt (Chan et al., 2012, Yang et al., 2009), targeting E3 ligase of Akt may serve as a promising strategy to tame cancer glycolysis.
Skp2 is an F-box protein, constituting one of the four subunits of the Skp1-Cullin-1-F-Box (SCF) ubiquitin E3 ligase complex. Earlier studies showed that Skp2 regulates apoptosis, cell cycle progression and proliferation by promoting ubiquitination and degradation of p27 (Nakayama et al., 2004). However, our recent report attributed a critical role of the Skp2 SCF complex in triggering non-proteolytic K63-linked ubiquitination of Akt (Chan et al., 2012). Overexpression of Skp2 is frequently observed in human cancers and ectopic expression of Skp2 promotes tumorigenesis in a prostate tumor xenograft model (Hershko, 2008, Lin et al., 2009). Moreover, Skp2 overexpression is found to promote cancer invasion and metastasis, whereas its deficiency inhibits these processes (Chan et al., 2010a). Using transgenic mouse models, we and others have illustrated that Skp2 is required for cancer development in multiple tumor-promoting conditions, and that Skp2 deficiency triggers p53-independent, p27-dependent cellular senescence/apoptosis but inhibits Akt-mediated glycolysis (Chan et al., 2012; Lin et al., 2010; Wang et al., 2010). Collectively, these studies suggest targeting Skp2 is a promising strategy for cancer treatment, thereby calling for an urgent need to develop specific Skp2 inhibitors.
In this study, we aimed to develop a specific Skp2 small molecule inhibitor and tested its efficacy on tumor suppression. Using structure-based high-throughput virtual screening technologies (Zhang, 2011), we have identified 25 compounds that potentially interact with Skp2 to prevent its binding to Skp1. Further experiments showed that one compound, called compound #25, indeed binds to Skp2, prevents Skp2-Skp1 interaction and inhibits Skp2 SCF E3 ligase activity, which consequently suppresses survival of cancer cells and cancer stem cells. More importantly, compound #25 also exhibits potent anti-tumor activity in animal studies. Our study therefore offers convincing evidence that pharmacological inactivation of Skp2 is a promising approach for cancer treatment.
Result
Identification of Skp2 small molecule inhibitors using high-throughput in silico screening approaches
The crystal structure of Skp2-SCF complex (PDB entry code: 2AST) has demonstrated that Skp2 directly interacts with Skp1 via its F-box domain and indirectly associates with Cullin-1 and Rbx1 (Schulman et al., 2000; Zheng et al., 2002). Along the large Skp2-Skp1 interaction interface (2,980Å2), some residues contribute more significantly to their binding than others; these residues are called hot spots. Based on our molecular visualization, literature reports and hot spot analysis, we found 19 residues on Skp2 in contact with Skp1.We grouped these critical Skp2-Skp1 contact sites into two pocket-like and distinct regions (Figure 1A). The first region (referred as pocket 1) is close to the N-terminus of Skp2 and is actually within the F-box motif including Trp97, Phe109, Glu116, Lys119 and Trp127. The second region (referred as pocket 2), in proximity to the C-terminus of Skp2, is formed by the first LRR region and some residues from the F-box (Figure 1A). Therefore inhibitors binding to these two pockets would potentially prevent the Skp2-Skp1 complex formation.
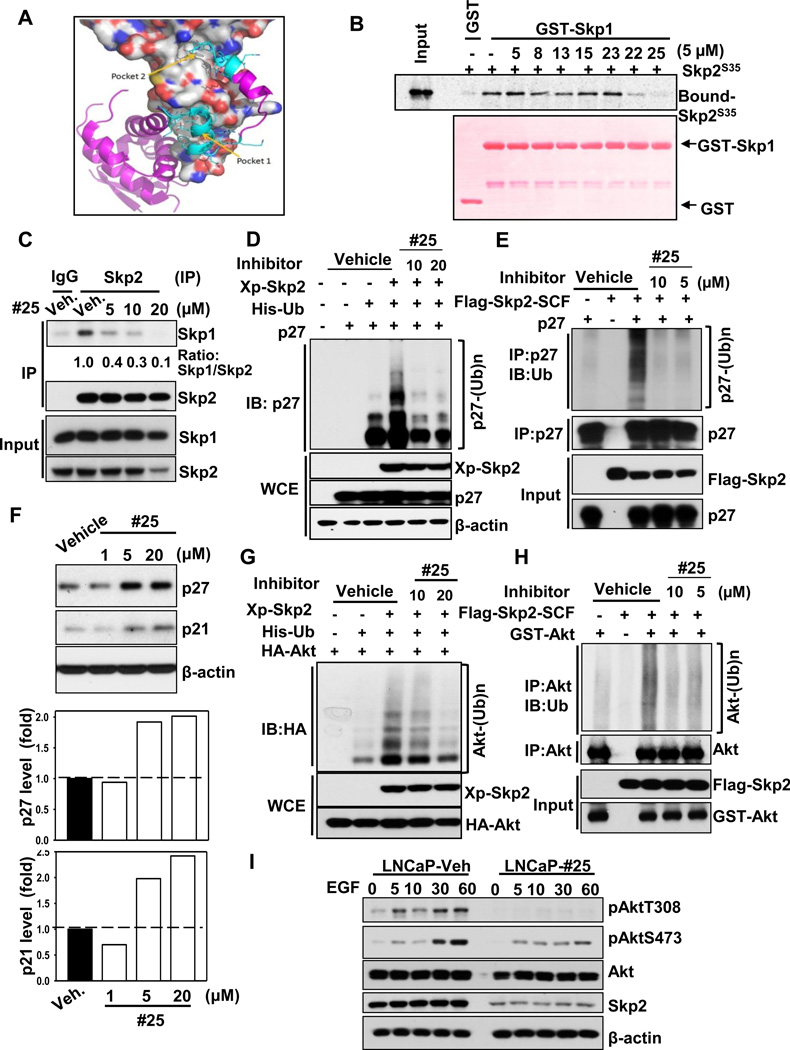
Figure 1. Identification of the Skp2 inhibitor which impairs Skp2 SCF E3 ligase activity via preventing Skp2-Skp1 binding.
(A) The identified potential binding pockets on the interface of Skp2-Skp1 complex. Skp2 is shown as molecular surface (grey for carbon, blue for nitrogen, and red for oxygen atoms). Skp1 is displayed in purple/cyan ribbon and stick form. The cyan region represents residues of Skp1 interacting with Skp2 in the two pockets. (B) In vitro Skp2-Skp1 binding assay with or without compound #25. (C) In vivo Skp2-Skp1 binding assay with or without compound #25 in PC3 cells. (D) In vivo p27 ubiquitination assay in 293T cells transfected with p27, His-Ub, along with Xp-Skp2 in the presence of DMSO or compound #25. WCE indicates whole cell extracts. (E) In vitro Skp2-mediated p27 ubiquitination assay was performed with or without Flag-Skp2-SCF or p27 in the presence of DMSO or compound #25. (F) PC3 cells were treated with DMSO or compound #25 at different doses for 24h and harvested for immunoblotting (IB) assay. (G) In vivo Akt ubiquitination assay in 293T cells transfected with various constructs in the presence of DMSO or compound #25. (H) In vitro Skp2-mediated Akt ubiquitination assay was performed with or without Flag-Skp2-SCF or GST-Akt in the presence of DMSO or compound #25 (I) LNCaP cells were serum starved in the absence or presence of compound #25 for 24h, stimulated with or without EGF and harvested for IB assay. See also Figure S1.
Based on the above predicted potential binding pockets, we performed high-throughput virtual screening with our rigorously validated hit identification workflow (Figure S1A) (Ahad et al., 2011; Yadav et al., 2012). We screened 120,000 diverse and commercially available chemical compounds using our HiPCDock program (Zhang and Du-Cuny, 2009; Zhang et al., 2008), and selected 25 hits that were predicted to bind to Skp2 (Figure S1A). Among them, 16 hits were predicted to interact with pocket 1, nine hits bind to pocket 2 and two hits dock on both pockets. During the selections of these hits, we considered not only the predicted binding affinities, but also various other drug-like properties such as solubility and molecular weight. We propose that by interacting with either (or both) of these two sites, our hits may block the binding of Skp1 to Skp2 and thereby resulting in deconstruction of the Skp2 SCF complex (Figure 1A).
Skp2 inhibitors prevent Skp2-Skp1 interactions and Skp2 SCF E3 ligase activity in vitro
To test our hypothesis generated in silico, we performed in vitro GST-pull down assay to assess which compounds from our 25 hits can prevent Skp2 and Skp1 interactions. We found that seven of them demonstrated strong inhibition of Skp2-Skp1 complex formation at 20µM (data not shown). Next, these seven compounds were subjected to in vitro Skp2-Skp1 binding assay at lower concentration (5µM) in order to identify more potent Skp2 inhibitors (Figure 1B). Strikingly, we revealed that SZL-P1–41 (referred to as compound #25) could completely prevent Skp2-Skp1 interactions at this concentration (Figure 1B). Consistently, compound #25 impeded in vivo binding of Skp2 to Skp1 in a dose-dependent manner (Figure 1C). It should be noted that another compound SZL-P1–35 (referred to as compound #22) also reduced Skp2-Skp1 complex formation, albeit to a lesser extent compared to compound #25 (Figure 1B). As the interaction between Skp2 and Skp1 is a prerequisite for the integrity of Skp2 SCF complex and is crucial for executing its E3 ligase activity (Schulman et al., 2000; Zheng et al., 2002), our data illustrated that the small molecules identified through in silico approach indeed prevent Skp2-Skp1 association.
The canonical role of the Skp2 SCF complex is primarily to trigger ubiquitination and degradation of its protein substrates (Chan et al., 2010b; Frescas and Pagano, 2008). Among all canonical Skp2 substrates, p27 is the most well characterized target whose deficiency can rescue the phenotypes observed in Skp2−/− mice, indicating that p27 is a physiological substrate for Skp2 (Nakayama et al., 2004). Since compound #25 displayed the highest efficacy in preventing Skp2-Skp1 complex formation (Figure 1B), we further determined its effect on Skp2 SCF E3 ligase activity. In vivo and in vitro p27 ubiquitination assays revealed that compound #25 inhibited Skp2-mediated p27 ubiquitination (Figures 1D and 1E). Consistent with the p27 ubiquitination assay, we found that treatment with compound #25 induced endogenous p27 protein expression in prostate cancer cells (Figures 1F and 3E), recapitulating the phenotype upon genetic Skp2 ablation (Carrano et al., 1999). Likewise, we found that compound #25 also induced expression of p21 (Figures 1F and 3E), another Skp2 substrate (Bornstein et al., 2003).
Figure 3. The Skp2 inhibitor specific diminishes E3-ligase activity of Skp2-SCF complex, but not of other F-box SCF complex.
(A–D) 293T cells transfected with various constructs in the presence of DMSO or compound #25 was treated with MG132 for 6 h followed by in vivo ubiquitination assay. (E) LNCaP cells were treated with DMSO, Skp2 inhibitors, or MLN4924 for 24h, and harvested for IB assay. See also Figures S4.
Akt is a Skp2 substrate undergoing K63-linked polyubiquitination (Chan et al., 2012). Consistent with the genetic evidence that Skp2 regulates Akt ubiquitination, compound #25 also inhibited Skp2-mediated Akt ubiquitination in vivo and in vitro (Figures 1G and 1H). Since Skp2 is required for EGF-mediated Akt phosphorylation and activation by triggering Akt ubiquitination, we next determined whether our Skp2 inhibitor attenuated the process. As expected, compound #25 profoundly inhibited EGF-mediated Akt phosphorylation and activation in various cancer cell lines (Figure 1I and Figures S1B and S1C). However, administration of compound #25, similar to Skp2 deficiency, displayed no noticeable impact on ERK activation (Figures S1C and S1D), underscoring the important role of Skp2 in selectively regulating EGF-mediated Akt signaling. It should be noted that compound #25 not only abrogated Skp2-Skp1 complex formation and Skp2 E3 ligase activity, but also downregulated Skp2 protein expression at higher dosage (20 µM) (Figures 1C, 1I and 3E and Figures S1B and S1C). Moreover, we uncovered that compound #25 primarily caused downregulation of Skp2 at the protein level, not mRNA level, and this reduction was partially rescued by proteasome inhibitor (Figures S1E and S1F). Collectively, our data indicate that the pharmacologic Skp2 inhibition stabilizes p27 and p21 expression and inhibits EGF-mediated Akt activation.
The physical binding of Skp2 inhibitor to Skp2 prevents Skp2-Skp1 complex formation
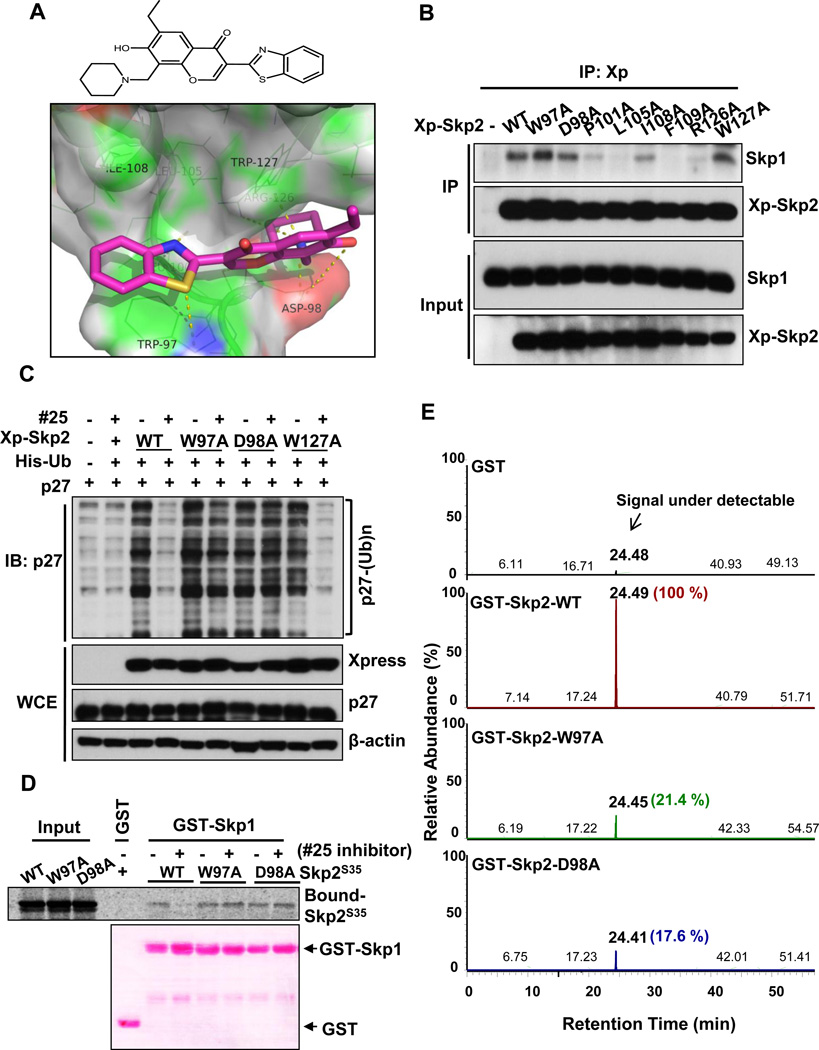
The chemical structure of #25 disclosed in Figure 2A is characterized by 1H, 13C NMR and LC-MS/MS analysis (Figure S2). To understand how compound #25 interacts with Skp2 and prevents Skp2-Skp1 complex formation, we analyzed our molecular docking results. The benzothiazole substructure of #25 is predicted to interact with residue Trp97, mainly via aromatic stacking and polar contacts (Figure 2A). Interestingly, it is also close to a hydrophobic pocket consisting of Trp97, Leu105, Ile108, Phe109 and Trp127 (Figure 2A). Potentially, interaction with this hydrophobic pocket can increase the binding. The other critical part of the inhibitor is the chromone moiety, which is extended through the pocket formed by residues Asp98 and Trp127 and has contacts with these residues via either hydrogen bonds or hydrophobic/aromatic interactions. Interestingly, the ethyl group on the phenol ring does not contribute significantly to the inhibitor binding to Skp2; however, it extends out into the region where Skp1 occupies when it binds to Skp2. Therefore this moiety is critical for preventing Skp1-Skp2 interactions, and removal of this ethyl group indeed abolished activity, in agreement with our structure-activity relationship studies (see SAR analysis section below). Additionally, the piperidine group of #25 is critical for the inhibitor binding as it interacts closely with Asp98 and Trp127.
Figure 2. Skp2 inhibitor directly interacts with Skp2 at Trp97 and Asp 98 residues.
(A) Chemical structure of compound #25. 3-(1,3-benzothiazol-2-yl)-6-ethyl-7-hydroxy-8-(1-piperidinylmethyl)-4H-chromen-4-one (upper panel). The docking between compound #25 (magenta sticks) and predicted pocket 1 of Skp2 (surface representation). The residues in green lines form hydrogen bonding/hydrophobic/aromatic stacking interactions with compound #25. The yellow dashed lines represent hydrogen bonds between compound #25 and Skp2 (lower panel). (B) In vivo Skp2-Skp1 binding assay in 293T cells transfected with Skp2 or its various mutants. (C) In vivo p27 ubiquitination assay in 293T cells transfected with various constructs in the presence of DMSO or compound #25. (D) In vitro binding of Skp1 with Skp2 WT or various mutants in the presence of DMSO or compound #25. (E) The extracted ion chromatography spectra demonstrating the quantity of compound #25 bound to GST alone, GST-Skp2 WT, W97A mutant and D98A mutant (retention time ~ 24 min.). The identity of the bound compound was further confirmed by MS/MS analysis as shown in Figure S3. See also Figures S2 and S3.
We found that our pocket 2 inhibitors (such as inhibitor #5) have very different core scaffolds. They bind to pocket 2 mainly via interactions with Trp128, Ser132, Trp137, Gln158, and the large leucine-rich hydrophobic pocket formed by the first and second LRRs. The potent Skp2 inhibitor (inhibitor #25 in Figure 1B) was predicted to bind to pocket 1, but not pocket 2, indicating that the pocket 1 within the F-box motif is probably more critical to the Skp2-Skp1 interaction. This notion is in agreement with previous reports underscoring the importance of F-box motif in Skp2 SCF complex formation as well as Skp2 E3 ligase activity (Kim et al., 2003; von der Lehr et al., 2003). Based on our modeling predictions, residues Trp97, Asp98, Arg126 and Trp127 from Skp2 directly interact with compound #25 (Figure 2A). While Pro101, Leu105, Ile108 and Phe109 residues are involved in Skp1 binding, but not significantly contributing to the binding of #25. We next performed site-directed mutagenesis to mutate these residues to alanine (Ala) and determined which residues of Skp2 are critical for Skp2-Skp1 binding and Skp2 E3 ligase activity. Of these residues, only the Skp2 Trp97Ala, Asp98Ala, and Trp127Ala mutants still retained their binding capacity to Skp1, whereas the rest of other Skp2 mutants lost their interaction with Skp1 (Figure 2B). The nature of the Arg126Ala mutant, which lost its ability to bind to Skp1, precluded us to further examine the inhibitory effect of compound #25 on its E3 ligase activity and binding to Skp1. For this reason, we focused on Skp2 Trp97Ala, Asp98Ala, and Trp127Ala to determine the activity of compound #25 on their E3 ligase activity and interaction with Skp1. While compound #25 abrogated E3 ligase activity of WT Skp2 and Skp2 Trp127Ala mutant, as determined by in vivo p27 ubiquitination assay, it failed to inhibit E3 ligase activity of Skp2 Trp97Ala and Asp98Ala mutants, (Figure 2C). Furthermore, in vitro Skp2-Skp1 binding assays revealed that although compound #25 prevented the interaction between WT Skp2 and Skp1, it was unable to inhibit the binding of Skp2 Trp97Ala and Asp98Ala mutants to Skp1 (Figure 2D). These results suggest that Trp97 and Asp98 residues of Skp2 are critical sites for the binding of Skp2 to compound #25.
To corroborate this notion, we performed the in vitro binding assay to compare the binding affinity of compound #25 to WT Skp2 and Skp2 mutants (Trp97Ala and Asp98Ala) using LC-MS/MS analysis. Of note, since the nature of recombinant Skp2 protein was unstable in the absence of Skp1 (Li et al., 2005), the GST-Skp2 WT, GST-Skp2 Trp97Ala and GST-Skp2Asp98Ala mutants subjected to LC-MS/MS analysis were co-expressed, purified and preformed complex with Skp1 (Figure S3A). In the preexisting Skp2-Skp1 complex, we clearly detected that compound #25 readily bound to WT Skp2, but failed to interact with the Trp97Ala and Asp98Ala mutants (Figure 2E and Figure S3B). Accordingly, our results underscore that the binding of compound #25 to Skp2 at Trp97 and Asp98 is essential for blocking Skp2 E3 ligase activity.
Skp2 inhibitor specifically suppresses the Skp2 SCF complex E3 ligase activity
To evaluate the specificity of the compound #25, we determined whether compound #25 affects E3 ligase activity of other F-box proteins, such as Fbw7 and β-TrCP. Although compound #25 displayed a profound effect on Skp2-mediated ubiquitination of p27 and Akt (Figures 1D, 1E, 1G, and 1F), it showed no appreciable effects on Fbw7-mediated ubiquitination of c-Jun and MCL-1 or β-TrCP-mediated ubiquitination of Snail and IκBα (Figures 3A–3D). Consequently, compound #25 selectively stabilized expression of Skp2 substrates including p27, p21 and Notch1, but not Fbw7 substrates, such as c-Jun and MCL-1 and β-TrCP substrates, such as Snail, IκBα and β-catenin (Figure 3E and Figure S4A). In contrast, treatment of MLN4924, a potent inhibitor for Nedd8 activating enzyme (NAE) that suppresses activity of Cullin-RING based ubiquitin ligases (CRLs) (Soucy et al., 2009), attenuated Fbw7-mediated c-Jun ubiquitination (Figure S4B). Additionally, substrates of SCF complexes, such as c-Jun, p27 and p21, were globally induced by MLN4924 (Figure 3E). Of note, MLN4924 also induced expression of heat shock protein 27 (Hsp27), indicating that MLN4924 elicits heat shock response through globally inhibiting intracellular proteolytic response (Figure 3E). These results underscore that compound #25 selectively targets E3 ligase activity of the Skp2-SCF complex but not other SCF complexes.
Skp2 inhibitor restricts cancer cell survival through triggering p53-independent cellular senescence and inhibiting aerobic glycolysis
Skp2 deficiency has a significant impact on cancer cell proliferation and survival by triggering cellular senescence, apoptosis, or inactivation of glycolysis (Chan et al., 2011; Chan et al., 2012; Lin et al., 2010; Yokoi et al., 2003). To test the therapeutic values of our Skp2 inhibitor, we determined the impact of compound #25 on cancer cell proliferation and survival. Indeed, compound #25 displayed potent effects on cell viability of prostate cancer cell lines; including PC-3 and LNCaP cells, but only slightly affected cell viability of normal prostate epithelial PNT1A cells (Figure 4A and Figure S5A). Our results indicate that compound #25 has a great selectivity on proliferation of prostate cancer cells over normal cells. We extended this finding to other cancer cell lines by showing that compound #25 favorably triggered cytotoxicity in tumorigenic lung cancer cells over non-tumorigenic lung fibroblasts (Figure 4B and Figure S5A). Moreover, we found that it elicited a potent suppressive effect on the viability of other human-tumor derived cell lines, including H3255 (lung), H1299 (lung), Hep3B (liver) and U2OS (osteocarcinoma), although different cancer cell types exhibited variable sensitivity to the Skp2 inhibitor (Figure S5A).
Figure 4. Inhibition of E3-ligase activity of Skp2-SCF complex results in cancer cell death, glycolysis defects and cellular senescence.
(A) Prostate cancer cells and normal epithelial cells (PNT1A) were treated with various doses of compound #25, followed by cell survival assay. (B) Lung cancer cells and normal fibroblasts (IMR90) were treated with various doses of compound #25, followed by cell survival assay. (C) PC3 cells were treated with or without compound #25 for 4 days and harvested for senescence assay. (D, E) Lactate production was measured in PC3 (D) or LNCaP (E) cells treated with DMSO, LY294002 or compound #25. (F) Apoptosis rate was determined in PC3 cells treated with DMSO or compound #25. (G, H) PC3 cells with or without Skp2 knockdown (G) or PC3 cells stably expressed with Skp2 WT, W97A or D98A mutants (H) were treated with various doses of compound #25, followed by cell survival assay. Cell survival percentage of each stable cell lines treated with various doses of compound #25 was normalized to that treated with DMSO. Results are presented as mean values ±S.D. * indicates p<0.05, ** indicates p<0.01. See also Figures S5 and S6.
We next determined the underlying mechanism by which compound #25 regulates cancer cell proliferation and survival. Consistent with the genetic Skp2 deficiency, our compound #25 caused cell cycle arrest at G2/M phase (Figures S5B–5D). Also, it triggered cellular senescence in p53-deficient prostate cancer cells, indicating that pharmacological Skp2 inactivation induced a p53-independent senescence response (Figure 4C). Moreover, Skp2 inhibition suppressed glycolysis in two prostate cancer cell lines similar to Akt inactivation, as determined by lactate production (Figures 4D and 4E). In addition, compound #25 caused higher apoptosis rates in cancer cells, consistent with the observations in Skp2-deficient cells (Figure 4F and Figures S6A and S6B). These results, along with previous reports, illustrate that genetic and pharmacological Skp2 inhibition attenuates cancer cell proliferation and survival possibly through regulating p53-independent cellular senescence, aerobic glycolysis or apoptosis.
To determine whether the biological effect of compound #25 is attributed to Skp2 inhibition, we examined the effect of compound #25 on cell survival in control- and Skp2-knockdown prostate cancer cells. While compound #25 displayed potent inhibitory effect on viability of control-knockdown cells in a dose-dependent manner, its inhibitory effect is compromised upon Skp2 silencing (Figure 4G and Figure S6C). We further demonstrated that ectopic expression of Skp2 Trp97Ala or Skp2 Asp98Ala mutants, which is resistant to the binding of compound #25, attenuated the inhibitory effect of compound #25 on cell viability (Figure 4H and Figure S6D). Taken together, our results indicate that the action of compound #25 on cancer cell viability acts through its Skp2 inhibition and binding.
Structure-activity relationships (SARs) of Skp2 inhibitor
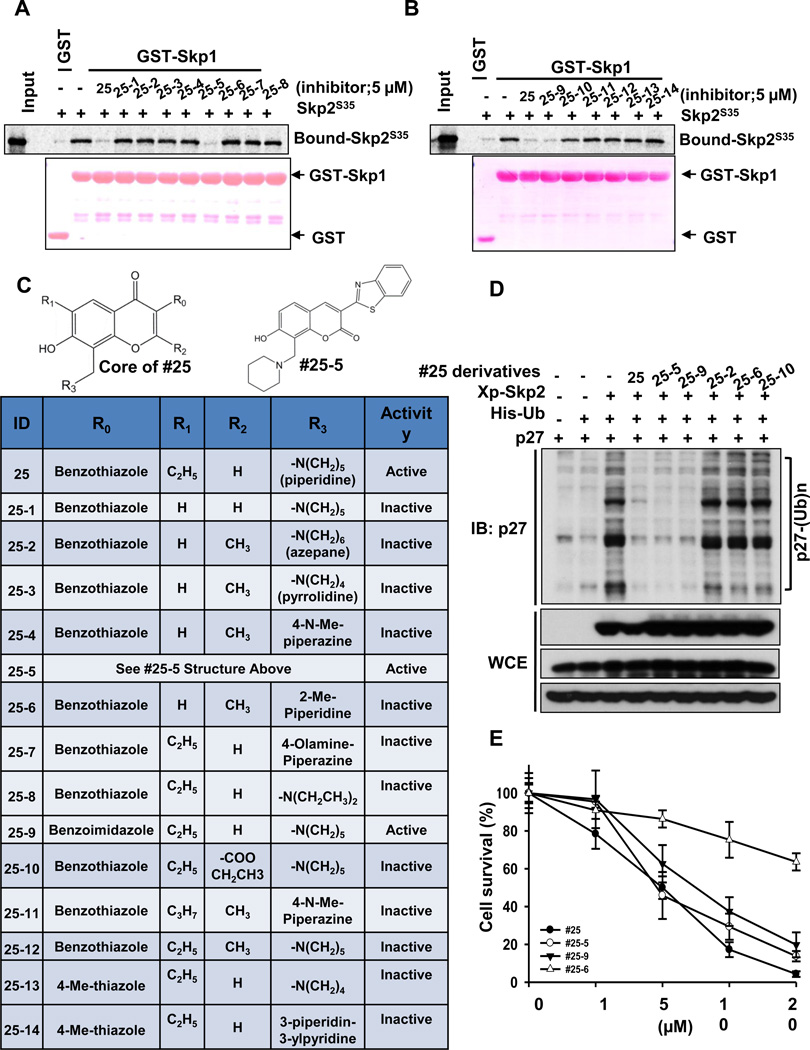
To explore the SARs of our Skp2 inhibitor, we have identified a series of compound #25 derivatives and evaluated their effects on in vitro Skp2-Skp1 binding (Figures 5A–5C). Our data demonstrated that the benzothiazole at the R0 position of the chromone core is necessary for binding, in agreement with our docking studies where it forms pi-stacking interactions with residue Trp97. The change of this group to a small thiazole ring caused the loss of activities (comparing #25 to #25-13 and #25-14 in Figures 5B and 5C), but modifications to a comparable moiety such as benzoimidazole retained the activities (#25-9). Apparently, the ethyl group on the R1 position is critical as the activities are completely abolished when it is changed to hydrogen (e.g., #25 compared to #25-1 in Figures 5A and 5C). This is consistent with our docking observations where this group occupies the space where the Skp1 chain resides in the SCF complex. Our SAR analysis indicated that any change to a big group (e.g., from hydrogen to methyl or ethyl ester) at the R2 position is detrimental to the inhibitor binding (e.g. #25 compared to #25-10 and #25-12 in Figures 5B and 5C). This is also in agreement with our structural observations where the R2 group has close contacts with Skp2 and therefore a small group is favorable to binding. Additionally, the chromone core of compound #25 can be altered to chromen-2-one, as compound #25-5 exhibited comparable ability to the lead compound (#25) in preventing Skp2-Skp1 interactions (Figures 5A and 5C). We also found that modifications on the R3 position, such as from the lead compound #25 to #25-7 or #25-8, destroyed the inhibitor activities. This is not surprising because in our docking models, we observed that the piperidine ring is in a channel formed by residues Asp98 and Trp127. It is also in proximity to residue Arg126. Therefore, a no-substituted ring structure is preferred (Figures 5A and 5C). This also supports our site-directed mutagenesis data that Asp98 is the critical binding residue for Skp2 inhibitors (Figures 2 and 5A–5C). It is worth mentioning that the lack of the ethyl group on #25-5 did not affect the activity significantly (Figures 5A and 5C) because this compound has a slightly altered binding pose due to its different core scaffold. As a matter of fact, it seems that the piperidine moiety in #25-5 may partially serve the function of the ethyl group to repel Skp1 away (data not shown).
Figure 5. The structure-activity relationship (SAR) of compound #25 derivatives.
(A, B) In vitro Skp2-Skp1 binding assay in the presence of DMSO, compound #25 or its derivatives. (C) SAR of compound #25 and its derivatives. #25-5 is illustrated separately due to its unique core structure. (D) In vivo p27 ubiquitination assay in 293T cells transfected with various constructs in the presence of DMSO, compound #25 or its derivatives with MG132 treatment. (E) PC3 cells were treated with various doses of compound #25 or its derivatives, followed by cell survival assay.
Based on the SAR analysis, both compounds #25-5 and #25-9 disrupted in vitro Skp2-Skp1 binding as efficiently as lead compound #25 (Figures 5A–5C). To determine whether these two derivatives exhibited the same biological efficacy as compound #25, we compared their roles in Skp2 E3 ligase and cancer cell survival. As expected, they profoundly inhibited Skp2-mediated p27 ubiquitination and cancer cell survival as compound #25 did, whereas other derivatives failed to do so (Figures 5D and 5E). These data along with mutagenesis results (Figures 2 and 5) collectively illustrate that the stable interactions with the Trp97 and Asp98 residues is indispensible for the activities of Skp2 inhibitors in suppressing Skp2-Skp1 binding, Skp2 E3 ligase activity, and cancer cell survival (Figures 2 and 5).
Skp2 inactivation inhibits cancer stem cell populations and self-renewal capability
Like stem cells, cancer stem cells (CSCs, also known as cancer initiating cells) express similar surface markers and possess asymmetric cell division and self-renewal capability. CSCs have recently emerged to play roles in cancer initiation and progression (Rajan and Srinivasan, 2008). However, how CSC’s populations and self-renewal ability are regulated remains largely unclear. A recent report illustrated glycine decarboxylase drives CSC properties and tumorigenesis by promoting aerobic glycolysis (Zhang et al., 2012). In addition to glycolysis, cellular senescence through telomere shortening has been linked to restrain CSC populations (Marian et al., 2010). In light of these observations, we reasoned that targeting aerobic glycolysis pathways and inducing cellular senescence may reduce CSC self-renewal and favor chemotherapy sensitivity.
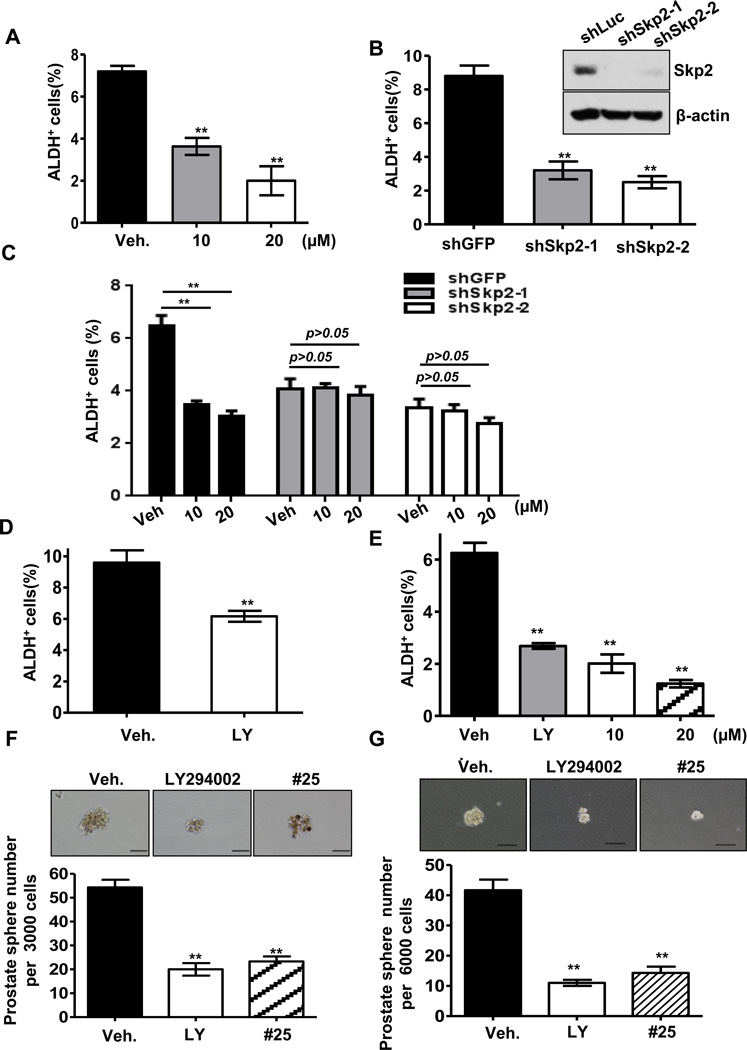
As Skp2 regulates p53-independent cellular senescence and Akt-mediated aerobic glycolysis, we determined whether Skp2 inactivation has any effect on CSC populations and their self-renewal capability. Both Skp2 knockdown and pharmacological Skp2 inactivation in prostate cancer cells reduced CSC populations, similar to what inhibition of the PI3K-Akt pathway did (Figures 6A–6E). Moreover, we found while compound #25 reduced the population of prostate CSCs in a dose dependent manner, it failed to do so upon Skp2 silencing. This data underscores the fact that Skp2-regulated CSC pool size strictly depends on Skp2 (Figure 6C). The prostate sphere formation assay also revealed that Skp2 inhibition by genetic and pharmacological approaches markedly suppressed self-renewal ability of prostate CSCs, so did Akt inactivation (Figures 6F and 6G). These results uncover a crucial role of Skp2 in regulating CSC population and self-renewal capability.
Figure 6. Skp2 inactivation diminishes cancer stem cell properties and heightens cancer cell sensitivity to chemotherapy.
(A) Populations of ALDH+ cells were determined by FACS analysis in PC3 cells treated with vehicle or compound #25. (B) Populations of ALDH+ cells were determined by FACS analysis in PC3 cells with control or Skp2 knockdown. (C) Populations of ALDH+ cells were determined by FACS analysis in PC3 cells with control or Skp2 knockdown treated with vehicle or compound #25. (D) Populations of ALDH+ cells were determined by FACS analysis in PC3 cells with DMSO or LY294002. (E) Populations of ALDH+ cells were determined by FACS analysis in LNCaP cells treated with DMSO, LY294002 or compound #25. (F, G) Prostate sphere- formation assay in PC3 (F) and LNCaP (G) cells treated with DMSO, LY294002 or compound #25. Results are presented as mean values ±S.D. ** indicates p<0.01.
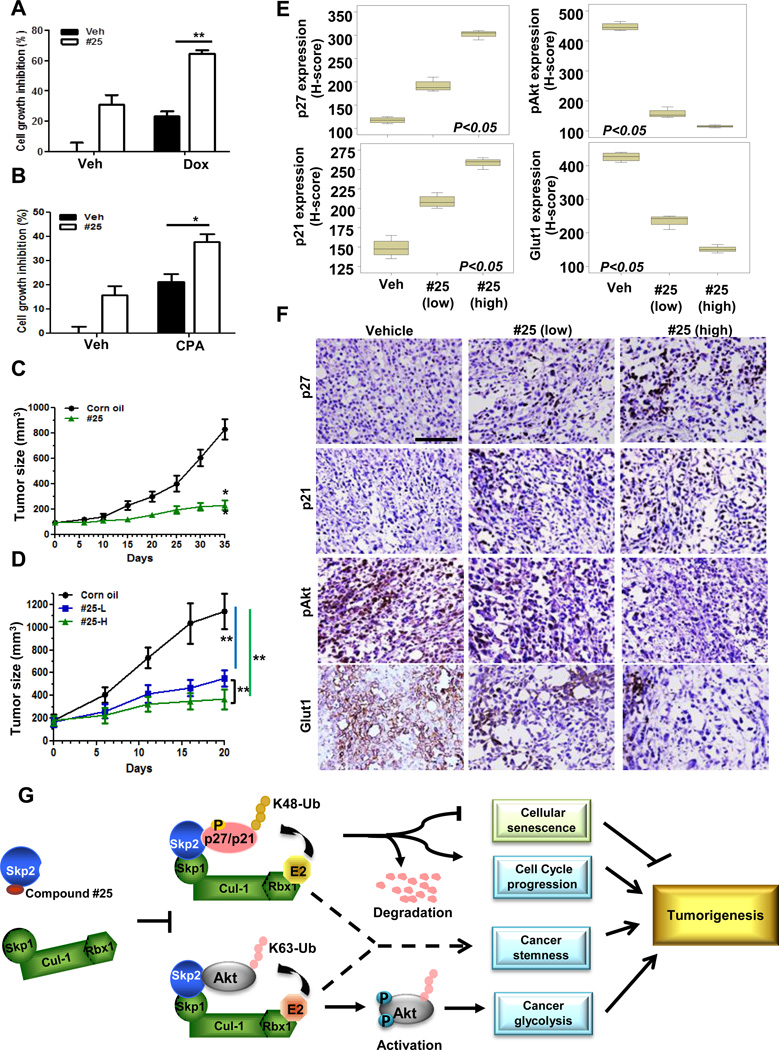
The existence of CSCs is the major cause for cancer cell resistance to chemotherapy (Koch et al., 2010). As genetic and pharmacological Skp2 inactivation effectively restricts CSCs properties in prostate cancer cells (Figure 6), we reasoned that our Skp2 inhibitor may sensitize cancer cell response to chemotherapy agents, such as doxorubicin (Dox) and cyclophosphamide (CPA). As expected, Skp2 inhibition increased the efficacy of Dox and CPA on prostate cancer cells (Figures 7A and 7B), indicating that our Skp2 inhibitor may overcome chemo-resistance via eradicating CSC populations.
Figure 7. Skp2 inhibitor suppresses tumor growth in human tumor xenografts.
(A, B) PC3 cells in the absence or presence of compound #25 were treated with Dox (A) or CPA (B), followed by cell survival assay. (C, D) Nude mice bearing A549 (C) or PC3 (D) tumor xenografts were administration with or without compound #25 via IP injection Mean tumor volumes ± s.d. are shown. n=6 mice per group. (E, F) The quantification results (E) and representative images (F) of histological analysis of p27, p21, pAkt, and Glut1 in PC3-induced tumor xenografts. “Low” indicates 40 mg/kg; “high” indicates 80 mg/kg of compound #25 were injected into mice. Scale bar indicates 100 µm. (G) The working model depicts how Skp2 inhibitor prevents Skp2-SCF complex formation and results in tumor suppression. Results are represented as mean values ±S.D. * indicates p<0.05, ** indicates p<0.01. See also Figure S7.
Pharmacological Skp2 inactivation exhibits strong anti-tumor activities in vivo
Escape of cellular senescence and promotion of aerobic glycolysis are two critical hallmarks for cancer progression and cancer drug resistance, and targeting these two events are therefore ideal approaches for cancer treatment. Since we have shown that compound #25 effectively triggers senescence and inhibits glycolysis, we then examined the efficacy of compound #25 on suppression of preexisting tumors in vivo. Figures 7C and 7D demonstrated that compound #25 displayed a potent effect on inhibiting prostate and lung tumor growth in vivo. Importantly, compound #25 restricted tumor growth in a dose-dependent fashion. Of note, compound #25 resulted in a dose-dependent decrease in Skp2 levels in tumor tissues, correlated with in vivo upregulation of p27, p21 and apoptosis, and downregulation of cancer cell proliferation, Akt activity and Glut1 (glucose transporter 1) expression, which are both critical regulators for glycolysis (Figures 7D–7F and Figures S7A and S7B). The pharmacokinetic analysis further revealed that the concentration of compound #25 reached maximum levels in blood 1h after IP administration (Cmax was 1428±1185 ng/ml), maintained up to 70 % of the compound for 6h, but quickly declined afterward with a terminal concentration of 126±48 ng/ml 24h after administration (Figure S7C). Of note, compound #25 indeed reached tumors within 1h with a concentration of 4787±1967 ng/g and slowly declined in tumor tissues and about 1164±494 ng/ml of compound #25 remained in tumor tissues 24h after initial administration (Figure S7C). Our results collectively provide convincing pharmacological evidence that Skp2 targeting represents a promising strategy and open an avenue for the treatment of human cancers.
Discussion
Characterization of modes of action of the specific Skp2 inhibitor
Skp2 is frequently upregulated in human cancer and plays a critical role in cancer development by targeting p27 protein degradation or activating Akt signaling, and therefore is regarded as a drug target. Recently, small molecules targeting the p27 pathways were identified: a small molecule cpdA targeting p27 ubiquitination (Chen et al., 2008), small molecules interrupting Skp2-p27 binding (Wu et al., 2012). Although cpdA could cause cell growth defects in vitro, it is unclear how cpdA regulates p27 ubiquitination and whether it has any direct effect on Skp2 activity. Small molecule inhibitors identified by Timothy Cardozo’s group were shown to interrupt Skp2-p27 binding but retain the Skp1-Skp2 complex formation, suggesting these inhibitors presumably will not affect Skp2 E3 ligase activity. Furthermore, the biological efficacy on tumor growth of either cpdA or the inhibitors identified by Timothy Cardozo’s group remains elusive. In our study, we found that compound #25 is a specific Skp2 inhibitor by directly binding to the F-box motif of Skp2, thus preventing Skp2-Skp1 interactions and subsequently abrogating Skp2 E3 ligase activity toward ubiquitination of p27 and Akt in vivo. Interestingly, our data revealed that Skp2 inhibitor #25 when used at higher doses not only prevents Skp1-Skp2 complex formation, but also induces Skp2 degradation in a manner partially dependent on the proteasome, indicating that the Skp2 degradation induced by the Skp2 inhibitor is likely caused by its effect on preventing Skp1-Skp2 complex formation. This notion is consistent with the fact that recombinant Skp2 protein becomes unstable in the absence of Skp1 (Li et al., 2005). Remarkably, compound #25 does not affect activity of other F-Box proteins including Fbw7 and β-TrCP. Future studies are required to characterize the impact of the Skp2 inhibitor on other F-box proteins.
It should be noted that while Skp2 inhibitor #25 could prevent the assembly of Skp2-Skp1 complex in vitro and in vivo (Figure 1B and 1C), it failed to disrupt the preexisting Skp2-Skp1 complex in vitro (data now shown). However, we noticed that in the preformed Skp2-Skp1 complex in vitro, compound #25 could still specifically bind to Skp2 (Figure 2E and Figure S3) and inactivate Skp2 activity as determined by in vitro ubiquitination of p27 and Akt (Figures 1E and 1H). Moreover, we found that compound #25 binds to Trp97 and Asp98 residues of Skp2 and such binding is required for its actions (Figures 2C and 4H). Collectively, our results suggest that two modes of action by Skp2 inhibitor #25 are likely operated in cells. We propose that the primary mechanism of Skp2 inhibitor #25 on Skp2 E3 ligase activity is to prevent the assembly of newly synthesized Skp2-Skp1 complex by binding to Skp2. Another mode of #25 action against Skp2 activity is through its binding to preformed Skp1–Skp2 complex. Nevertheless, both actions of compound #25 on the assembly of newly synthesized Skp2-Skp1 complex or preexisting Skp2-Skp1 complex are attributed to specific Skp2-compound #25 binding, thereby inactivating its E3 ligase activity.
Molecular docking and SAR analysis reveals crucial structural features for Skp2 inhibitor activity
The high-throughput virtual screening employed in the current study allowed us to identify compound #25 as a specific Skp2 inhibitor and reveals its structural binding mechanism (Figure 1 and S1A) to Skp2. Based on the prediction; it was observed that Trp97 is the essential residue which directly interacts with the benzothiazole substructure of compound #25. We also found that Leu105, Ile108, Phe109, and Trp127 form a hydrophobic pocket that is critical for Skp1 binding and may be used to design more potent inhibitors than compound #25. In support of this notion, our mutagenesis data showed that the mutations of these residues to alanine abolished the Skp2-Skp1 interaction. In agreement with the prediction by molecular modeling, Skp2 Trp97Ala and Asp98Ala mutants failed to interact with compound #25, thereby displaying resistance to our inhibitors. Furthermore, derivatives of compound #25 with a bulky group at the R1 but not the R2 position are favorable to the inhibitory activities of interfering with Skp2-Skp1 complex formation and Skp2 activity. The docking and SAR analysis of Skp2 inhibitors not only is consistent with our experimental notion that Trp97 and Asp98 residues are indispensible for the binding of inhibitors, but also provides important information to guide our future chemical optimization of lead compound #25.
Skp2 regulates CSC population and functions
CSCs are regarded as seeds for cancer initiation, progression, and relapse after chemotherapy (Rajan and Srinivasan, 2008). Thus, orchestrating the populations and functions of CSCs may help thwart cancer progression and improve the efficacy of current chemotherapy agents. Our findings using the genetic (Skp2 knockdown) and pharmacological (Skp2 inhibitors) approaches demonstrated that Skp2 inactivation reduces CSC populations and their ability to form prostate spheres (Figure 6), suggesting that Skp2 is a previously uncharacterized, key regulator for the maintenance of CSCs. Given that Skp2 targeting attenuates aerobic glycolysis and induces cellular senescence in cancer cells, our study implies that targeting glycolysis and inducing cellular senescence pathways may represent attractive strategies for CSC elimination.
Pharmacological Skp2 inhibition in treatment of cancer and other diseases
We discovered for the first time specific Skp2 inhibitors targeting Skp2 SCF complex formation by using in silico high-throughput screening of a large chemical database. These inhibitors were validated for their effects on preventing Skp2-Skp1 interaction, inhibiting Skp2 SCF E3 ligase activity toward ubiquitination of p27 and Akt, and repressing in vivo tumor growth. Importantly, we also revealed a critical function of Skp2 in governing CSC properties (Figure 7G). Our study provides proof of principle that pharmacological Skp2 inactivation could profoundly restrict cancer developments in various cancer models and confer sensitivity to chemotherapy, supporting the notion that Skp2 is a promising drug target for human cancers.
Not only does Skp2 regulate glucose metabolism and therefore endows cancer development, but also it plays an essential role in other metabolic disorder such as obesity, evidenced by a previous report demonstrating that Skp2 ablation protect mice from developing obesity through reducing adipocyte proliferation (Sakai et al., 2007). Having shown the promise of the Skp2 inhibitor in suppressing glycolysis, it would be interesting to test whether Skp2 inhibitors could be applied for anti-obesity therapy in the future. As such, our current study opens up an avenue in multiple therapeutic areas that can translate basic findings into the clinical setting, thereby offering great opportunity and impact on treatment of cancer as well as other metabolic disorders.
Experimental Procedures
Cell culture and reagents
293T, PC3, A549, H460, H1299, Hep3B and U2OS cells were cultured in DMEM containing 10% FBS while LNCaP and H3255 were cultured in RPMI-1640 medium containing 10% FBS. (His)6-ubiquitin, GST-Akt, and HA-Akt constructs was described previously (Yang et al., 2009). Flag-Skp2 was a gift from Dr. W. Wei. GST-Skp2 Trp97Ala (W97A) and GST-Skp2 Asp98Ala (D98A) mutants were generated in pGEXSkp2ΔN-Skp1Δ, a gift from Dr. B.A. Schulman, by site-directed mutagenesis assay according to manufacturers’ standard procedures (Stratagene). Compound #25 and its analytical data including 1H, 13C NMR and LC-MS/MS analysis were obtained from ChemBridge. LY294002 was purchased from Cell signaling.
LC-MS/MS analysis of the in vitro binding between Skp2 and Skp2 inhibitor
GST-Skp2 W97A and GST-Skp2 D98A mutants used for demonstrating #25-Skp2 interaction (Figure 2E) were generated in pGEXSkp2ΔN-Skp1Δ bicistronic coexpression constructs. Subsequently, the GST-Skp2 WT, GST-Skp2 W97A and GST-Skp2 D98A mutants were co-expressed, purified and formed complex with Skp1 (Δ38–44 a.a.) as described previously (Li et al., 2005). The purified proteins were incubated with compound #25 for overnight at 4°C, and the GST tags were thrombin-cleaved and pulled-down by glutathione sepharose beads (Invitrogen). The untagged protein samples were pH-adjusted by adding 50 mM ammonium bicarbonate and digested by adding 200 ng modified trypsin (sequencing grade, Promega) for 18 hrs at 37°C. The resulting peptides and compound were analyzed by LC-MS/MS on an Orbitrap-Elite mass spectrometer (Thermo Scientific, Waltham MA). Proteins were identified by database searching of the fragment spectra against the SwissProt (EBI) protein database using Mascot (v 2.3, Matrix Science, London, UK). Typical search settings were: mass tolerances, 10 ppm precursor, 0.8d fragments; variable modifications, methionine sulfoxide, pyro-glutamate formation; up to 2 missed cleavages. The extracted ion chromatograms (XICs) for the bound compounds were examined and peak heights were analyzed and quantified by using Qual Browser (Thermo). The identity of the bound compound shown in XIC (Figure 2E) was further confirmed by MS/MS analysis as shown in Figure S3B.
In vivo drug treatment in the preclinical tumor model
Nude mice bearing A549 xenograft tumors (around 100 mm3) were administrated with vehicle or 80 mg/kg of compound #25 through intraperitoneal (IP) injection, while mice bearing PC3 xenograft tumors (around 180 mm3) were administrated with vehicle, 40 mg/kg or 80 mg/kg of compound #25, respectively, on a daily basis. Tumor size was measured weekly with a caliper, and tumor volume was calculated by using the standard formula: L × W2 × 0.52, where L and W are the length and width, respectively. For mouse pharmacokinetic studies, nude mice bearing PC3 xenograft tumors (around 200 mm3) were administrated with 80 mg/kg of compound #25 through IP injection with a single dosage. Plasma and tumor samples were then harvested from euthanized mice at the indicated time and flash-frozen in liquid nitrogen. 100 µl of plasma were incubated with 2× volume of acetonitrile, mixed and then centrifuged at maximum speed for 3–5 minutes. Tumor tissues were weighed, added with methanol to a wt/vol concentration of 25 mg/mL, and homogenized on ice using a handheld tissue homogenizer. 100 µl of tissue homogenate were mixed with 100 µl of 0.2% formic acid and 100µl of acetonitrile for 5 min., incubated on ice for an additional 10 min and centrifuged. Afterward, 200 µl of clear supernatant from plasma or tumor homogenate were subjected to a HPLC vial for analysis by LC/MS/MS on a Waters Acquity Triple Quadrupole Mass Spectrometer to determine the concentration of compound #25.
Supplementary Material
Highlights.
Integrated computational approaches identified a specific Skp2 inhibitor
The Skp2 inhibitor selectively impairs Skp2 activity and its functions
The Skp2 inactivation restricts cancer stemness and potentiates chemotherapy sensitivity
The Skp2 inhibitor exhibits potent anti-tumor activity
Acknowledgments
We thank Dr. BA Schulman for reagents and Dr. David Hawke at Proteomics Facility of U. T. MD Anderson Cancer Center (MDACC) for the support of LC-MS/MS analysis. The pharmacokinetic analysis is supported in part by the MDACC Support Grant CA016672 with special thanks to the Pharmacology and Analytical Facility. This work was supported by the MDACC Trust Scholar Award, the MDACC Prostate SPORE Development Award (P50 CA140388), NIH grants, and a CPRIT grant to H.K.L., the US Department of Defense Concept Awards (BC085871), US National Institute of Health P41 grant (5P41GM079588-03), Grant # IRG-08-061-01 from the American Cancer Society, MD ACC Prostate SPORE Career Award, and MDACC-UT Austin CTT-TI3D grants to S.Z., the MDACC Breast SPORE Career Development Award and Susan G Komen Foundation Postdoctoral Fellowship Award (PDF12230504) to C. H. C. and the grant from the Department of Health in Taiwan (DOH99-TD-C-111-004) to C.F.L.
Reference
- Ahad AM, Zuohe S, Du-Cuny L, Moses SA, Zhou LL, Zhang S, Powis G, Meuillet EJ, Mash EA. Development of sulfonamide AKT PH domain inhibitors. Bioorg Med Chem. 2011;19:2046–2054. doi: 10.1016/j.bmc.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chan CH, Gao Y, Moten A, Lin HK. Novel ARF/p53-independent senescence pathways in cancer repression. J Mol Med (Berl) 2011;89:857–867. doi: 10.1007/s00109-011-0766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010a;12:457–467. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. Scientific World Journal. 2010b;10:1001–1015. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du-Cuny L, Song Z, Moses S, Powis G, Mash EA, Meuillet EJ, Zhang S. Computational modeling of novel inhibitors targeting the Akt pleckstrin homology domain. Bioorg Med Chem. 2009;17:6983–6992. doi: 10.1016/j.bmc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1424. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Koch U, Krause M, Baumann M. Cancer stem cells at the crossroads of current cancer therapy failures--radiation oncology perspective. Semin Cancer Biol. 2010;20:116–124. doi: 10.1016/j.semcancer.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Li T, Pavletich NP, Schulman BA, Zheng N. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 2005;398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Mickey BE, Wright WE, Shay JW, Bachoo RM. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Rajan P, Srinivasan R. Targeting cancer stem cells in cancer prevention and therapy. Stem Cell Rev. 2008;4:211–216. doi: 10.1007/s12015-008-9037-x. [DOI] [PubMed] [Google Scholar]
- Robey RB, Hay N. Is Akt the "Warburg kinase"?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Sakaue H, Nakamura T, Okada M, Matsuki Y, Watanabe E, Hiramatsu R, Nakayama K, Nakayama KI, Kasuga M. Skp2 controls adipocyte proliferation during the development of obesity. J Biol Chem. 2007;282:2038–2046. doi: 10.1074/jbc.M608144200. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nat Genet. 2010;42:83–88. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–1524. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VR, Prasad S, Gupta SC, Sung B, Phatak SS, Zhang S, Aggarwal BB. 3-Formylchromone interacts with cysteine 38 in p65 protein and with cysteine 179 in IkappaBalpha kinase, leading to down-regulation of nuclear factor-kappaB (NF-kappaB)-regulated gene products and sensitization of tumor cells. J Biol Chem. 2012;287:245–256. doi: 10.1074/jbc.M111.274613. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Yasui K, Iizasa T, Takahashi T, Fujisawa T, Inazawa J. Down-regulation of SKP2 induces apoptosis in lung-cancer cells. Cancer Sci. 2003;94:344–349. doi: 10.1111/j.1349-7006.2003.tb01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Computer-aided drug discovery and development. Methods Mol Biol. 2011;716:23–38. doi: 10.1007/978-1-61779-012-6_2. [DOI] [PubMed] [Google Scholar]
- Zhang S, Du-Cuny L. Development and evaluation of a new statistical model for structure-based high-throughput virtual screening. Int J Bioinform Res Appl. 2009;5:269–279. doi: 10.1504/IJBRA.2009.026419. [DOI] [PubMed] [Google Scholar]
- Zhang S, Kumar K, Jiang X, Wallqvist A, Reifman J. DOVIS: an implementation for high-throughput virtual screening using AutoDock. BMC Bioinformatics. 2008;9:126. doi: 10.1186/1471-2105-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wei L, Bastow K, Zheng W, Brossi A, Lee KH, Tropsha A. Antitumor agents 252. Application of validated QSAR models to database mining: discovery of novel tylophorine derivatives as potential anticancer agents. J Comput Aided Mol Des. 2007;21:97–112. doi: 10.1007/s10822-007-9102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.