Abstract
Autophagy, an evolutionarily conserved lysosomal degradation process, has drawn an increasing amount of attention in recent years for its role in a variety of human diseases, such as cancer. Notably, autophagy plays an important role in regulating several survival and death signaling pathways that determine cell fate in cancer. To date, substantial evidence has demonstrated that some key autophagic mediators, such as autophagy-related genes (ATGs), PI3K, mTOR, p53, and Beclin-1, may play crucial roles in modulating autophagic activity in cancer initiation and progression. Because autophagy-modulating agents such as rapamycin and chloroquine have already been used clinically to treat cancer, it is conceivable that targeting autophagic pathways may provide a new opportunity for discovery and development of more novel cancer therapeutics. With a deeper understanding of the regulatory mechanisms governing autophagy, we will have a better opportunity to facilitate the exploitation of autophagy as a target for therapeutic intervention in cancer. This review discusses the current status of targeting autophagic pathways as a potential cancer therapy.
Keywords: Autophagy, cancer, cell death, survival, drug discovery
Autophagy, a term derived from the Greek words “auto” (self) and “phagy” (to eat), refers to an evolutionarily conserved, multi-step, lysosomal degradation process in which a cell degrades long-lived proteins and damaged organelles[1],[2]. Three forms of autophagy have been identified based upon the mode of delivery to the lysosome, namely macroautophagy, microautophagy, and chaperone-mediated autophagy[3],[4]. Macroautophagy (hereafter referred to as autophagy) is a major regulated catabolic process that involves the delivery of cytoplasmic cargo sequestered inside double-membrane vesicles to the lysosome. Autophagy is strictly regulated by a number of autophagy-related genes (ATGs) that were originally discovered in yeast (Figure 1)[5]. To date, over 35 distinct ATGs have been identified in yeast, and even more ATGs are probably expressed in mammals[6],[7]. Under most conditions, autophagy promotes cell survival by allowing cells to adapt to stressful conditions; thus, cells are provided with the energy required for minimal cellular functions even when nutrients are scarce[8]. Alternatively, many studies have demonstrated that autophagy plays a pro-death role in type II programmed cell death (type II PCD) but not in apoptosis (type I PCD)[9]. Therefore, depending on the cell type and context, autophagy appears to play opposite roles in determining cell fate. Autophagic cell fate is regulated by several key mediators [e.g., Beclin-1, phosphatidylinositol 3 kinase (PI3K), mammalian target of rapamycin (mTOR), Bcl-2, and p53], which have an astonishing number of links to many human diseases, most notably cancer[10],[11]. In cancer cells, autophagy can either serve as a temporary survival mechanism, which may provide a means of recycling macromolecules, or lead to cell death if autophagy is excessively induced by cellular stresses[12],[13]. Therefore, autophagy may play a two-faced role, acting either as a guardian or as an executioner in cancer, depending on the stage of cancer initiation and progression or on the surrounding cellular environment[14].
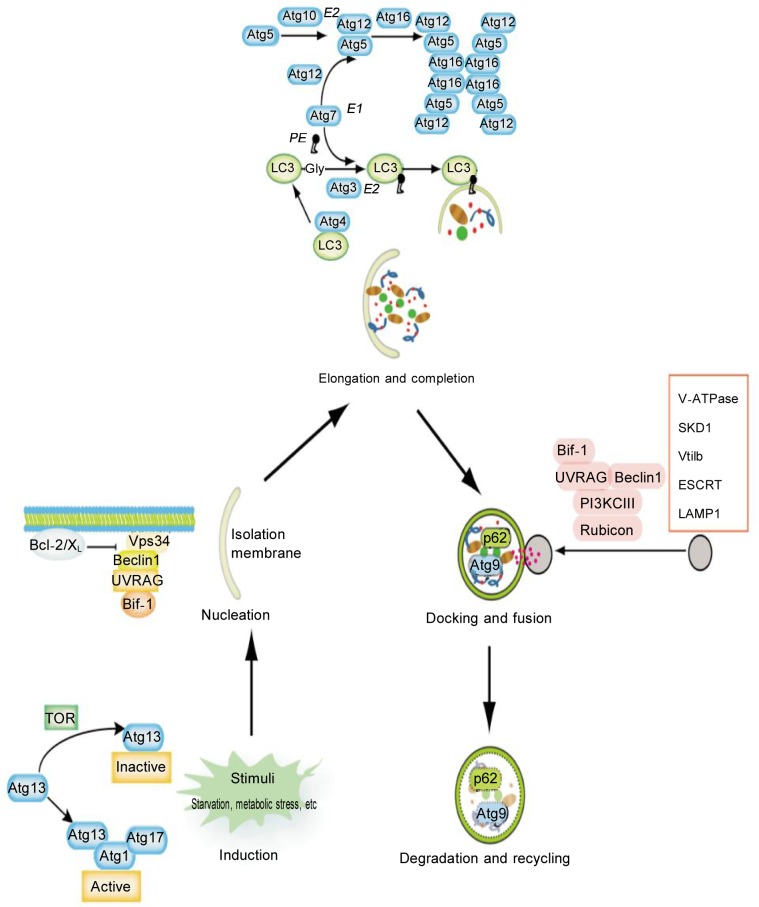
Figure 1. Multiple stages of autophagy and the involved molecular regulators.
Autophagy is stimulated by nutrient deprivation, hypoxia, cytokines, hormones, and DNA damage. The early stages of activation require ATG1 and ATG13, which in turn can be inhibited by mTOR. Vesicle nucleation depends on Beclin-1-class III PI3K-Vps15 core complexes and other proteins. Vesicle elongation and completion are mediated by the Atg16L complex and LC3. Docking and fusion refer to the maturation of autolysosomes and are promoted by Rab7, LAMP1, LAMP2, SKD1, Vtil 1b, and the ESCRT complex. In the last stage, autophagosomal cargoes are digested and then nutrients and energy are recycled.
Molecular Pathways of Autophagy Regulation in Cancer
Autophagy is a complicated process that involves input from many upstream regulatory signaling pathways[15]. Although the regulatory mechanisms of autophagy are partially known, the exact function of autophagy in cancer is still controversial. When analyzing the intricate relationship between autophagy and cancer, a common challenge is to determine whether autophagy protects cell survival or contributes to cell death[16]. To resolve the role of autophagy in cancer cell fate, several hypotheses have been put forward. One hypothesis proposes that the role of autophagy varies depending on the stage of tumor development. For instance, autophagy limits tumor formation in early stages but favors tumor cell survival, invasion, and metastasis after tumors have formed[15],[16]. Another hypothesis suggests that autophagy can affect tumorigenesis in a cell- or tissue-specific manner[15],[17]. Therefore, at the molecular level, autophagy plays either a pro-survival or a pro-death role by regulating tumor suppressor genes or oncogenes. These autophagic pro-survival or pro-death genes, and the corresponding proteins, can integrate into cancer cell signaling networks and ultimately regulate cell survival or death[18].
ATGs play a key role in the formation of autophagosomes and regulation of autophagic activity; furthermore, they are closely linked to cancer initiation and progression. Silencing some essential ATGs, such as ATG3, ATG4, Beclin-1/ATG6, ATG10, and ATG12, can sensitize cancer cells to a wide spectrum of stressful conditions[19]. Additionally, targeting selected protein kinases involved in autophagy regulation with small molecule kinase inhibitors may be another feasible approach in cancer treatment. A number of protein kinases regulate the induction of autophagy following nutrient deprivation or other cellular stresses. The following protein kinases have been reported to activate protective autophagy in cancer cells as a response to cytotoxic agents, including AMP-activated protein kinase (AMPK), glycogen synthase kinase 3 (GSK3) beta, extracellular signal-regulated kinases 1 and 2 (ERK1/2), and eukaryotic elongation factor-2 kinase (eEF-2K)[20]–[22].
mTOR, an evolutionarily conserved serine/threonine kinase, serves as the main negative regulator of autophagy in cancer cells. mTOR forms two complexes in mammalian cells. Only mammalian target of rapamycin complex 1 (mTORC1) is sensitive to inhibition by rapamycin; therefore, we focus on the role of mTORC1 in autophagy[23]. Three major mTORC1-inducing pathways have been elucidated, including the PI3K-Akt pathway and the MAPK/ERK pathway, consisting of Ras-proto-oncogene serine/threonine-protein kinase (Raf-1), mitogen-activated protein kinase 1/2 (MEK1/2), and extracellular signal-regulated kinase 1/2 (ERK1/2). The LKB1-AMPK pathway, consisting of liver kinase B1 and AMPK, can inhibit mTORC1[24]. The TSC2/TSC1 complex, which has a tumor suppressor function in various cancers, is a key point upstream of mTORC1 since TSC2/TSC1 can suppress mTORC1 by inactivating the mTORC1-interacting protein, Rheb[25],[26]. Upon PI3K activation, Akt phosphorylation of TSC2 destabilizes TSC2 and disrupts its interaction with TSC1 to abolish the negative regulatory effect of the TSC2/TSC1 complex on mTORC1[27]. Phosphorylation of TSC2 by AMPK can increase the GAP activity of TSC2, stabilize the TSC2/TSC1 complex, and inactivate Rheb, resulting in the inactivation of mTORC1 and the initiation of autophagy[25]. In mammals, two homologs of ATG1, namely uncoordinated 51-like kinase 1 (ULK1) and ULK2, mammalian autophagy-related protein 13 (mATG13), and scaffold protein FIP200 have been identified. Under nutrient starvation conditions, mTORC1 disrupts the binding of ATG13 with ULK and destabilizes ULK, thereby inhibiting the ULK-dependent phosphorylation of FIP200 and autophagy induction by phosphorylation of ULK and ATG13[28]. Moreover, mTORC1 regulates autophagy by mediating protein translation and cell growth through phosphorylation of 4E-binding protein 1 (4E-BP1) and p70S6K. Phosphorylation of 4E-BP1 leads to its dissociation from eukaryotic translation initiation factor 4E (eIF4E) and up-regulates cap-dependent translation. Phosphorylation of p70S6K by mTORC1 enhances p70S6K activity and allows it to phosphorylate downstream targets[2]. Once activated, p70S6K phosphorylates eukaryotic elongation factor 2 kinase (eEF2K) to relieve elongation factor 2 (eEF2) from inhibition by eEF2K in addition to promoting autophagy[29]. DEPTOR, an inhibitor of mTORC1 and mT0RC2, inhibits mTORC1 and mT0RC2 by directly binding to them both. DEPTOR is subjected to proteasome-dependent degradation upon serum stimulation to ensure mTOR activation.
p53, a well characterized human tumor suppressor gene involved in genotoxic stress response and DNA damage repair, also participates in autophagy regulation[30]. Intriguingly, the role of p53 in autophagy seems to be paradoxical depending on its Subcellular localization, which may dictate whether p53 contributes to cancer cell survival or death[31]. In the nucleus, p53 can activate AMPK to inhibit mTOR and induce the autophagic process. Also, p53 can promote autophagy through targeting multiple genes that code for pro-autophagic modulators, including DAPK-1, damage-regulated autophagy modulator (DRAM), pro-apoptotic Bcl-2 proteins (e.g., Bad, Bax, BNIP3, and PUMA), Sestrin1/2, and TSC2[15]. Notably, Sestrin1 and Sestrin2, which are usually expressed under conditions of DNA damage and oxidative stresses, are negative regulators of mTORC1 and execute their function through activation of AMPK and TSC2; thus, Sestrin1/2 establish a connection between p53 and autophagy through mTORC1[32]. Opposite to nuclear p53, cytoplasmic p53 inhibits autophagy by mTORC1 activation. Tasdemir et al.[33] have shown that depletion of p53 in mice induces autophagy. The autophagy induced by loss of p53 promotes the survival of p53-deficient cells to sustain high ATP levels under conditions of hypoxia and nutrient depletion[34]. Therefore, p53 signaling controls autophagy in an ambiguous fashion that depends on its Subcellular localization and plays a two-sided role in cancer.
Beclin-1, the mammalian homolog of ATG6 and a Bcl-2 interacting coiled-coil protein, is essential for the formation of double-membrane autophagosomes, which are required in the initial step of autophagy[35]. Beclin-1 can promote interaction of Bcl-2 with other autophagy regulators, such as Vps34 (PI3K), p150, UVRAG, Bif1, ATG14L, and Rubicon, to form huge protein complexes[36]. Additionally, Beclin-1 is a haploinsufficient tumor suppressor gene. Beclin-1 can enhance autophagy by combining with PI3KIII in the initiating stage of autophagy. UVRAG, a major positive mediator of Beclin-1, can directly and markedly enhance PI3KIII lipid kinase activity, thus, facilitating autophagy. Through mediating the Beclin-1/PI3KIII complex, UVRAG can promote autophagy and inhibit tumorigenesis. Bif-1, another positive mediator of Beclin-1, can interact with Beclin-1 through UVRAG to regulate autophagy and suppress tumorigenesis[37]. Following dissociation of Beclin-1 from Bcl-2, autophagy may be activated depending on whether Bcl-2 has been phosphorylated by the starvation-activated c-JUN N-terminal kinase (JNK)[38]. The tumor suppressor function of Beclin-1 is supported by the identification of its mediators in tumorigenesis[39]. Bcl-2 inhibits autophagy through interacting with Beclin-1 as Beclin-1 contains a BH3 domain that facilitates the interaction of Beclin-1 with Bcl-2[40]. By interacting with Beclin-1, Bcl-2 blocks the interaction of Beclin-1 with PI3KIII, decreases PI3KIII activity, and down-regulates autophagy. Overall, Beclin-1 may enhance autophagy and inhibit tumorigenesis by forming signaling complexes mediated by positive and negative regulators, which suggests a crucial role for Beclin-1 in cancer.
Mounting evidence has demonstrated that mitochondria, the main source of reactive oxygen species (ROS) in cells, may orchestrate the autophagic process in cancer initiation and progression. For instance, ROS play multifaceted roles as a “molecular switch” in the regulation of several core autophagic pathways (e.g., ATG4-ATG8/LC3, Beclin-1, PTEN, p53, PI3K-Akt-mTOR, and MAPK signaling) that may jointly seal the fate of cancer cells. MAPKs and p21-activated kinases (PAK), two classes of downstream signaling molecules regulated by ROS, are thought to be the major signaling pathways for driving cancer cell metastasis[41],[42].
In summary, all of the aforementioned survival/death signaling pathways involved in ATGs, the mTOR subnetwork, the Beclin-1 interactome, p53 signaling, and ROS may play crucial roles in autophagy-related cancer signaling networks. This suggests that autophagic pathways could be promising new targets in cancer drug development, which we will discuss in the following section.
Autophagy-modulating Agents for Cancer Treatment
Evidence suggests that induction of autophagy may help transform tumor phenotypes during cancer therapy. At this time, several novel strategies are being used to target autophagic signaling pathways for drug discovery in cancer treatment because a number of autophagy-inducing drugs have been identified as potential cancer therapeutic agents[43]–[45]. Several autophagy-inducing agents are already being used to treat different human cancers and should be further explored both at the bench and in the clinic.
Tumor cells can use autophagy to supply nutrients and energy, and promote tumor survival when nutrients are limited. Therefore, some drugs have been developed to block autophagic processes so as to suppress tumor progression. Autophagy process can be divided into several phases, including the initiation period, docking and fusion of autophagosomes with lysosomes, and catalytic degradation of cytoplasmic materials inside autolysosomes. PI3K inhibitors, including 3-methylade-nine (3-MA), wortmannin, and LY294002, can interfere with or block autolysosome formation and result in the inhibition of autophagy[46]. Also, it has been observed that cancer cells undergo increased autophagy and are more sensitive to lysosomotrophic agents. Bafilomycin A1, vinblastine, and Nuokaodazuo can inhibit the fusion process to interrupt autophagy[47]. Bafilomycin A1, a type of macrolide antibiotic derived from Streptomyces griseus, has been reported to block the fusion of autophagosomes with lysosomes in tumor cells. Two anti-malarial drugs, hydroxychloroquine (HCQ) and chloroquine (CQ), can inhibit lysosomal acidification and prevent the degradation of autophagosomes, thereby suppressing autophagy in Myc-driven lymphoma and increasing the antitumor effects of cyclophosphamide[48]–[50]. In imatinib-resistant BCR-ABL-positive chronic myeloid leukemia (CML) cell lines, CQ enhanced cell death by inhibiting autophagy and strengthened the activity of vorinostat, an histone deacetylase (HDAC) inhibitor[51],[52]. In combination with the anti-malarial drug quinacrine, CQ remarkably sensitized gastrointestinal stromal tumor cells to imatinib, both in vitro and in vivo, and reinforced the efficiency of quinacrine[53]. CQ has recently been reported to be able to inhibit therapy-induced autophagy and to increase cell death in established tumors, leading to tumor regression[54].
Nevertheless, autophagy is not only a survival response that opposes growth factor and nutrient deprivation but also an important mechanism for tumor cell suicide. Recently, an increasing amount of data have suggested that autophagy, as a mechanism of type II PCD, may present new opportunities for developing alternative anti-cancer therapies. Tamoxifen, an antagonist of estrogen receptor (ER), has a high binding affinity for the microsomal antiestrogen binding site (AEBS), a hetero-oligomeric complex involved in cholesterol metabolism. Tamoxifen and other AEBS ligands induce breast cancer cell autophagy through inducing Sterol accumulation[55],[56]. These data indicate a therapeutic implication for selective AEBS ligands in breast cancer management and reveal a mechanism that may explain the induction of autophagy in MCF-7 cells by tamoxifen and other selective ER modulators[57],[58]. Imatinib (Gleevec), an inhibitor of tyrosine kinases, can induce autophagy in multidrug-resistant Kaposi's sarcoma cells[59],[60]. HDAC inhibitors, such as suberoyla-nilide hydroxamic acid (SAHA), have been reported to be able to induce autophagy and cell death in HeLa cells independent of caspase-dependent apoptosis[61]–[63]; thus, initiation of autophagic cell death by SAHA has clear therapeutic implications for apoptosis-defective tumors. It is well known that mTOR is a major regulator of cell growth that has also been implicated in tumorigenesis[64],[65]. The tumor suppressing action of rapamycin, an inhibitor of mTOR, is linked to induction of autophagic cell death. In addition to these agents, there are also other interesting examples of autophagy-inducing agents from traditional Chinese medicine. One such traditional Chinese compound, arsenic trioxide (As2O3), has been reported to be able to induce apoptosis through cytochrome c release and Caspase activatiori[66],[67]. Interestingly, recent studies showed that treatment of human T-lymphocytic leukemia cells with As2O3 led to cytotoxicity through inducing autophagy. A Bcl-2 family member, Bcl-2-adenovirus E1B 19-kDa-interacting protein 3 (BNIP3), was reported to play a pivotal role in As2O3-induced autophagic cell death in malignant glioma cells[68],[69]. Additionally, Polygonatum cyrtonema lectin (PCL) was shown to be able to induce autophagic cell death via a mitochondria-mediated ROS-p38-p53 pathway in human melanoma A375 cells[70],[71]. Based on the aforementioned examples, autophagy may play an important role in the cytotoxic effects of these compounds that could spark new autophagy-targeted cancer therapeutic strategies[72],[73].
Additionally, DNA damage agents have been found to be able to induce autophagy in tumor cells. For example, temozolomide (TMZ), an alkylating agent, is widely used to treat primary and recurrent high-grade gliomas. The cytotoxicity of TMZ is thought to result from the formation of O-6-methylguanine in DNA, which mispairs with thymine during DNA replication and triggers futile cycles of the mismatch repair system and subsequent DNA damage. It was shown that TMZ induces autophagy and that pharmacologic inhibition of autophagy could influence cellular outcome.
Much work is needed to determine how modulators of autophagy impact cancer initiation, progression, and therapeutic response, and to determine exactly why targeting autophagic signaling pathways may be a valuable strategy for cancer drug development.
Concluding Remarks and Future Directions
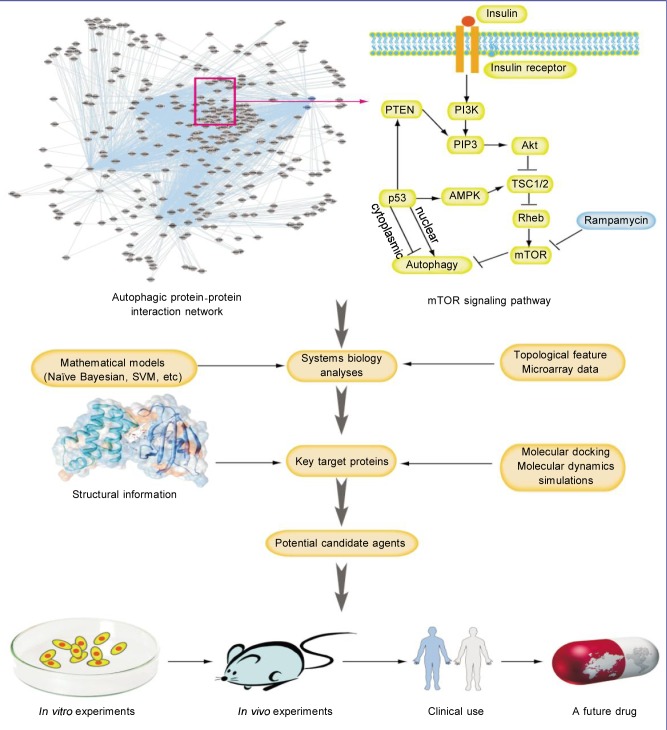
Autophagy plays a dual role in the regulation of pro-survival and pro-death signaling pathways in a variety of diseases, including cancer. Several key autophagic mediators, including ATGs, PI3K, mTOR, p53, Beclin-1 interacome, and ROS, have been demonstrated to play pivotal roles in the complex autophagic network in cancer cells. However, much work is needed to determine the intricate molecular mechanisms of autophagy in cancer, to define how crucial modulators of autophagy in cancer impacts cancer initiation and progression, and to elucidate why targeting autophagic signaling pathways is promising for cancer therapeutics. Furthermore, recent biological insights can provide a fertile foundation for launching this next round of small-molecule drug discovery. These discoveries are being driven by an abundance of structural information on the potential targets; therefore, X-ray crystallography, nuclear magnetic resonance (NMR), and structural bioinformatics-docking techniques will be invaluable in the efforts to target autophagic pathways for drug discovery. More importantly, there is an increasing emergence of sophisticated mathematical models, such as the Naive Bayesian framework and support vector machine (SVM), for the disruption of protein-protein interactions (PPIs). The best hope for targeting autophagy as a therapeutic intervention may lie in the discovery of agents that are able to target the altered autophagy-regulating signaling pathways, or even the autophagic network, rather than targeting the individual genes or proteins. A better understanding of the autophagic PPI network will provide useful insights into how these hub proteins and autophagy-related signaling pathways can be exploited as potential therapeutic targets for treatment of human diseases (Figure 2). Due to the complex, two-sided nature of autophagy, establishing the dual role of autophagy in tumor survival vs. death may assist in determining therapeutic potential. Inhibiting autophagy may enhance the efficacy of currently used anti-cancer drugs and radiotherapy. In addition, promoting autophagy may induce cancer cell death with a high threshold to apoptosis. Therefore, both strategies have significant potential to be translated into ongoing clinical trials that may provide more valuable information regarding whether targeting autophagic pathways in tumor cells would be a promising avenue for cancer therapeutics.
Figure 2. Autophagy network-based identification of novel targets for drug discovery.
Some sophisticated mathematical models have been used to disrupt protein-protein interactions. In addition, with increasing accuracy, small molecules that inhibit or promote protein-protein interactions (PPIs) can be screened as potential candidate drugs. Thus, the autophagic PPI network can provide more novel insights into how these hub proteins and their autophagic pathways can play key roles as potential drug targets in cancer treatment.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu B, Cheng Y, Liu Q, et al. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol Sin. 2010;31:1154–1164. doi: 10.1038/aps.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine B, Kraemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ. Cell biology: regulated self-cannibalism. Nature. 2004;431:31–32. doi: 10.1038/431031a. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 9.Hannigan AM, Gorski SM. Macroautophagy: the key ingredient to a healthy diet? Autophagy. 2009;5:140–151. doi: 10.4161/auto.5.2.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubinsztein DC, Gestwicki JE, Murphy LO, et al. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 12.Kevin M, Luo SQ, David CR. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang SY, Yu QJ, Zhang RD, et al. Core signaling pathways of survival/death in autophagy-related cancer networks. Int J Biochem Cell Biol. 2011;43:1263–1266. doi: 10.1016/j.biocel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 15.Morselli E, Galluzzi L, Kepp O, et al. Anti- and pro-tumor functions of autophagy. Biochim Biophys Acta. 2009;1793:1524–1532. doi: 10.1016/j.bbamcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 17.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalby KN, Tekedereli I, Lopez-Berestein G, et al. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6:322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elisabeth AC, Pietri P, Marja J. Apoptosis and autophagy: targeting autophagy signaling in cancer cells—‘trick or treats’? FEBS J. 2009;276:6084–6096. doi: 10.1111/j.1742-4658.2009.07332.x. [DOI] [PubMed] [Google Scholar]
- 20.Bhutia SK, Kegelman TP, Das SK, et al. Astrocyte elevated gene-1 activates AMPK in response to cellular metabolic stress and promotes protective autophagy. Autophagy. 2011;7:547–548. doi: 10.4161/auto.7.5.15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Takahashi Y, Cheng E, et al. GSK-3beta promotes cell survival by modulating Bif-1-dependent autophagy and cell death. J Cell Sci. 2010;123:861–870. doi: 10.1242/jcs.060475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu WK, Cho CH, Lee CW, et al. Macroautophagy and ERK phosphorylation counteract the antiproliferative effect of proteasome inhibitor in gastric cancer cells. Autophagy. 2010;6:228–238. doi: 10.4161/auto.6.2.11042. [DOI] [PubMed] [Google Scholar]
- 23.Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 24.Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning BD, Tee AR, Logsdon MN, et al. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 26.Inoki K, Li Y, Zhu T, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, Chen Z, Erdjument-Bromage H, et al. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 29.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 30.Krejci O, Wunderlich M, Geiger H, et al. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111:2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ertmer A, Huber V, Gilch S, et al. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21:936–942. doi: 10.1038/sj.leu.2404606. [DOI] [PubMed] [Google Scholar]
- 32.Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53- induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 33.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young AR, Narita M, Ferreira M, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 36.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–137. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furuya N, Yu J, Byfield M, et al. The evolutionarily conserved domain of Beclin-1 is required for Vps34 binding, autophagy and tumor-suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 38.Maiuri MC, Criollo A, Tasdemir E, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 39.Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazure NM, Pouysségur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Y, Ren X, Zhang Y, et al. eEF-2 kinase dictates crosstalk between autophagy and apoptosis induced by Akt inhibition, thereby modulating cytotoxicity of novel Akt inhibitor MK-2206. Cancer Res. 2011;71:2654–2663. doi: 10.1158/0008-5472.CAN-10-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turcotte S, Giaccia AJ. Targeting cancer cells through autophagy for anticancer therapy. Curr Opin Cell Biol. 2010;22:246–251. doi: 10.1016/j.ceb.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Rehman SK, Zhang W, et al. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010;1806:220–229. doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi H, Kondo Y, Fujiwara K, et al. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 47.Fujiwara K, Iwado E, Mills GB, et al. Akt inhibitor shows anticancer and radiosensitizing effects in malignant glioma cells by inducing autophagy. Int J Oncol. 2007;31:753–760. [PubMed] [Google Scholar]
- 48.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maclean KH, Dorsey FC, Cleveland JL, et al. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981;90:665–669. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glaumann H, Ahlberg J. Comparison of different autophagic vacuoles with regard to ultrastructure, enzymatic composition, and degradation capacity—formation of crinosomes. Exp Mol Pathol. 1987;47:346–362. doi: 10.1016/0014-4800(87)90018-9. [DOI] [PubMed] [Google Scholar]
- 52.Bellodi C, Lidonnici MR, Hamilton A, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A, Roy S, Lazar AJ, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci USA. 2010;107:14333–14338. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Y, Qiu F, Ye YC, et al. Autophagy inhibits reactive oxygen species-mediated apoptosis via activating p38-nuclear factor-kappa B survival pathways in oridonin-treated murine fibrosarcoma L929 cells. FEBS J. 2009;276:1291–1306. doi: 10.1111/j.1742-4658.2008.06864.x. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Y, Li H, Ren X, et al. Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One. 2010;5:e9715. doi: 10.1371/journal.pone.0009715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Medina P, Payré B, Boubekeur N, et al. Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism. Cell Death Differ. 2009;16:1372–1384. doi: 10.1038/cdd.2009.62. [DOI] [PubMed] [Google Scholar]
- 58.Younes A. Therapeutic activity of mTOR-inhibitors in mantle cell lymphoma: clues but no clear answers. Autophagy. 2008;4:707–709. doi: 10.4161/auto.6232. [DOI] [PubMed] [Google Scholar]
- 59.Kim KW, Hwang M, Moretti L, et al. Autophagy upregulation by inhibitors of caspase-3 and mTOR enhances radiotherapy in a mouse model of lung cancer. Autophagy. 2008;4:659–668. doi: 10.4161/auto.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanzawa T, Zhang L, Xiao L, et al. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 61.Fu LL, Zhou CC, Yao S, et al. Plant lectins: targeting programmed cell death pathways as anti-tumor agents. Int J Biochem Cell Biol. 2011;43:1442–1449. doi: 10.1016/j.biocel.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Shao Y, Gao Z, Marks PA, et al. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu JJ, Lin M, Yu JY, et al. Targeting apoptotic and autophagic pathways for cancer therapeutics. Cancer Lett. 2011;300:105–114. doi: 10.1016/j.canlet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Wu JM, Li J, et al. Polygonatum cyrtonema lectin induces murine fibrosarcoma L929 cell apoptosis and autophagy via blocking Ras-Raf and PI3K-Akt signaling pathways. Biochimie. 2010;92:1934–1938. doi: 10.1016/j.biochi.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 66.Wang SY, Yu QJ, Bao JK, et al. Polygonatum cyrtonema lectin: a potential antineoplastic drug targeting programmed cell death pathways. Biochem Biophys Res Commun. 2011;406:497–500. doi: 10.1016/j.bbrc.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 67.Miller WH, Jr, Schipper HM, Lee JS, et al. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62:3893–3903. [PubMed] [Google Scholar]
- 68.Liu B, Cheng Y, Bian HJ, et al. Molecular mechanisms of polygonatum cyrtonema lectin induced apoptosis and autophagy in cancer cells. Autophagy. 2009;5:253–255. doi: 10.4161/auto.5.2.7561. [DOI] [PubMed] [Google Scholar]
- 69.Kanzawa T, Zhang L, Xiao L, et al. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 70.Liu B, Cheng Y, Zhang B, et al. Polygonatum cyrtonema lectin induces apoptosis and autophagy in human melanoma A375 cells through a mitochondria-mediated ROS-p38-p53 pathway. Cancer Lett. 2009;275:54–60. doi: 10.1016/j.canlet.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 71.Liu B, Min MW, Bao JK. Induction of apoptosis by Concanavalin A and its molecular mechanisms in cancer cells. Autophagy. 2009;5:432–433. doi: 10.4161/auto.5.3.7924. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X, Chen LX, Ouyang L, et al. Plant natural compounds: targeting pathways of autophagy as anti-cancer therapeutic agents. Cell Proliferation. 2012 doi: 10.1111/j.1365-2184.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu LL, Wen X, Bao JK, et al. MicroRNA-modulated autophagic signaling networks in cancer. Int J Biochem Cell Biol. 2012;44:733–736. doi: 10.1016/j.biocel.2012.02.004. [DOI] [PubMed] [Google Scholar]