Abstract
During DNA replication, mismatches and small loops in the DNA resulting from insertions or deletions are repaired by the mismatch repair (MMR) machinery. Proliferating cell nuclear antigen (PCNA) plays an important role in both mismatch-recognition and resynthesis stages of MMR. Previously, two mutant forms of PCNA were identified that cause defects in MMR with little, if any, other defects. The C22Y mutant PCNA protein completely blocks MutSα-dependent MMR, and the C81R mutant PCNA protein partially blocks both MutSα-dependent and MutSβ-dependent MMR. In order to understand the structural and mechanistic basis by which these two amino acid substitutions in PCNA proteins block MMR, we solved the X-ray crystal structures of both mutant proteins and carried out further biochemical studies. We found that these amino acid substitutions lead to subtle, distinct structural changes in PCNA. The C22Y substitution alters the positions of the α-helices lining the central hole of the PCNA ring, whereas the C81R substitution creates a distortion in an extended loop near the PCNA subunit interface. We conclude that the structural integrity of the α-helices lining the central hole and this loop are both necessary to form productive complexes with MutS α and mismatch-containing DNA.
INTRODUCTION
Inaccurate DNA replication can result in base-base mismatches and small loops arising from insertions or deletions. These mismatches and loops are recognized and repaired by the mismatch repair (MMR) machinery. The mechanisms of MMR in E. coli have been studied extensively and are relatively well understood [1–6]. The MutS protein is a homodimer that recognizes base-base mismatches and small nucleotide insertions and deletions. The MutL protein is a homodimer that interacts with MutS in an ATP-dependent manner to initiate MMR [7–11]. The MutH endonuclease is then activated and generates a nick in the newly synthesized, unmethylated DNA strand of a hemimethylated duplex [12]. Subsequent steps include unwinding and degradation of the newly synthesized DNA strand and filling the resulting gap by DNA polymerase III [1–6].
The mechanisms of MMR in eukaryotes are more complicated and are not as well understood. In yeast, there are six MutS homologs designated Msh1 to Msh6; in mammals, there are five: Msh2 to Msh6. These proteins function as heterodimers, such as MutSβ (comprised of Msh2 and Msh6) and MutSβ (comprised of Msh2 and Msh3), that have partially overlapping mismatch recognition functions [13–15]. In addition to the MutS homologs, there are several MutL homologs, including Mlh1, Mlh2, Mlh3, and Pms1, which also function as heterodimers. The best characterized of these is MutLα (Mlh1/Pms1 in yeast and Mlh1/Pms2 in humans), which functions with both MutSα and MutSβ [16–20]. Mutations in both MutS and MutL homologs that disrupt mismatch repair cause sporadic and hereditary human cancers, including hereditary nonpolyposis colorectal cancer (HNPCC) [2, 21–25]. Other key proteins involved in the subsequent excision and resynthesis steps include exonuclease I (ExoI) [26–30], DNA polymerase delta (pol δ) [31], replication protein A (RPA) [32, 33], replication factor C (RFC) [34], and proliferating cell nuclear antigen (PCNA) [35–37].
The PCNA clamp is an essential replication accessory protein that forms a ring-shaped homotrimer and encircles duplex DNA [38]. In addition to serving as a processivity factor for DNA polymerases, it interacts with a wide variety of proteins and plays important roles in DNA replication, repair, recombination, translesion synthesis, chromatin remodeling, sister chromatid cohesion, and cell cycle regulation [39–44]. During MMR, PCNA functions in the initiation and mismatch recognition stage as well as the excision and resynthesis stage. The role of PCNA in the initial stage of MMR is not well understood. PCNA interacts with both MutSα and MutSβ and is thought to facilitate their recruitment to mismatches [37, 45–49]. Moreover, it has been suggested that PCNA plays a role in strand discrimination, i.e., the recognition of the newly synthesized daughter strand [35, 50, 51].
Various PCNA mutant alleles have been identified that lead to elevated mutation rates [36, 52–54]. Genetic studies have shown that two of these mutant alleles, pol30-201 and pol30-204, specifically disrupt the MMR pathway with little, if any, effect on other DNA metabolic processes [54]. The pol30-201 allele, which encodes the C22Y mutant PCNA protein, completely blocks MutSα-dependent MMR, and the pol30-204 allele, which encodes the C81R mutant PCNA protein, partially blocks both MutSα-dependent and MutSβ-dependent MMR [54]. In order to understand the structural and mechanistic basis by which these mutant PCNA proteins disrupt MMR, we solved the X-ray crystal structures of both mutant proteins and carried out related biochemical studies. We found that these two amino acid substitutions lead to distinct structural changes in PCNA. The C22Y substitution alters the positions of the α-helices lining the inside of the PCNA ring, whereas the C81R substitution disrupts an extended loop located near the PCNA subunit interface. We conclude that the structural integrity of the α-helices lining the central hole and the structural integrity of this loop are both necessary to form productive complexes with MutSα and mismatch-containing DNA.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The wild-type and the C22Y and C81R mutant PCNA proteins from S. cerevisiae were overexpressed as N-terminally His6-tagged proteins in E. coli and were purified as described previously [55]. Replication factor C (RFC) from S. cerevisiae was over-expressed in E. coli and purified as previously described [56]. DNA polymerase delta (pol δ) from S. cerevisiae was over-expressed in S. cerevisiae and purified as previously described [57]. MutSα from S. cerevisiae was over-expressed in E. coli and purified as described previously [58].
DNA and nucleotide substrates
The template strand used to measure pol δ activity was a 68-mer oligodeoxynucleotide with the sequence 5'- GAC GGC ATT GGA TCG ACC TCX AGT TGG TTG GAC GGG TGC GAG GCT GGC TAC CTG CGA TGA GGA CTA GC with biotins on both ends. The X represents the position of a non-damaged G or an abasic site. The primer strand was a 31-mer oligodeoxynucleotide with the sequence 5'-TCG CAG GTA GCC AGC CTC GCA CCC GTC CAA C. The primer strand was 5'-32P-end-labeled with T4 polynucleotide kinase and (γ-32P)ATP and annealed to the template strand at 1 µM in 25 mM TrisCl, pH 7.5, and 100 mM NaCl at 90°C for 2 min and slowly cooled to 30°C. A mixture of all four dNTPs (10 mM each) was purchased from New England Biolabs.
Two 37-mer duplex DNA substrates were used in the sedimentation analysis, one with a G/C pair at position 19 (the homoduplex) and one with a G/T mispair at position 19 (the heteroduplex). The DNA substrates were formed by annealing a top strand to a bottom strand. The top strand, which had a Cy3 fluorescent tag on the 3' end, had the sequence: 5'-ATT TCC TTC AGC AGA TAG GAA CCA TAC TGA TTC ACA T. The bottom strand had the sequence: 5'-ATG TGA ATC AGT ATG GTT XCT ATC TGC TGA AGG AAA T, where the X represents the position of either a C in the case of the homoduplex or a T in the case of the heteroduplex. The annealing reactions were carried out as described above.
Crystallization of the C22Y and C81R mutant PCNA proteins
The C22Y mutant PCNA protein and the C81R mutant PCNA protein were crystallized using the hanging drop method with 400 nl drops prepared using a Mosquito Crystallization Robot (TTP Labtech). The best diffracting C22Y mutant PCNA protein crystals were obtained by combining an equal volume of protein (30 mg/ml) with a reservoir containing 1.6 M ammonium sulfate and 0.1 M citric acid. Crystals formed after 3 days at 18°C. The best diffracting C81R mutant PCNA protein crystals were obtained by combining an equal volume of protein (20 mg/ml) with a reservoir containing 20% PEG1000, 0.2 M MgCl2 hexahydrate, and 0.1 M sodium cacodylate trihydrate, pH 6.5. Crystals formed after 3 days at 18°C.
Data Collection and Structural Determination
The C22Y and C81R mutant PCNA protein crystals were soaked in a mother-liquor solution containing 10% (v/v) glycerol prior to flash-cooling in liquid nitrogen. In the case of the C22Y mutant PCNA protein, data were collected using a Rigaku rotating anode X-ray source. In the case of the C81R mutant PCNA protein, data were collected at the 4.2.2 synchrotron beamline at the Advanced Light Source in the Ernest Orlando Lawrence Berkeley National Laboratory. The data were processed and scaled using d*TREK [59]. For the C22Y mutant protein crystals, the space group was determined to be P212121, and there was one PCNA trimer in the asymmetric unit. For the C81R mutant protein crystals, the space group was determined to be P213, and there was one PCNA subunit in the asymmetric unit. Molecular replacement was performed using the known structure of wild-type PCNA [PDB 1PLQ] with PHASER [60]. Refinement was done using REFMAC5 from CCP4 [61] and PHENIX [62]. All model building was carried out using Coot [63]. The 2Fo-Fc omit maps were generated in PHENIX using the fully refined structure by setting the occupancies of the relevant residues to zero and introducing random shifts in the positions of the atoms by up to 0.5 Å and running a few rounds of refinement.
PCNA trimer stability assays
For non-denaturing polyacrylamide gel electrophoresis (PAGE), the wild-type and C22Y mutant and C81R mutant PCNA proteins (0.1 to 1.0 mg/ml) were incubated in 25 mM TrisCl, pH 7.4, 150 mM NaCl, 0.01% bromophenol blue, and 10% glycerol for 5 min and then run on a TrisCl pre-cast 4–20% gradient non-denaturing polyacrylamide gel (Bio-Rad) at 4°C at a current of 25 mA using 25 mM Tris, pH 8.3, and 0.2 M glycine running buffer. Protein bands were visualized using Coomassie blue staining. For size exclusion chromatography, wild-type and mutant PCNA proteins were diluted to various concentrations (0.01 to 10 mg/mL) in 25 mM Tris, pH 7.4, 150 mM NaCl, 5% glycerol and run on a 120 ml HiLoad 16/60 Superdex 200 PG column (GE Healthcare). The size exclusion chromatography column was calibrated using thyroglobin (670 kDa), γ-globin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa), which eluted at 51.9 ml, 69.5 ml, 85.5 ml, 98.6 ml, and 118.0 ml, respectively.
Polymerase delta activity assays
Running start assays were performed as described previously [64]. Reactions were carried out in the absence of PCNA and in the presence of 90 nM wild-type or mutant PCNA proteins (trimer concentration), and contained 20 nM pol δ, 25 nM DNA, and 100 µM of each of the four dNTPs. Reactions were stopped after 30 min, and the extension products were analyzed on a 15% polyacrylamide sequencing gel containing 8 M urea.
Enzyme-linked immunosorbent assays
The wells of a 96 well EIA/RIA plate (Corning) were coated with 0.75 µg of MutSα in PBS (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl) for two hours. The wells were then washed four times with PBS, 0.2% Tween-20, blocked for one hour with PBS with 5% milk, and washed again. Various amounts of the wild-type, the C22Y mutant, or the C81R mutant PCNA proteins (1 to 20 µg) in 100 µl of PBS with 5% milk were then added to the wells and incubated for one hour. After washing, a 1:1000 dilution of rabbit polyclonal anti-PCNA antibody in PBS with 5% milk was added to the wells and incubated for 30 minutes. The wells were washed again, and a 1:10,000 dilution of goat anti-rabbit antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch) in PBS with 5% milk was added and incubated for 30 minutes. The plate was then washed, and 0.8 mg/mL of O-phenylenediamine (Aldrich) in 0.05 M phosphate-citrate buffer with 0.03% sodium perborate (Sigma) was added. Absorbance was measured at 450nm after various amounts of time (5 to 35 minutes) using an iMark microplate reader (BioRad). Parallel control reactions using bovine serum albumin instead of MutSα were carried out and these background absorbance values were subtracted from the absorbance of each sample at the corresponding PCNA protein concentration. All steps were performed at 25°C.
Sedimentation assays
Samples (100 µl) were prepared with 300 nM MutSα, 300 nM of either the wild-type or mutant PCNA proteins (trimer concentration), and 300 nM of either the homoduplex or heteroduplex DNA in 1xTBS buffer. The samples were incubated on ice for 30 minutes prior to loading on a 5 ml glycerol gradients (15–30%) and were then spun for 20 h at 45,000 rpm at 4°C in a Thermo Sorvall WX ultracentrifuge using an AH-651 swing bucket rotor. Sixteen 300 µl aliquots were collected from the bottom of each gradient, were concentrated using Millipore Amicon® Ultra 10K centrifugal filters, and were analyzed by SDS PAGE using 4–15% pre-cast gradient gels (BioRad). The Cy3-labeled DNA in each fraction was visualized using a BioRad ChemiDoc-MP Imaging System after which the gels were silver stained according to the BioRad Polyacrylamide Gel Staining Procedure.
RESULTS
We have examined two mutant forms of PCNA that are known to cause defects in MMR with little if any defects in other DNA metabolic processes [54]. The C22Y mutant PCNA protein completely blocks MutSα-dependent MMR, and the C81R mutant PCNA protein partially blocks both MutSα-dependent and MutSβ-dependent MMR. In order to understand the structural and mechanistic basis by which they disrupt MMR, we solved the X-ray crystal structures of both mutant proteins and analyzed their biochemical properties.
Structure of the C22Y mutant PCNA protein
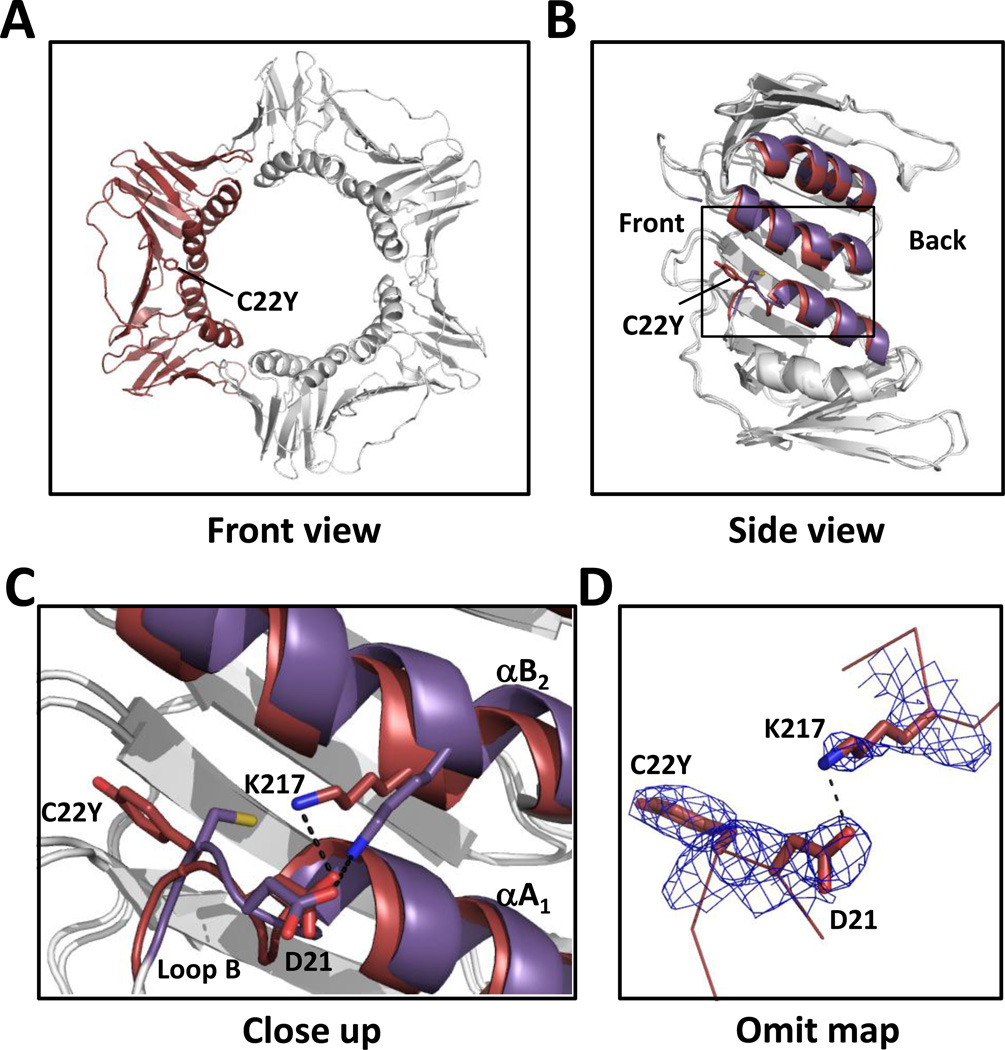
We first determined the X-ray crystal structure of the C22Y mutant PCNA protein to a resolution of 2.7 Å (Table 1). Overall, its structure resembles that of the wild-type PCNA protein with each subunit comprised of two domains, an N-terminal domain (residues 1–117) and a C-terminal domain (residues 135–258) linked by a long, inter-domain connector loop (residues 118–134) (Fig. 1A). The subunits are arranged in a head-to-tail fashion to form a ring-shaped trimer. Residue 22 is located in the N-terminal domain on loop B (residues 21–25). This loop follows α- helix AS (residues 9–20), which along with α-helices B1, A2, and B2, form the inside ring of the PCNA trimer (Fig. 1B). Because of its larger size, the substituted tyrosine side chain is unable to occupy the same position as the wild-type cysteine side chain. Consequently, the tyrosine side chain points toward the front of the PCNA ring rather than toward α-helix B2 (Fig. 1C, 1D). This rearrangement induces a set of other structural changes that ultimately alter the positions of both α-helix A2 and α-helix B2. First, the α-carbon of residue 22 is shifted 1.5 Å from its position in the wild-type protein structure. This in turn causes the α-carbon of Asp-21 to move 1.1 Å and the δ-oxygen of Asp-21 to move 1.6 Å from their positions in the wild-type protein structure. This causes the ε-nitrogen of Lys-217, which is located in α-helix B2 and forms a hydrogen bond with the δ-oxygen of Asp-21, to move 3.4 Å, and this causes the α-carbons in α-helix B2 (residues 209–221) to move up to 2.1 Å from their positions in the structure of the wild-type protein and the α-carbons in α-helix A2 (residues 141–153) to move up to 1.8 Å from their positions in the wild-type protein structure. The changes in the positions of the two α-helices in the C-terminal domain are the most notable structural alterations in the C22Y mutant PCNA protein.
Table 1.
Data collection and refinement statistics.
| C22Y mutant protein | C81R mutant protein | |
|---|---|---|
| (A) Data collection statistics | ||
| Resolution (Å) a | 20.9–2.7 (2.8–2.7) | 21.5–3.0 (3.1–3.0) |
| Wavelength (Å) | 1.54 | 1.00 |
| Space Group | P212121 | P213 |
| Cell (Å) | a = 85.9, b = 90.6, c = 140.6 | a=b=c=121.8 |
| Completeness (%) a | 100 (100) | 100 (100) |
| Redundancy a | 7.1 (7.2) | 19.9 (20.2) |
| <I/σ(I)> a | 7.0 (2.0) | 9.1 (2.0) |
| Rmerge (%) a | 13.3 (65.9) | 14.2 (77.3) |
| (B) Refinement statistics | ||
| Resolution range (Å) | 19.9–2.7 | 20.5–3.0 |
| R (%) | 20.5 | 19.8 |
| Rfree (%) | 28.7 | 25.4 |
| rms bonds (Å) | 0.017 | 0.013 |
| rms angles (°) | 1.78 | 1.57 |
| Number of protein atoms | 6026 (761 residues) | 1975 (254 residues) |
| Number of water molecules | 0 | 0 |
| Ramachandran analysis (%) | ||
| Most favored | 92.5 | 92.0 |
| Allowed | 6.3 | 8.0 |
| PDB ID code | 4L6P | 4L60 |
Values in parentheses are for the highest resolution shell
Figure 1.
Structure of the C22Y mutant PCNA protein. (A) Front view of the C22Y mutant PCNA protein trimer with one of the subunits shown in red. (B) Side view of one subunit of the C22Y mutant PCNA protein with the α-helices A2, B2, and A1 shown in red. The wild-type PCNA structure is overlaid with the same α-helices shown in purple. (C) Close up of the region near the C22Y substitution. The mutant PCNA protein is shown in red and the wild-type PCNA protein is shown in purple. (D) 2Fo-Fc omit map contoured at 1σ and carved at 1.7 Å around the substituted tyrosine at residue 22, Asp-21, and Lys-217.
Structure of the C81R mutant PCNA protein
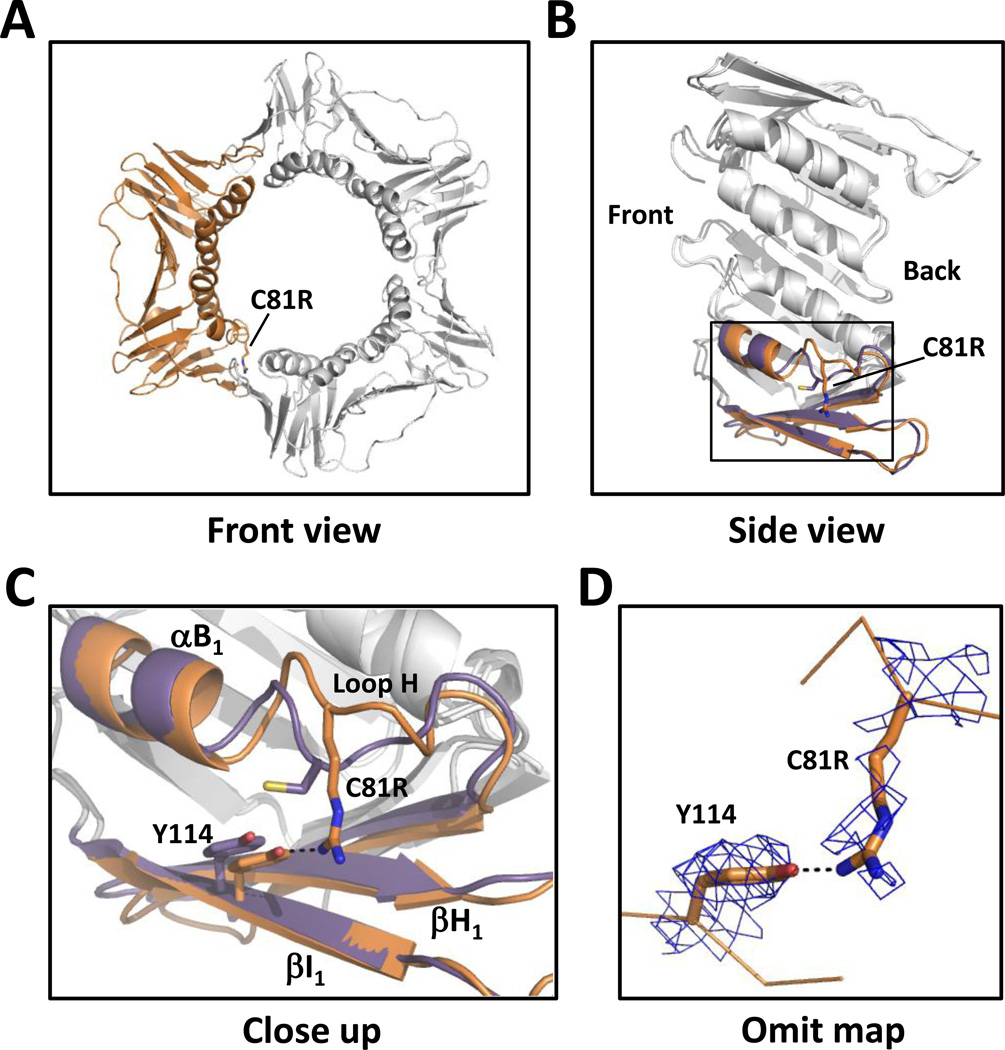
We next determined the X-ray crystal structure of the C81R mutant PCNA protein to a resolution of 3.0 Å (Table 1 and Fig. 2A). Residue 81 is located in the N-terminal domain on loop H (residues 80–86). This loop follows α-helix B1 (residues 72-79) and is near the subunit interface (Fig. 2B). The substituted arginine side chain points toward β-strand I1 (residues 109–117), which comprises the subunit interface, and forms a new hydrogen bond with the η-oxygen of Tyr-114 (Fig. 2C, 2D). This interaction causes the α-carbon of Tyr-114, located in β-strand I1, to move 0.5 Å from its position in the wild-type protein structure. This is a minor change and does not alter the β strands comprising the subunit interface in any substantive way. By contrast, the interaction between the substituted arginine at residue 81 and Tyr-114 causes the α-carbon of residue 81 to move 3.2 Å from its position in the wild-type protein structure thereby distorting the structure of loop H. This distortion of loop H is the only notable structural alteration in the C81R mutant PCNA protein.
Figure 2.
Structure of the C81R mutant PCNA protein. (A) Front view of the C81R mutant PCNA protein trimer with one of the subunits shown in orange. (B) Side view of one subunit of the C81R mutant PCNA protein with the α-helix B1 and the β-strands H1 and I1 shown in orange. The wild-type PCNA structure is overlaid with the same α-helix and β-strands shown in purple. (C) Close up of the region near the C81R substitution. The mutant PCNA protein is shown in orange and the wild-type PCNA protein is shown in purple. (D) 2Fo-Fc omit map contoured at 1 σ and carved at 1.7 Å around the substituted arginine at residue 81 and Tyr-114.
Stability of the mutant PCNA proteins
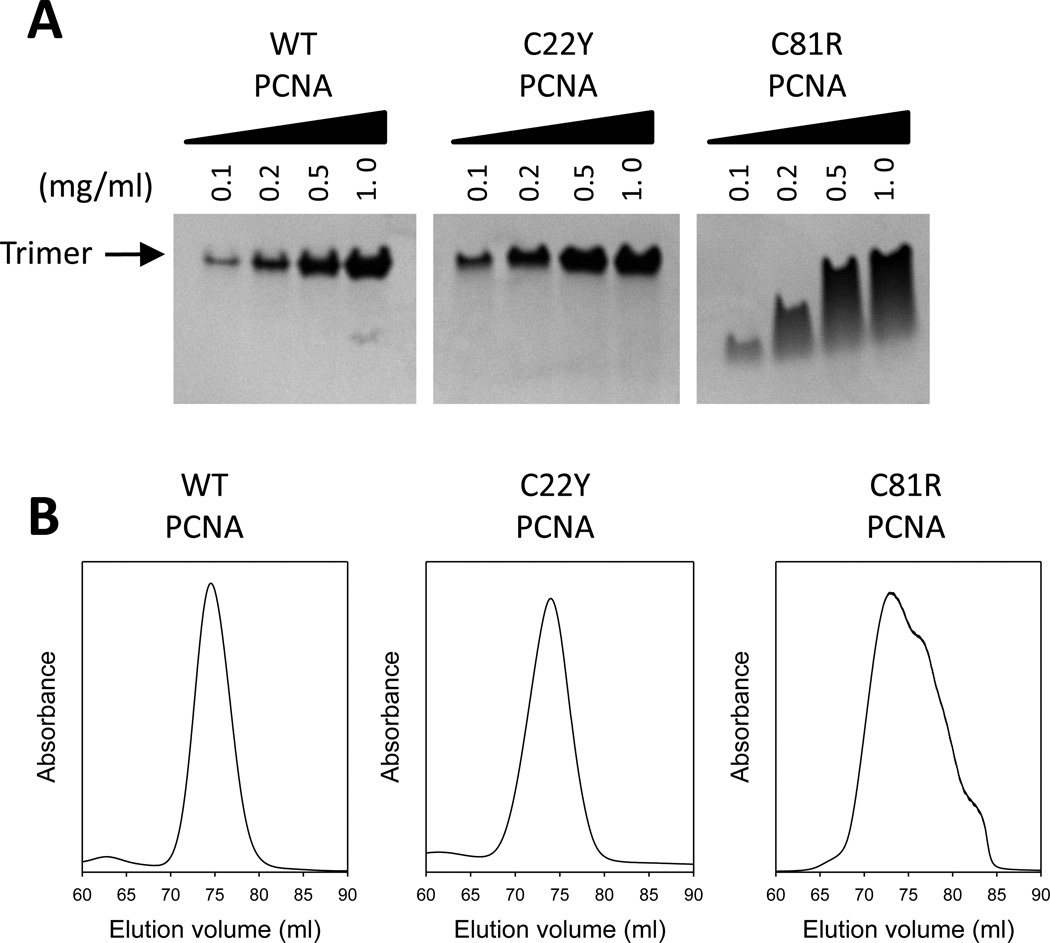
It was previously suggested that the C81R mutant PCNA protein was a monomer in vitro [54]. This, however, seemed inconsistent with the structure of the C81R mutant PCNA protein, which showed it to be a trimer with no significant structural perturbations to the β strands comprising the subunit interface. Thus, we re-examined the stability of the mutant PCNA trimers. First, we used native polyacrylamide gel electrophoresis (PAGE) to determine the oligomeric form of the wild-type and the two mutant PCNA proteins at various concentrations (Fig. 3A). Both the wildtype and the C22Y mutant PCNA proteins were trimers at all concentrations tested (0.1 to 1.0 mg/ml). By contrast, the C81R mutant PCNA protein did not form stable trimers as indicated by the higher mobility species at low concentrations (0.1 and 0.2 mg/ml) and the smeared gel bands at higher concentrations (0.5 and 1.0 mg/ml).
Figure 3.
Trimer stability of the PCNA mutant proteins. (A) Analysis of the PCNA proteins by native gel electrophoresis. Solutions containing the wild-type or mutant PCNA proteins (0.1 to 1.0 mg/ml) were run on a non-denaturing polyacrylamide gradient gel (4 to 20%) and Coomassie stained. The position of the PCNA trimer is indicated. (B) Analysis of the PCNA proteins by size exclusion chromatography. The elution profile of a size exclusion chromatography column in which a solution of the wild-type PCNA protein, the C22Y mutant PCNA proteins, and the C81R mutant PCNA protein (10 mg/ml) were run are shown.
In order to further assess PCNA trimer stability, we analyzed the proteins by size exclusion chromatography (Fig. 3B). When loaded onto the size exclusion column at high concentration (10 mg/ml), the wild-type PCNA protein and the C22Y mutant protein eluted in a narrow peak as expected for the 90-kDa trimer. When the C81R mutant protein was loaded at high concentration (10 mg/ml), it eluted in a broad peak corresponding to a mixture of mostly trimer with some dimer present. At lower protein concentration (0.01 mg/ml), the C81R mutant protein eluted as the 30-kDa monomer (Fig. S1 in the Supporting Information). Taken together, both the native PAGE and size exclusion chromatography results show that the C81R mutant PCNA protein forms trimers in vitro at higher protein concentrations, but that these trimers are less stable than in the cases of either the wild-type PCNA protein or the C22Y mutant PCNA protein. Thus distortions to loop H, such as those occurring with the C81R mutant PCNA protein, do impact the stability of the PCNA trimer. It should be noted, however, that this mutant PCNA protein supports normal cell growth and likely forms stable trimers in vivo.
Impact of the mutant PCNA proteins on DNA polymerase δ activity
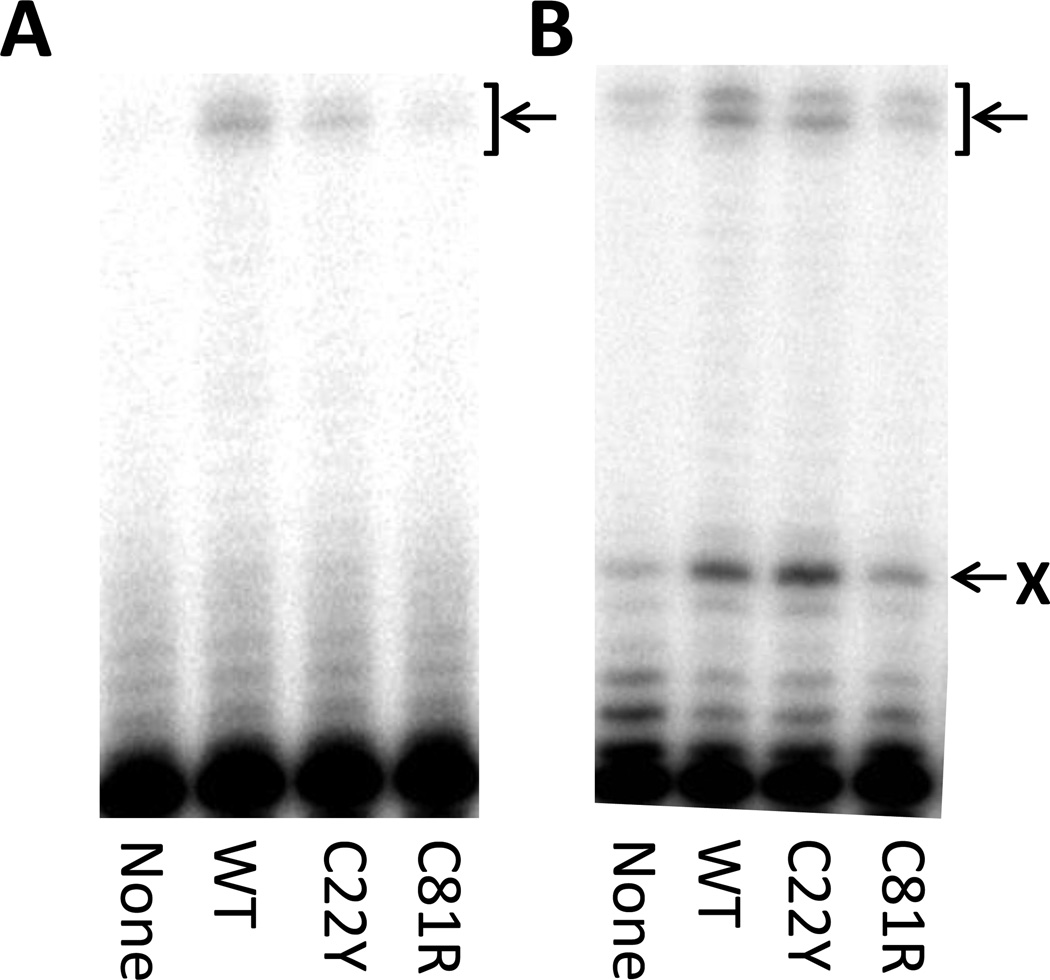
DNA polymerase δ (pol δ) is responsible for the majority of lagging strand synthesis during normal DNA replication and is also involved in base excision repair, nucleotide excision repair, mismatch repair, and double strand break repair [65–68]. To determine whether the two mutant PCNA proteins can stimulate DNA synthesis by pol δ, we used running start experiments to measure pol δ activity in the absence and presence of the wild-type and mutant PCNA proteins on non-damaged DNA (Fig. 4A). The wild-type PCNA protein stimulates DNA synthesis by pol δ with ~5-fold more full-length runoff products formed in the presence of the wild-type PCNA protein than in its absence. The C22Y mutant PCNA protein and the C81R mutant PCNA protein stimulated DNA synthesis by pol δ to differing extents. In the presence of the C22Y mutant protein, ~3-fold more full-length runoff products were observed than in its absence. In the presence of the C81R mutant protein, ~2-fold more full-length runoff products were observed.
Figure 4.
DNA synthesis by pol δ in the presence of the PCNA proteins. (A) An autoradiogram of the products of pol δ-catalyzed DNA synthesis on a non-damaged DNA substrate in the presence of no PCNA, the wild-type PCNA protein, the C22Y mutant PCNA protein, and the C81R mutant PCNA protein. The position of the fully extended, runoff product is indicated with an arrow. (B) An autoradiogram of the products of pol δ-catalyzed DNA synthesis on an abasic site-containing DNA substrate in the presence of no PCNA, the wild-type PCNA protein, the C22Y mutant PCNA protein, and the C81R mutant PCNA protein. The position of the abasic site is indicated by an X, and the position of the fully extended, runoff product is indicated with an arrow.
PCNA also facilitates the bypass of abasic sites by pol δ, and we examined if this activity was affected by the mutant PCNA proteins (Fig. 4B). In the presence of the wild-type PCNA protein, ~5-fold more extension products resulting from incorporation opposite the abasic site were observed than in the absence of PCNA. Moreover, ~4-fold more full-length runoff products were observed in the presence of the wild-type PCNA protein. Similarly, in the presence of the C22Y mutant PCNA protein, there were ~6-fold more extension products resulting from incorporation opposite the abasic site and ~3-fold more full-length runoff products than in the absence of PCNA. In the presence of the C81R mutant PCNA protein, there were ~2-fold more extension products resulting from incorporation opposite the abasic site and no increase in the amount of full-length products than in the absence of PCNA.
Taken together, these experiments show that, like the wild-type PCNA protein, the C22Y mutant PCNA protein fully supports pol δ function in both normal and translesion DNA synthesis, whereas the C81R mutant PCNA protein only partially supports pol δ function. It should be noted that under these assay conditions, the concentration of PCNA was low (0.01 mg/ml), and most of the C81R mutant PCNA protein was not expected to be trimeric. Thus, the reduced ability of the C81R mutant PCNA protein to stimulate the activity of pol δ is very likely due to this mutant protein not forming stable trimers in these assays.
Interactions of the mutant PCNA proteins with MutSα
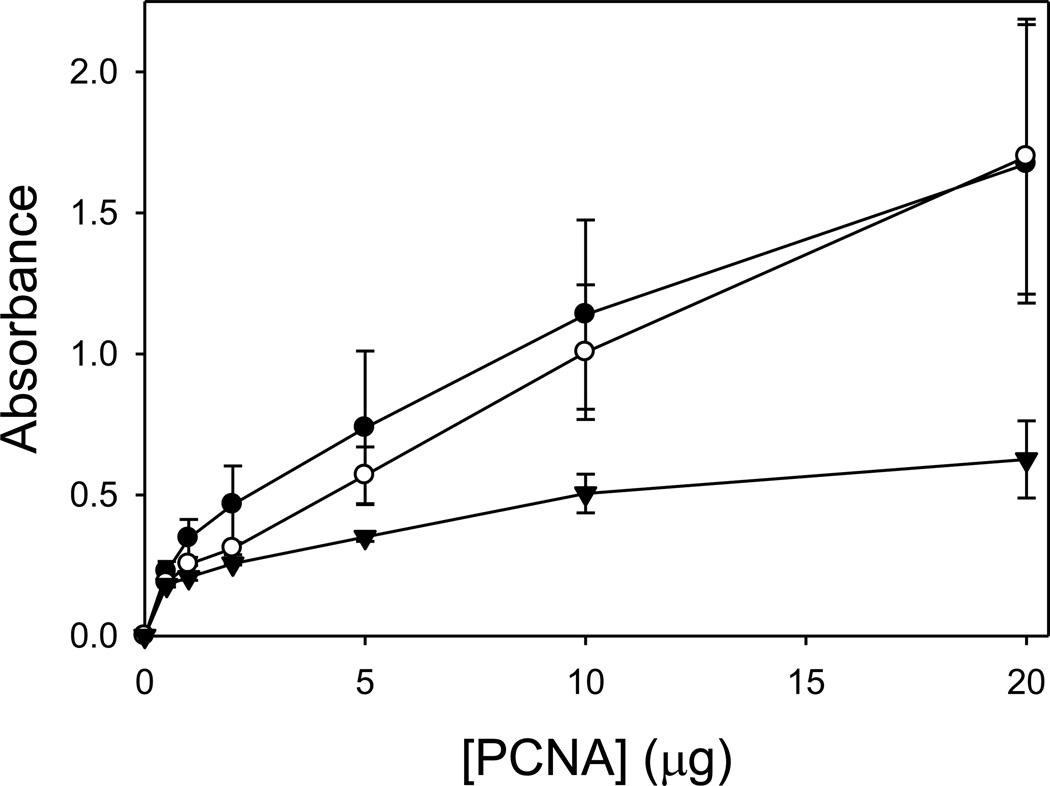
Since PCNA is known to interact with MutSα during MMR and since the C22Y and C81R mutant PCNA proteins are defective in MMR, we examined the ability of these mutant proteins to bind MutSα. We used an enzyme-linked immunosorbent assay (ELISA) to monitor this interaction (Fig. 5). A fixed concentration of the MutS α was immobilized in a microtiter plate and titrated with various concentrations of the wild-type PCNA protein, the C22Y mutant PCNA protein, or the C81R mutant PCNA proteins. The absorbance signal is proportional to the amount of PCNA bound to MutSα. No significant difference was observed between the binding of the wild-type PCNA protein and the C22Y mutant PCNA protein to MutSα. The C81R mutant PCNA protein, by contrast, exhibited weaker binding than the wild-type and the C22Y mutant PCNA proteins, though still significantly greater than the background. These results differ somewhat from previously published co-sedimentation studies that showed that the C22Y mutant PCNA protein binds MutS α in vitro, but the C81R mutant PCNA proteins does not [54]. It should be pointed out again that the concentrations of PCNA used in both the ELISA assays reported here and the co-sedimentation assay reported previously are less than 0.2 mg/ml. Under these conditions, most of the C81R mutant protein is not expected to be trimeric. Thus, the apparent weaker binding of the C81R mutant PCNA protein to MutSα is very likely due to this mutant protein not forming stable trimers in these assays.
Figure 5.
Interactions of the PCNA proteins with MutSα. The results of an ELISA assay showing the interactions of the wild-type PCNA (●), the C22Y mutant PCNA protein (○), and the C81R mutant PCNA protein (▼) with MutSα. Control experiments using BSA instead of MutSα have been subtracted from each of the values.
Interactions of the mutant PCNA proteins with MutSα and DNA
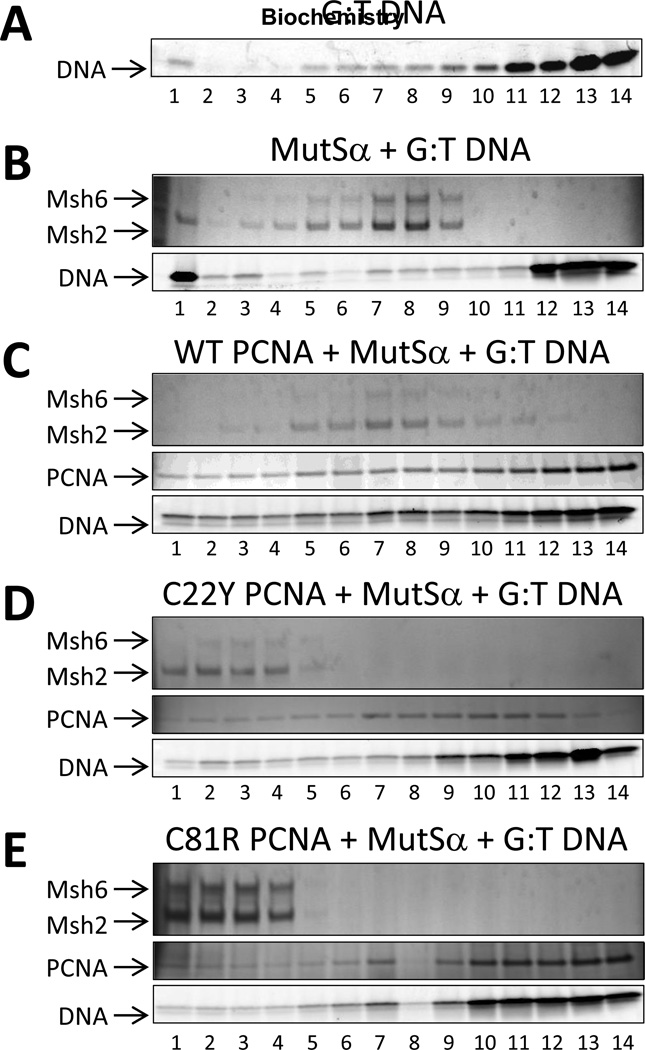
It has previously been shown that PCNA, MutSα, and mismatch-containing DNA form a stable ternary complex [37]. We carried out sedimentation analysis to determine if these amino acid substitutions in PCNA affect its ability to form ternary complexes with MutSα and DNA. The DNA contained a G:T mismatched base-pair flanked on both sides by 18 matched base-pairs. In the absence of PCNA, MutSα was found mainly in fractions 5 to 9 along with DNA (Fig. 6B), suggesting that a single MutSα protein binds to the centrally positioned G:T mismatch. In the presence of the wild-type PCNA protein, MutSα was found mainly in fractions 5 to 9 along with PCNA and DNA (Fig. 6C), suggesting that a single MutSα protein and a single PCNA protein binds to the mismatch. Interestingly, in the presence of either the C22Y mutant PCNA protein or the C81R mutant PCNA protein, MutSα was found mainly in fractions 1 to 4 along with the mutant PCNA proteins and DNA (Fig. 6D and Fig. 6E). This means that the MutSα-containing complex is larger in the presence of the mutant PCNA proteins than in the presence of the wild-type PCNA protein. This may be due to the fact that in the case of the mutant PCNA proteins, MutSα no longer specifically binds the centrally positioned G:T mismatch, and more than one MutSα protein can bind the same DNA molecule. The precise nature of these larger complexes with the mutant PCNA proteins remains unclear, and further structural analysis will be required to understand how and why they form. Nevertheless, these results convincingly demonstrate that the complexes of the mutant PCNA proteins, MutSα, and mismatch-containing DNA are aberrant. Moreover, these aberrant complexes are strictly dependent both on the presence of the mutant PCNA proteins and on the presence of a mismatch in the DNA, as they do not form in the absence of DNA (Fig. S2D, S2E, and S2F in the Supporting Information) or in the presence of fully matched G:C DNA (Fig. S2H, S2I, and S2J in the Supporting Information).
Figure 6.
Sedimentation analysis of the interactions of the PCNA proteins with MutSα and DNA. (A) Fractions of a glycerol gradient (15–30%) containing the heteroduplex DNA in the absence of PCNA and MutS α (Msh2-Msh6) were analyzed by denaturing polyacrylamide gradient gel electrophoresis (4–15%). The fractions ranged from 1 (the bottom of the gradient) to 14 (the top of the gradient). The DNA substrate was visualized by Cy3 fluorescence. (B) Fractions of a glycerol gradient containing MutS α and heteroduplex DNA in the absence of PCNA were analyzed by denaturing polyacrylamide gradient gel electrophoresis (4–15%). The proteins were visualized by silver staining. (C) Fractions of a glycerol gradient containing MutSα and heteroduplex DNA in the presence of the wild-type PCNA protein were analyzed by denaturing polyacrylamide gradient gel electrophoresis. (D) Fractions of a glycerol gradient containing MutSα and heteroduplex DNA in the presence of the C22Y mutant PCNA protein were analyzed by denaturing polyacrylamide gradient gel electrophoresis. (E) Fractions of a glycerol gradient containing MutSα and heteroduplex DNA in the presence of the C81R mutant PCNA protein were analyzed by denaturing polyacrylamide gradient gel electrophoresis.
DISCUSSION
PCNA plays a critical role in many aspects of DNA metabolism and the maintenance of genome stability. It interacts with a wide variety of proteins, recruits them, and coordinates their activities at sites of DNA synthesis. It functions in DNA replication, translesion synthesis, base excision repair, nucleotide excision repair, mismatch repair, homologous recombination, chromatin remodeling, sister chromatid cohesion, and cell cycle regulation [39–44]. Genetic studies, especially in yeast, have identified a number of mutations in PCNA that disrupt one or more of these processes. For example, simple amino acid substitutions in PCNA have been identified that interfere with translesion synthesis [69, 70], error-free postreplication repair [71], MMR [36, 52–54], and chromatin remodeling [72, 73]. In this study, we focused on two important substitutions that block MMR.
A systematic study of amino acid substitutions in PCNA that interfere with MMR has shown that many of the changes impact cell growth at non-permissive temperatures and are sensitive to DNA damaging agents such as methyl methanesulfonate, ultraviolet radiation, and hydroxyurea [54]. Most of these substitutions in PCNA cause an increase in spontaneous mutations in assays measuring the frequency of reversion mutations of a one-nucleotide insertion in the hom3-10 allele, the frequency of reversion mutations of a four-nucleotide insertion in the lys2-bgl allele, and the frequency of forward mutations in the CAN1 gene. Two of these amino acid substitutions in PCNA, the C22Y substitution (encoded by the pol30-201 allele) and the C81R substitution (encoded by the pol30-204 allele), specifically impact MMR and do not cause notable defects in other DNA replication and repair processes [54]. Neither substitution causes temperature-dependent growth defects or increased sensitivity to DNA damaging agents. Both substitutions lead to an increase in the frequency of reversion mutations in the hom3-10 and lys2-bgl alleles and in the frequency of forward mutations in the CAN1 gene. Genetic analysis of these mutant forms of PCNA in combination with the msh2Δ, msh3Δ, and msh6Δ mutations, which disrupt MutSα or MutSβ, imply that the C22Y mutant PCNA protein completely blocks MutSα-dependent MMR and the C81R mutant PCNA protein partially blocks both MutSα-dependent and MutSβ-dependent MMR [54].
In the present study, we examined the structural changes in PCNA induced by these two amino acid substitutions to understand the basis for their specific defects in MMR. Interestingly, these two substitutions caused two distinct structural alterations in PCNA. The C22Y mutant PCNA protein has shifts in the α-helices that line the central hole of the PCNA ring that encircles DNA. This mutant protein forms stable trimers and stimulates DNA synthesis by pol δ. The C81R mutant PCNA protein has a localized distortion in loop H which follows α-helix B1 near the PCNA subunit interface. This mutant protein forms less stable trimers compared to the wild-type and the C22Y mutant PCNA proteins. Consequently, this mutant protein exhibits only a moderate stimulation of DNA synthesis by pol δ, which is still higher than in the absence of PCNA.
Since both PCNA mutant proteins are known to block MutSα-dependent MMR, we examined their ability to interact with MutSα. The C22Y mutant PCNA protein interacts with MutSα with the same affinity as does the wild-type PCNA. The C81R mutant PCNA protein also interacts with MutSα, but does so with lower affinity compared to the wild-type and C22Y mutant proteins. Again, this apparent weaker binding is likely due to the instability of the C81R mutant PCNA protein trimers, and we believe that the C81R mutant PCNA protein trimers that do form under these experimental conditions still bind MutSα. This is in contrast to the previous suggestion that this mutant PCNA protein does not interact with MutSα [54]. It should be noted that the conformational changes induced by these amino acid substitutions do not perturb the hydrophobic binding pocket on PCNA which binds the canonical PCNA-interacting protein (PIP) motif in the N-terminal region of the Msh6 subunit of MutSα. Overall, these findings suggest that the disruption of MMR by these PCNA mutant proteins does not arise from substantial defects in the interaction with MutSα, especially in the case of the C22Y mutant PCNA protein.
We then examined the ability of the PCNA mutant proteins to form ternary complexes with MutS α and mismatch-containing DNA. Surprisingly, we found that despite relatively subtle changes in the structures of the C22Y and C81R mutant PCNA proteins, they both formed aberrant complexes with MutSα and DNA. In the presence of the mutant PCNA proteins and a mismatch, the MutSα-containing complexes were larger than in the presence of the wild-type PCNA protein. A possible explanation for this is that improper mismatch recognition leads to multiple MutS α proteins on the DNA substrate. Understanding why these higher-ordered complexes form awaits further structural studies. However, we conclude that these complexes are aberrant and that the formation of proper, productive complexes between MutSα and mismatch-containing DNA depends on both the structural integrity of the α-helices lining the central hole of the PCNA ring and the structural integrity of loop H near the PCNA subunit interface.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Christine Kondratick, Dr. John Pryor, Dr. Maria Spies, Dr. Marc Wold, and Kyle Powers for valuable discussions. We also thank Lokesh Gakhar and Nicole Sipfle both for technical assistance in data collection and refinement and for discussions.
ABBREVIATIONS
- BSA
bovine serum albumin
- ELISA
enzyme-linked immunosorbent assay
- HNPCC
hereditary nonpolyposis colorectal cancer
- MMR
mismatch repair
- PAGE
polyacrylamide gel electrophoresis
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA-interacting protein
- Pol
polymerase
- RFC
replication factor C
- RPA
replication protein A
- TLS
translesion synthesis
Footnotes
The project described was supported by Award Number GM081433 from the National Institute of General Medical Sciences to M.T.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
SUPPORTING INFORMATION
(1) Size exclusion chromatography experiments and (2) sedimentation analysis experiments are provided as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Iyer RR, et al. DNA mismatch repair: Functions and mechanisms. Chemical Reviews. 2006;106(2):302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annual Review of Biochemistry. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nature Reviews Molecular Cell Biology. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 4.Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Research. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 5.Kunz C, Saito Y, Schaer P. DNA Repair in Mammalian Cells. Cellular and Molecular Life Sciences. 2009;66(6):1021–1038. doi: 10.1007/s00018-009-8739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkel TA, Erie DA. DNA mismatch repair. Annual Review of Biochemistry. 2005:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 7.Grilley M, et al. ISOLATION AND CHARACTERIZATION OF THE ESCHERICHIACOLI MUTL GENE-PRODUCT. Journal of Biological Chemistry. 1989;264(2):1000–1004. [PubMed] [Google Scholar]
- 8.Galio L, Bouquet C, Brooks P. ATP hydrolysis-dependent formation of a dynamic ternary nucleoprotein complex with MutS and MutL. Nucleic Acids Research. 1999;27(11):2325–2331. doi: 10.1093/nar/27.11.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spampinato C, Modrich P. The MutL ATPase is required for mismatch repair. Journal of Biological Chemistry. 2000;275(13):9863–9869. doi: 10.1074/jbc.275.13.9863. [DOI] [PubMed] [Google Scholar]
- 10.Schofield MJ, et al. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. Journal of Biological Chemistry. 2001;276(30):28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 11.Acharya S, et al. The coordinated functions of the E-coli MutS and MutL proteins in mismatch repair. Molecular Cell. 2003;12(1):233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 12.Au KG, Welsh K, Modrich P. INITIATION OF METHYL-DIRECTED MISMATCH REPAIR. Journal of Biological Chemistry. 1992;267(17):12142–12148. [PubMed] [Google Scholar]
- 13.Marsischky GT, et al. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes & Development. 1996;10(4):407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 14.Sia EA, et al. Microsatellite instability in yeast: Dependence on repeat unit size and DNA mismatch repair genes. Molecular and Cellular Biology. 1997;17(5):2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genschel J, et al. Isolation of MutS beta from human cells and comparison of the mismatch repair specificities of MutS beta and MutS alpha. Journal of Biological Chemistry. 1998;273(31):19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 16.Li GM, Modrich P. RESTORATION OF MISMATCH REPAIR TO NUCLEAR EXTRACTS OF H6 COLORECTAL TUMOR-CELLS BY A HETERODIMER OF HUMAN MUTL HOMOLOGS. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(6):1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habraken Y, et al. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Current Biology. 1997;7(10):790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 18.Habraken Y, et al. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. Journal of Biological Chemistry. 1998;273(16):9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 19.Bowers J, et al. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. Journal of Molecular Biology. 2001;306(5):957–968. doi: 10.1006/jmbi.2001.4467. [DOI] [PubMed] [Google Scholar]
- 20.Raschle M, et al. Mutations within the hMLH1 and hPMS2 subunits of the human MutL alpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutS alpha. Journal of Biological Chemistry. 2002;277(24):21810–21820. doi: 10.1074/jbc.M108787200. [DOI] [PubMed] [Google Scholar]
- 21.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Current Opinion in Genetics & Development. 1999;9(1):89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 22.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. Journal of Medical Genetics. 1999;36(11):801–818. [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodner RD. MISMATCH REPAIR - MECHANISMS AND RELATIONSHIP TO CANCER SUSCEPTIBILITY. Trends in Biochemical Sciences. 1995;20(10):397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 24.de la Chapelle A. Genetic predisposition to colorectal cancer. Nature Reviews Cancer. 2004;4(10):769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 25.Rowley PT. Inherited susceptibility to colorectal cancer. Annual Review of Medicine. 2005:539–554. doi: 10.1146/annurev.med.56.061704.135235. [DOI] [PubMed] [Google Scholar]
- 26.Tishkoff DX, et al. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Research. 1998;58(22):5027–5031. [PubMed] [Google Scholar]
- 27.Tishkoff DX, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(14):7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand P, et al. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(24):14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmutte C, et al. Human exonuclease I interacts with the mismatch repair protein hMSH21. Cancer Research. 1998;58(20):4537–4542. [PubMed] [Google Scholar]
- 30.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5 ' and 3 ' mismatch repair. Journal of Biological Chemistry. 2002;277(15):13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 31.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. Journal of Biological Chemistry. 1997;272(16):10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 32.Lin YL, et al. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein a is required for DNA replication and mismatch repair but not for nucleotide excision repair. Journal of Biological Chemistry. 1998;273(3):1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 33.Ramilo C, et al. Partial reconstitution of human DNA mismatch repair in vitro: Characterization of the role of human replication protein A. Molecular and Cellular Biology. 2002;22(7):2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dzantiev L, et al. Defined human system that supports bidirectional mismatchprovoked excision. Molecular Cell. 2004;15(1):31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Umar A, et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87(1):65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RE, et al. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. Journal of Biological Chemistry. 1996;271(45):27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 37.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nature Genetics. 2000;26(3):375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 38.Krishna TSR, et al. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 39.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. Journal of Cell Science. 2003;116(15):3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 40.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Naryzhny SN. Proliferating cell nuclear antigen: a proteomics view. Cellular and Molecular Life Sciences. 2008;65(23):3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurimoto T. PCNA binding proteins. Frontiers in Bioscience. 1999;4:d849–d858. doi: 10.2741/tsurimoto. (CITED APRIL 7, 2000) [DOI] [PubMed] [Google Scholar]
- 43.Zhuang ZH, Ai YX. Processivity factor of DNA polymerase and its expanding role in normal and translesion DNA synthesis. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804(5):1081–1093. doi: 10.1016/j.bbapap.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dieckman LM, Freudenthal BD, Washington MT. PCNA Structure and Function: Insights from Structures of PCNA Complexes and Post-translationally Modified PCNA. Sub-cellular biochemistry. 2012;62:281–99. doi: 10.1007/978-94-007-4572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark AB, et al. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. Journal of Biological Chemistry. 2000;275(47):36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 46.Kleczkowska HE, et al. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes & Development. 2001;15(6):724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau PJ, Kolodner RD. Transfer of the MSH2 center dot MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. Journal of Biological Chemistry. 2003;278(1):14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- 48.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiale Msh6 is an unstructured tether to PCNA. Molecular Cell. 2007;26(4):565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hombauer H, et al. Visualization of Eukaryotic DNA Mismatch Repair Reveals Distinct Recognition and Repair Intermediates. Cell. 2011;147(5):1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pluciennik A, et al. PCNA function in the activation and strand direction of MutL alpha endonuclease in mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(37):16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu LY, et al. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Research. 1998;26(5):1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C, et al. Saccharomyces carevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Molecular and Cellular Biology. 1999;19(11):7801–7815. doi: 10.1128/mcb.19.11.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokoska RJ, et al. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics. 1999;151(2):511–519. doi: 10.1093/genetics/151.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Molecular and Cellular Biology. 2002;22(19):6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freudenthal BD, et al. Structure of a Mutant Form of Proliferating Cell Nuclear Antigen That Blocks Translesion DNA Synthesis. Biochemistry. 2008;47(50):13354–13361. doi: 10.1021/bi8017762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finkelstein J, et al. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Analytical Biochemistry. 2003;319(1):78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 57.Dieckman LM, et al. Pre-Steady State Kinetic Studies of the Fidelity of Nucleotide Incorporation by Yeast DNA Polymerase delta. Biochemistry. 2010;49(34):7344–7350. doi: 10.1021/bi100556m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antony E, Hingorani MM. Mismatch recognitioh-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42(25):7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallographica Section D-Biological Crystallography. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 60.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallographica Section D-Biological Crystallography. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 61.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Freudenthal BD, et al. Crystal structure of SUMO-modified proliferating cell nuclear antigen. J Mol Biol. 2011;406(1):9–17. doi: 10.1016/j.jmb.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burgers PMJ. Polymerase Dynamics at the Eukaryotic DNA Replication Fork. Journal of Biological Chemistry. 2009;284(7):4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends in Cell Biology. 2008;18(11):521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McElhinny SAN, et al. Division of labor at the eukaryotic replication fork. Molecular Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shcherbakova PV, Bebenek K, Kunkel TA. Functions of eukaryotic DNA polymerases. Sci Aging Knowledge Environ. 2003;2003(8):RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 69.Zhang HS, Gibbs PEM, Lawrence CW. The Saccharomyces cerevisiae rev6-1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen. Genetics. 2006;173(4):1983–1989. doi: 10.1534/genetics.106.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Northam MR, et al. A novel function of DNA polymerase zeta regulated by PCNA. Embo Journal. 2006;25(18):4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.TorresRamos CA, et al. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9676–9681. doi: 10.1073/pnas.93.18.9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang ZG, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408(6809):221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 73.Sharp JA, et al. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Current Biology. 2001;11(7):463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.