Abstract
Expression profiling is one of the most important tools for dissecting biological functions of genes and the upregulation or downregulation of gene expression is sufficient for recreating phenotypic differences. Expression divergence of genes significantly contributes to phenotypic variations. However, little is known on the molecular basis of expression divergence and evolution among rice genotypes with contrasting phenotypes. In this study, we have implemented an integrative approach using bioinformatics and experimental analyses to provide insights into genomic variation, expression divergence, and evolution between salinity-sensitive rice variety Nipponbare and tolerant rice line Pokkali under normal and high salinity stress conditions. We have detected thousands of differentially expressed genes between these two genotypes and thousands of up- or downregulated genes under high salinity stress. Many genes were first detected with expression evidence using custom microarray analysis. Some gene families were preferentially regulated by high salinity stress and might play key roles in stress-responsive biological processes. Genomic variations in promoter regions resulted from single nucleotide polymorphisms, indels (1–10 bp of insertion/deletion), and structural variations significantly contributed to the expression divergence and regulation. Our data also showed that tandem and segmental duplication, CACTA and hAT elements played roles in the evolution of gene expression divergence and regulation between these two contrasting genotypes under normal or high salinity stress conditions.
Keywords: custom microarray, gene set enrichment analysis, promoter motif, single nucleotide polymorphism, tandem and segmental duplication, transposition
Introduction
Salinity is one of the major environmental factors that limit crop production. Rice is moderately susceptible to salinity and most rice cultivars are severely injured when grown under field conditions with an electrical conductivity 6–10 dS m−1 and yield losses were estimated to be 30–50% (Zeng et al. 2002; Islam et al. 2007). The yield losses would be due to high concentrations of saline in the soil (5 dS m−1≈ 50 mM NaCl), which led to ion cytotoxicity, metabolic imbalances, and stress damage (Zhu 2001; Chinnusamy et al. 2005; Joseph and Mohanan 2013). With increasing population and reduced area of rice field, it is essential to breed and/or genetically engineer salinity-tolerant rice varieties suitable for planting in saline soil such as coastal areas. Indica rice varieties generally show higher level of tolerance when compared with japonica rice varieties (Lee et al. 2003). Furthermore, within either japonica or indica germplasms, they also exhibited significant differences in their response to high salinity stress (Gregorio et al. 2002; Lee et al. 2003; Ismail et al. 2007). These results suggested that valuable salinity stress-related genes could be found from rice germplasms, and these genes can be employed to improve rice varieties by molecular breeding. Therefore, the key is how to efficiently identify these genes and use them for rice genetic improvement.
Genome-wide expression analysis is an efficient tool to identify differentially expressed genes under high salinity stress conditions. Early rice microarray analysis was carried out using the custom microarrays including only 1,728 cDNAs from libraries of salt-stressed roots (Kawasaki et al. 2001). By using the chips, around 10% of probes were detected with differential expression regulation under salinity stress. After that, Affymetrix microarrays were used to investigate transcriptional profiling under salt stress. Walia et al. (2005) compared expression patterns of two contrasting indica rice lines FL478 (tolerance to high salinity) and IR29 (sensitive to high salinity) during vegetative growth stage. Their data showed that salinity stress induced a number of genes involved in flavonoid biosynthesis pathway only in IR29 but not in FL478 and cell wall-related genes were responsive in both genotypes. Cotsaftis et al (2011) analyzed the root-specific transcript profiling by comparing four indica rice lines under salinity stress. They have identified some genes and their families that are likely involved in the response to this stress. All of the aforementioned data were from indica rice. Genome-wide comparative expression was also carried out using both japonica and indica rice lines under salinity stress (Walia et al. 2007). However, they only analyzed the expression during panicle initiation stage. Salinity susceptibility to abiotic stress varies during rice life cycle. During germination stage, rice is relatively tolerant to high salinity stress and becomes very sensitive at two stages, including young seedling stage and early reproductive stage. Thus, no comparative expression data are available at young seedling stage between indica and japonica rice lines under salinity stress.
Under high salinity stress, diverse damages have been observed in plants such as the disruption of intracellular ion homeostasis, membrane dysfunction, inhibition of metabolic activity and reduction of photosynthesis, and production of ROS (reactive oxygen species) (Zhu 2001; Mahajan and Tuteja 2005). Some gene families have been observed to play crucial roles in these damages and their recovery or other stress-related biological processes. Here, seven different gene families were selected for transcriptional profiling, and they were named as high-affinity K+ transporter (HKT), Na_H_exchanger (NHX), calmodulin, calcineurin B-like protein and its target kinase (CIPK), vacuolar H(+)-translocating inorganic pyrophosphatase (VHP), dehydrin, and stress-responsive protein (SRP) families. The HKT family mediates important Na+ tolerance mechanisms in plants (Mäser et al. 2002; Ren et al. 2005; Horie et al. 2009). For example, OsHKT2;1 (previously named as OsHKT1) in rice was downregulated in 30 mM NaCl (Horie et al. 2001) and played a role in mediating Na+ influx into roots of K+-starved rice plants (Horie et al. 2007). The NHX family plays roles in maintaining cellular pH and Na+ homeostasis by moving Na+ either out of cells or into organelles in exchange for H− (Rodríguez-Rosales et al. 2009). Calmodulin proteins were also involved in salt stress and ion homeostasis as well as in transcriptional regulation under stress (Ranty et al. 2006; Reddy et al. 2011). CIPKs play important roles in plant calcium signaling in response to stresses (Kolukisaoglu et al. 2004). VHP is an electrogenic proton pump that translocates protons into vacuoles in plant cells (Silva and Geros 2009). Efficient Na+ exclusion is one of the major mechanisms for salinity tolerance (Munns and Tester 2008). The exclusion of Na+ is driven by the plasma membrane H(+)-ATPase and by the vacuolar membrane H(+)-ATPase and H(+)-pyrophosphatase. Dehydrins are a class of hydrophilic, thermostable stress proteins that belong to the Group II late embryogenesis abundant family. These genes are expressed during late embryogenesis or in vegetative tissues in response to abiotic stresses in some of the tested plants (Yang et al. 2012). SRP family members are proteins with Pfam domain (PF06219). Their functions are not yet determined. On the other hand, transcription factors also play important roles in high salinity stress. Currently, many members from different families of transcription factors have been proven with biological functions in plant salinity stress response. For example, AP2/EREBP transcription factors consist of four subfamilies including AP2, CBF/DREB, ERF, and RAV, and at least one member from these subfamilies have been involved in salinity stress (Zhang et al. 2009). Many transcription factors from other families also function in salinity stress-related biological processes such as WRKY and NAC (No apical meristem, Arabidopsis transcription activation factor, Cup-shaped cotyledon) transcription factors (Ramamoorthy et al. 2008; Nakashima et al. 2012).
In Affymetrix rice microarray chips, the arrays contain probes to query approximately 48,564 japonica and 1,260 indica transcripts. Compared with the release 7 of Michigan State University (MSU) rice genome annotation database (http://rice.plantbiology.msu.edu/ [last accessed October 28, 2013]; Ouyang et al. 2007; Kawahara et al. 2013), only <36,000 out of 56,081 annotated genes were probed in chips. No expression data are available for the remaining annotated genes using the chips. In this study, we have designed custom microarray chips with probes covering majority of annotated genes from the MSU database. The chips were then employed for comparative transcript profiling analysis between salinity-sensitive and tolerant rice lines under normal and high salinity stress conditions. We have also identified single nucleotide polymorphisms (SNPs) between these two genotypes at genome level and investigated their biological effects. We compared their genomic variation and expression divergence and analyzed overrepresented genes and their functions by using gene set enrichment analysis (GSEA; Mootha et al. 2003) program. We then investigated up-/downregulated genes by high salinity stress, specially focusing on these genes encoding various transcription factors and stress-related proteins. We also surveyed molecular basis and mechanisms underlying expression divergence and evolution. Our data showed that genes with large-effect SNPs between these two genotypes were limited, and they mainly function in protein and macromolecular modification by GSEA. We have detected around 10% of probes/genes with differential expression under high salinity stress. Both sensitive and tolerant genotypes showed significant gene expression divergence under normal and NaCl stress conditions. Genomic variations in promoter regions resulted from SNPs, indels (1–10 bp of insertion/deletion), and structural variations (SVs, including large scale deletions, insertions, duplications, inversions, and translocations) significantly contributed to the expression divergence and regulation. Some gene families were highly regulated by the abiotic stress and might play key roles in stress-responsive biological processes. Our data also showed that tandem and segmental duplication, CACTA and hAT elements significantly contributed to expression divergence and regulation between these two contrasting genotypes under normal conditions or high salinity stress.
Materials and Methods
Plant Materials, Growth Conditions, and NaCl Stress Treatments
Both Nipponbare (japonica) and Pokkali (indica) rice plants (Oryza sativa L.) were used for all experiments in this study. After seed germination, they were planted in greenhouse and were grown under natural light and temperature conditions. Two-week old seedlings were collected from pots and washed with water to remove all soil. The seedling plants were then treated with salinity by submerging their roots into 250 mM of sodium chloride (NaCl) solution. Samples were collected in 0- and 8-h intervals.
Custom Design of Agilent Microarray Chips, Hybridization, and Data Analysis
The publicly available eArray software (Agilent Technologies, Santa Clara, CA) was used to design custom 60-mer oligonucleotide probes. Positive and negative control probes were designed from six housekeeping genes encoding elongation factor 1-alpha, sucrose synthase, ubiquitin, CYP86, actin, and GAPDH (glyceraldehyde-3-phophate dehydrogenase). In rice, a total of 66,433 gene models were annotated in the MSU rice genome annotation database (http://rice.plantbiology.msu.edu/; Ouyang et al. 2007; Kawahara et al. 2013), from which 40,800 non-transposable element (non-TE) and 14,278 TE probes (60 bp each) were designed, and the remaining genes were not suitable for probe design due to the sequence homology of other genes. Therefore, we have designed a total of 55,078 probes for microarray analysis. These probe sequences were then submitted for manufacturing on 8 × 60 k format of chips. Hybridization was carried out using the standard Agilent protocol. Agilent GeneSpring GX 11.5 software was used for data analysis. All expression data in this work have been deposited into the NCBI GEO database (www.ncbi.nlm.nih.gov/geo, last accessed October 28, 2013) under the GEO accession number GSE48395.
Microarray Data Verification by Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
A total of 12 genes were randomly selected to verify our microarray expression data by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The Applied Biosystems Primer Express® software was used to design all primer sequences. Non-gene-specific primer sequences filtered by Blast searches were then excluded instead of newly designed ones. All the primer sequences used in this study are listed in supplementary table S1, Supplementary Material online. QIAGEN RNeasy Mini Kit was used for total RNA isolation. The first strand cDNA was synthesized using Invitrogen kit. The qRT-PCR analyses were carried out using the AB power SYBR Green PCR Master mix kit (Applied Biosystems, P/N 4367659) according to the manufacturer’s protocol. The amplification of an eEF-1a gene was used as an internal control to normalize the data and the corresponding sequences of these primers are listed in supplementary table S1, Supplementary Material online. The threshold cycle (CT) value was automatically calculated based on the changes in fluorescence of SYBR Green I dye in every cycle monitored by the ABI 7900 system software. The mRNA relative amount was estimated from the CT value according to our previous description (Jiang et al. 2007) and was used to evaluate expression level of analyzed genes.
Identification of Genes with Differential Expression between Nipponbare and Pokkali under Normal Conditions or High Salinity Stress
Differentially expressed genes between Nipponbare and Pokkali were identified by their expression abundance. Processed microarray data by computing geometric mean were used for statistical analyses. Genes with at least two times difference in their mRNA levels between these two genotypes were submitted for Student’s t-test. These genes with a statistical difference at P < 0.05 were regarded as differentially expressed genes between these two genotypes. A similar standard was also applied for the identification of up- or downregulated genes under high salinity stress conditions.
Determination of Overrepresented Genes and Their Gene Ontology Categories
Gene ontology (GO) assignments for rice genes were obtained from the release 7 of MSU rice genome annotation database (http://rice.plantbiology.msu.edu/; Ouyang et al. 2007; Kawahara et al. 2013). We analyzed all three top GO categories including biological processes (P), molecular functions (F), and cellular components (C) (Harris et al. 2004). GSEA was used to determine over- or underrepresented genes and their GO categories. The Z test was used to test whether tandem/segmental duplication or transposition by mobile elements significantly contributes to expression divergence between Nipponbare and Pokkali or expression regulation under high salinity stress.
Promoter Isolation and Sequencing
To evaluate the contribution of indels and SVs in promoter regions to expression divergence and regulation, 24 genes with expression divergence/regulation between Nipponbare and Pokkali under either normal or high salinity stress conditions were selected. Their 1-kb promoter fragments were amplified from genomic DNA samples by PCRs. Primer sequences were listed in supplementary table S1, Supplementary Material online. PCRs were carried out using Qiagen Taq DNA Polymerase with cat. no. 201205. PCR amplifications were performed in 20 μl reaction mixtures with 50 ng of DNA, 200 μM of each of dNTPs, 0.5 μM each of primers, 1.5 mM MgCl2, 1 unit of Taq DNA polymerase, and buffer provided by the supplier. The reactions were performed in the PTC200 (MJ Research) thermocycler. The program used for PCR was 94 °C for 2 min followed by 30 cycles at 94 °C for 30 s, 55–60 °C for 60 s, and 72 °C for 120 s. The reaction was terminated with a 5-min extension step at 72 °C. PCR products of expected sizes were purified from agarose gel using Qiagen Gel Extraction Kit. These DNA fragments were directly used for sequencing by DNA Sequencing Kit (PE Applied Biosystems). Sequencing reactions were carried out in PTC200 (MJ Research) thermocycler using the ABI cycle sequencing protocol: 30 cycles at 96 °C for 30 s, 50 °C for 15 s, and 60 °C for 4 min.
Detection of Promoter Motifs and Their Overrepresentation Analysis
Nipponbare promoter sequences 1-kb upstream of start codon of each gene were retrieved from the MSU rice genome annotation database (http://rice.plantbiology.msu.edu/ [last accessed October 28, 2013]; Ouyang et al. 2007; Kawahara et al. 2013). Promoter motifs were detected by submitting these promoter sequences to the PLACE database (Higo et al. 1999; http://www.dna.affrc.go.jp/PLACE/index.html, last accessed October 28, 2013). A small program was designed to investigate the frequency of a promoter motif. Overrepresented motifs were detected by the BioProspector software (Liu et al. 2001). Promoter logos were generated by the enoLOGOS program (Workman et al. 2005).
Genome Sequence and Annotation Databases
Nipponbare genomic, cDNA, and protein sequences were downloaded from the release 7 of MSU rice genome annotation database (http://rice.plantbiology.msu.edu/, last accessed October 28, 2013; Ouyang et al. 2007; Kawahara et al. 2013). Pokkali SNP data were downloaded from the OryzaSNP project (McNally et al. 2009; http://www.OryzaSNP.org, last accessed October 28, 2013). A small perl program was written to map all SNPs from Pokkali onto the Nipponbare genome so that we can compare the effect of SNPs on genes and their promoters at genome level.
Identification of Transcription Factors and NaCl Stress-Related Seven Gene Families at Genome Level
A total of 84 families of transcription factors (Pérez-Rodríguez et al. 2010) were selected for genome-wide identification. The GRASSIUS database was also used for the identification of rice transcription factors (Yilmaz et al. 2009). In addition to these, for genome-wide identification of these transcription factors or seven gene family members, their protein motif or domain sequences from seed members were obtained from the Pfam database (http://pfam.sanger.ac.uk/, last accessed October 28, 2013). Their seed amino acid sequences were used to construct hidden Markov model (HMM) profiles using HMMER 2.3.2 (http://hmmer.janelia.org/, last accessed October 28, 2013) with default values. Using the profile HMMs, we scanned the rice annotated protein database and searched for all putative transcription factors and seven gene family members. These members were further confirmed by motif and domain analysis. All identified genes encoding transcription factors and seven gene families are listed in supplementary tables S2 and S3, Supplementary Material online, respectively.
Genome-Wide Identification of Tandemly/Segmentally Duplicated or Mobile Element Related Genes
DNA, cDNA, and protein sequences downloaded from the release 7.0 of the MSU rice genome annotation database were used for genome-wide identification of tandem/segmental duplication and mobile element-related genes. Tandemly and segmentally duplicated rice genes were identified according to the description (Jiang, González, et al. 2013). Full-length long terminal repeat (LTR) retrotransposon elements were identified by executing the LTR_Finder program (Xu and Wang 2007). CACTA DNA transposon elements were identified by both BlastN and HMM searches. We first carried out BlastN searches using both terminal inverted repeats (TIRs) and subterminal repeats (TRs) of the elements from multiple species (Pereira et al. 1986; Wang et al. 2003; Wicker et al. 2003; and references therein). We also built a HMM profile using HMMER 2.3.2 (http://hmmer.janelia.org/, last accessed October 28, 2013) with default values using seed TIRs and TRs from multiple species to scan the rice genome. These searches generated two sets of sequences including 5′ and 3′ terminal regions, respectively. Similar strategies were used to identify rice hAT elements with query sequences from multiple species (Hehl et al. 1991; Huttley et al. 1995; Kempken and Windhofer 2001; Moon et al. 2006). Full-length CACTA and hAT elements were identified by matching these two terminal regions with <30,000 and 4,000 bp in length, respectively. Identification of retrogenes, Pack-MULE (Mutator-like transposable element), hAT (hobo-Ac-Tam3), and Helitron elements was carried out according to the description (Juretic et al. 2005; Wang et al. 2006; Sweredoski et al. 2008). These genes fully or partially located within a mobile element were regarded as mobile element-related genes.
Results
Genomic Variations between Nipponbare and Pokkali Revealed by SNPs
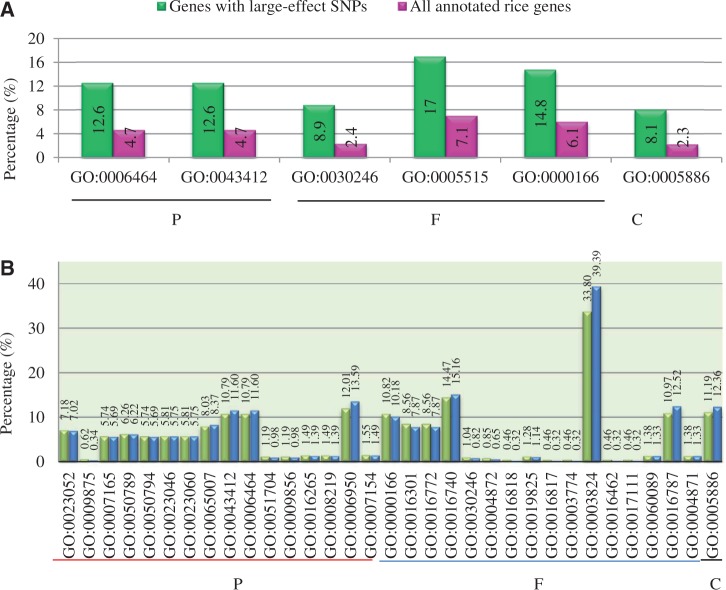
Nipponbare is a japonica rice and is sensitive to high salinity stress. Pokkali is an indica rice line and is tolerant to abiotic stress. The Nipponbare genome has been sequenced (Goff et al. 2002), and the SNP data are available for the Pokkali genome (McNally et al. 2009). On the basis of these data, we have identified a total of 131,080 SNPs located on coding regions. Up to 221 genes contain large-effect SNPs that lead to premature stop codons or changed start codons with shorter or longer length of open reading frames. GSEA showed that overrepresented genes with large-effect SNPs function mainly in carbohydrate/nucleotide/protein binding. Their proteins were mainly located in the plasma membrane by prediction and were involved in biological processes including protein/macromolecule modification by GSEA (fig. 1A). Expression analysis showed that only around 65% of them were detected with expression signal, whereas on average, up to 83% of annotated rice genes were expressed in this study (see later). Furthermore, their expression levels are relatively low when compared with other genes without large-effect SNPs. In addition, only a few genes among these large-effect SNP-containing genes were regulated by high salinity stress in their expression.
Fig. 1.—
The effects of SNPs on gene functions and GSEA. (A) GO term analysis of genes with large-effect SNPs by comparing with all annotated rice non-TE genes. Green and pink columns indicate the percentages of this GO term in SNP-affected proteins and total annotated rice proteins in the release 7 of MSU rice genome annotation database (http://rice.plantbiology.msu.edu/ [last accessed October 28, 2013]; Ouyang et al. 2007; Kawahara et al. 2013), respectively. (B) GO term analysis of genes with nonsynonymous substitutions by SNPs. Green and blue columns indicate the percentages of this GO term in SNP-affected proteins and total annotated rice proteins, respectively. Only GO terms with statistically significant differences at P < 0.05 were listed in (A) and (B). “P,” “F,” and “C” in (A) and (B) indicate the GO categories biological process, molecular function, and cellular component, respectively. GO term annotation in (A) and (B) refers to the following: GO:0006464 (protein modification process), GO:0043412 (macromolecule modification), GO:0030246 (carbohydrate binding), GO:0005515 (protein binding), GO:0000166 (nucleotide binding), GO:0005886 (plasma membrane), GO:0023052 (signaling), GO:0009875 (pollen–pistil interaction), GO:0007165 (signal transduction), GO:0050789 (regulation of biological process), GO:0050794 (regulation of cellular process), GO:0023046 (signaling process), GO:0023060 (signal transmission), GO:0065007 (biological regulation), GO:0051704 (multiorganism process), GO:0009856 (pollination), GO:0016265 (death), GO:0008219 (cell death), GO:0006950 (response to stress), GO:0007154 (cell communication), GO:0016301 (kinase activity), GO:0016772 (transferase activity, transferring phosphorus-containing groups), GO:0016740 (transferase activity), GO:0004872 (receptor activity), GO:0016818 (hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides), GO:0019825 (oxygen binding), GO:0016817 (hydrolase activity, acting on acid anhydrides), GO:0003774 (motor activity), GO:0003824 (catalytic activity), GO:0016462 (pyrophosphatase activity), GO:0017111 (nucleoside-triphosphatase activity), GO:0060089 (molecular transducer activity), GO:0016787 (hydrolase activity), and GO:0004871 (signal transducer activity).
Besides large-effect SNPs, other SNPs located on coding regions might contribute to either nonsynonymous or synonymous substitutions. As synonymous substitutions may not lead to any change in protein sequences, we only analyzed genes with nonsynonymous substitutions. Totally, we have detected 7,561 genes, in which nonsynonymous substitutions occurred. GO analysis showed that overrepresented proteins were mainly localized on plasma membrane. Their overrepresented molecular functions include kinase, pyrophosphatase, and transferase activities as well as catalytic, molecular transducer, motor, and receptor activities (fig. 1B). They mainly participate in biological regulation, protein modification, signaling, and stress response (fig. 1B). Expression analysis showed that similar percentages of genes were detectable between two groups of genes with either nonsynonymous or synonymous substitutions.
General Outlines of Expression Patterns in Nipponbare and Pokkali
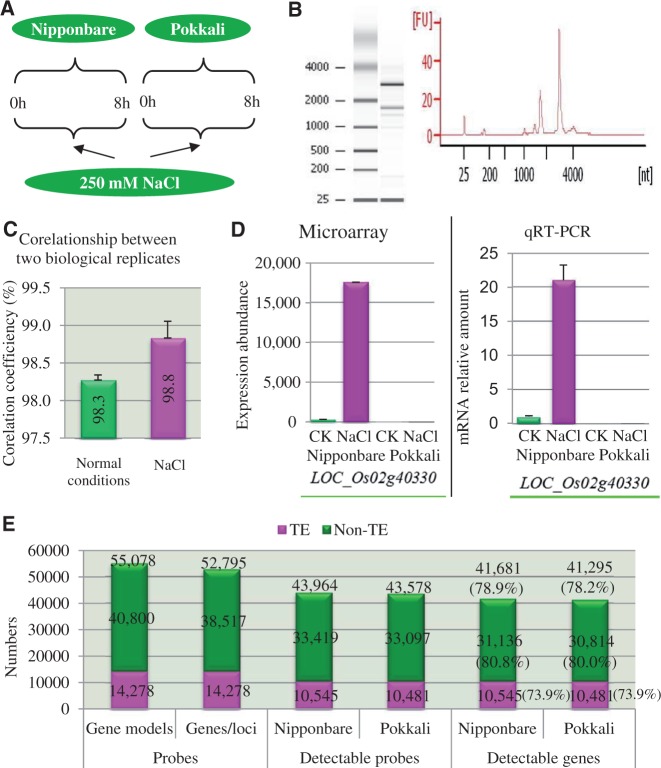
To figure out the difference in expression patterns between Nipponbare and Pokkali under normal and high salinity stress conditions, custom microarray chips were manufactured and used for comparative expression profiling. The expression analysis was carried out using 14-day-old whole seedlings from both Nipponbare and Pokkali rice lines. Two biological replicates were performed for both control and 250 mM NaCl treatments, resulting in a data set of eight microarrays (fig. 2A). Total RNA quality was analyzed by nanodrop reading and Agilent Bioanalyzer running (fig. 2B). The data quality was assessed by measuring the correlation coefficients (fig. 2C). The general coefficients between two biological replicates ranged from 98.3% (under normal growth conditions) to 98.8% (under NaCl treatments). We have randomly selected 459 probes, and each probe was printed into 15 or 16 different positions in the same chip for technical replicates. The resulted coefficients among technical replicates were measured from 98.9% to 99.6%. These analyses suggested the high quality of data obtained in this study. On the other hand, we have also validated our data by qRT-PCR (fig. 2D).
Fig. 2.—
Sampling and quality assessment of the microarray expression data and general expression profiling. (A) Sample collection of 14-day-old whole seedlings. (B) An example of total RNA quality analysis by nanodrop reading and Agilent Bioanalyzer running. (C) Coefficiency analysis. The general coefficients between two biological replicates ranged from 98.3% (under normal growth conditions) to 98.8% (under NaCl treatments). (D) An example of microarray expression data confirmed by qRT-PCR. A total of 12 genes were selected for qRT-PCR analysis. (E) Numbers of probes used in microarray chips and general expression profiling of genes encoding TEs and non-TEs.
In the release 7 of MSU rice genome annotation database (http://rice.plantbiology.msu.edu/ [last accessed October 28, 2013]; Ouyang et al. 2007; Kawahara et al. 2013), a total of 39,102 protein-coding transcripts have been annotated (non-TE). Out of the collection, 38,517 probes were generated and for the remaining 585 transcripts, no probe was generated due to duplicated sequences. Among 38,517 genes/loci, we have randomly selected 2,283 genes/loci with alternative splicing for analyzing expression divergence between different splicing models. Thus, a total of 40,800 probes were designed for non-TE genes/loci (fig. 2E). On the other hand, we have also designed 14,278 probes from these genes/loci encoding for TEs. Totally, we have designed 55,078 probes representing 38,517 non-TE and 14,278 TE genes. These probes have been designed for array manufacturing on Agilent 8 × 60k format.
Among a total of 55,078 probes from 52,795 genes/loci printed on the microarray chips, 43,964 probes were detectable in Nipponbare (fig. 2E). Among them, 33,419 probes were from non-TE genes, and the remaining 10,545 were from TE genes. In Pokkali, 43,578 probes were detected with expression signaling including 33,097 non-TE and 10,481 TE genes (fig. 2E). Because some genes contain alternative splicing, a total of 41,681 (78.9%) and 41,295 (78.2%) genes were expressed in Nipponbare and Pokkali, respectively (fig. 2E). No statistical difference was detected in their percentages of expressed genes between these two genotypes. Higher percentages of expressed genes were detected in non-TE genes when compared with TE genes in both genotypes.
Differentially Expressed Genes between Nipponbare and Pokkali
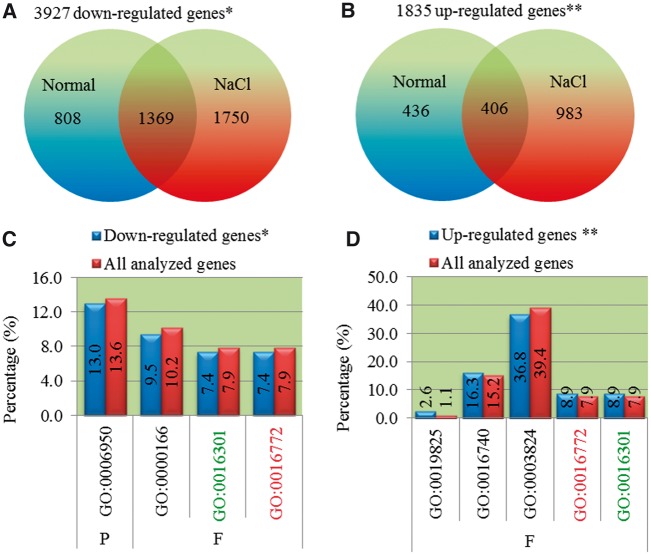
Based on the genome-wide SNP analysis between Nipponbare and Pokkali (fig. 1), limited divergence was observed in these two genomes. However, they exhibit obvious differences in their phenotypes. To understand their inside molecular bases, we further analyzed their expression divergence between these two genotypes under normal and high salinity stress conditions. Our data revealed 3,927 genes with at least two times lower expression abundance (downregulated) in Pokkali when compared with Nipponbare (fig. 3A). Among them, 808 and 1,750 genes were detected from normal and high salinity stress conditions, respectively. The remaining 1,369 genes showed differential expression in both growth conditions. On the other hand, only 1,835 genes were detected with at least two times higher expression (upregulated) in Pokkali when compared with Nipponbare (fig. 3B). Interestingly, under high salinity stress, much more differentially expressed genes were detected between these two genotypes than under normal growth conditions (fig. 3A and B).
Fig. 3.—
Differentially expressed genes between Nipponbare and Pokkali. (A) Differentially expressed genes with at least two times lower in Pokkali in their expression level when compared with Nipponbare (downregulated genes). (B) Differentially expressed genes with at least two times higher in Pokkali in their expression level when compared with Nipponbare (upregulated genes). (C) Over- or underrepresented GO terms by GSEA in downregulated genes as shown in (A). (D) Over- or underrepresented GO terms by GSEA in upregulated genes as shown in (B). GO terms highlighted by red and green fonts were detected in both down- and upregulated genes. “P” and “F” indicate the GO categories biological process and molecular function, respectively. GO term annotation in (A) and (B) refers to the following: GO:0006950, response to stress; GO:0000166, nucleotide binding; GO:0016301, kinase activity; GO:0016772, transferase activity (transferring phosphorus-containing groups); GO:0019825, oxygen binding; GO:0016740, transferase activity; GO:0003824, catalytic activity.
To evaluate whether these differentially expressed genes between these two genotypes were biased toward certain specific functions, we carried out the investigation on GO. We identified over- or underrepresented GO terms by GSEA. Among genes with lower expression abundance in Pokkali (fig. 3A), a total of four GO terms were underrepresented in Pokkali when compared with that in Nipponbare (fig. 3C). These GO terms included GO:0006950 (response to stress), GO:0000166 (nucleotide binding), GO:0016301 (kinase activity), and GO:0016772 (transferase activity, transferring phosphorus-containing groups). On the other hand, among genes with higher expression in Pokkali (fig. 3B), a total of four overrepresented GO terms were detected, which included GO:0019825 (oxygen binding), GO:0016740 (transferase activity), GO:0016301, and GO:0016772 (fig. 3D). Both GO:0016301 and GO:0016772 were underrepresented in genes with lower expression in Pokkali (fig. 3C) but were overrepresented in genes with higher expression in Pokkali (fig. 3D). The term GO:0003824 (catalytic activity) was the only term with underrepresentation in genes with higher expression in Pokkali (fig. 3D).
Differentially Regulated Genes under High Salinity Stress in Both Nipponbare and Pokkali
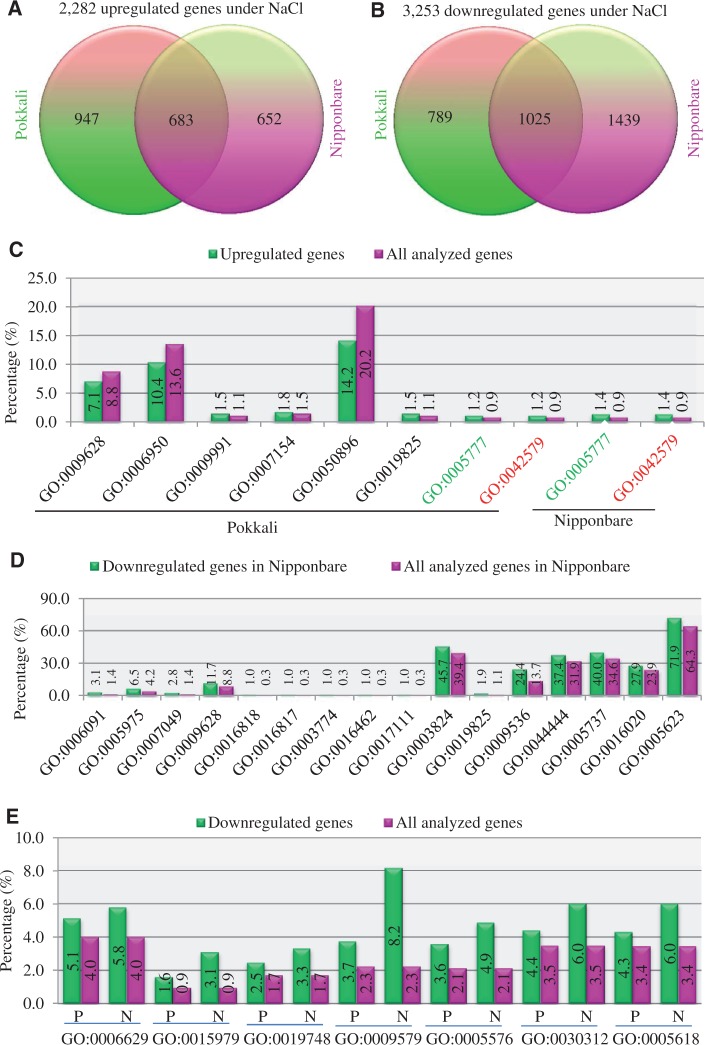
Nipponbare is a salinity-sensitive rice variety, whereas Pokkali is a salinity-tolerant line. We are interested in exploring the difference in gene expression under high salinity stress between these two genotypes. We have identified a total of 2,282 upregulated genes under high salinity stress (250 mM NaCl). Among them, 947 and 652 genes were detected only in Pokkali and Nipponbare, respectively. The remaining 683 genes were detected in both genotypes (fig. 4A). On the other hand, compared with upregulated genes, we have identified nearly double numbers of downregulated genes under high salinity stress (fig. 4B). Among the 3,253 downregulated genes, 789 and 1,439 genes were detected only in Pokkali and Nipponbare, respectively, indicating a higher percentage of downregulated genes in Nipponbare (fig. 4B). The remaining 1,025 genes were detected in both genotypes.
Fig. 4.—
Differentially regulated genes under high salinity stress in both Nipponbare and Pokkali. (A) and (B) Up- and downregulated genes under 250 mM NaCl, respectively. (C) Over- or underrepresented GO terms by GSEA in upregulated genes as shown in (A). GO terms highlighted by green and red fonts were these terms detected in both Pokkali and Nipponbare. (D) Overrepresented GO terms by GSEA in downregulated genes only in Nipponbare as shown in (B). (E) Overrepresented GO terms by GSEA in downregulated genes in both Nipponbare and Pokkali as shown in (B). GO term annotation in (C)–(E) refers to the following: GO:0009628, response to abiotic stimulus; GO:0006950, response to stress; GO:0009991, response to extracellular stimulus; GO:0007154, cell communication; GO:0050896, response to stimulus; GO:0019825, oxygen binding; GO:0005777, peroxisome; GO:0042579, microbody; GO:0006091, generation of precursor metabolites and energy; GO:0005975, carbohydrate metabolic process; GO:0007049, cell cycle; GO:0016818, hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides; GO:0016817, hydrolase activity, acting on acid anhydrides; GO:0003774, motor activity; GO:0016462, pyrophosphatase activity; GO:0017111, nucleoside-triphosphatase activity; GO:0003824, catalytic activity; GO:0009536, plastid; GO:0044444, cytoplasmic part; GO:0005737, cytoplasm; GO:0016020, membrane; GO:0005623, cell; GO:0006629, lipid metabolic process; GO:0015979, photosynthesis; GO:0019748, secondary metabolic process; GO:0009579, thylakoid; GO:0005576, extracellular region; GO:0030312, external encapsulating structure; GO:0005618, cell wall.
We were also interested in the biological functions of these up- or downregulated genes. We analyzed these genes regulated by high salinity stress only in either Nipponbare or Pokkali but not in both genotypes as we were interested in figuring out the difference between these two genotypes in response to high salinity stress. For 947 upregulated genes only in Pokkali, three GO terms, GO:0009628 (response to abiotic stimulus), GO:0006950 (response to stress), and GO:0050896 (response to stimulus), were underrepresented; the remaining five GO terms were overrepresented (fig. 4C). They are GO:0009991 (response to extracellular stimulus), GO:0007154 (cell communication), GO:0019825 (oxygen binding), GO:0005777 (peroxisome), and GO:0042579 (microbody). However, for 652 upregulated genes in Nipponbare, only two overrepresented GO terms were detected, and they were also presented in Pokkali (fig. 4C). For 1,439 downregulated genes in Nipponbare, 23 GO terms were detected, and all of them were overrepresented (fig. 4D and E). Among them, 16 overrepresented GO terms were Nipponbare specific (fig. 4D). They are GO:0006091 (generation of precursor metabolites and energy), GO:0005975 (carbohydrate metabolic process), GO:0007049 (cell cycle), GO:0009628 (response to abiotic stimulus), GO:0006629 (lipid metabolic process), GO:0016818 (hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides), GO:0016817 (hydrolase activity, acting on acid anhydrides), GO:0003774 (motor activity), GO:0016462 (pyrophosphatase activity), GO:0017111 (nucleoside-triphosphatase activity), GO:0003824 (catalytic activity), GO:0019825 (oxygen binding), GO:0009536 (plastid), GO:0044444 (cytoplasmic part), GO:0005737 (cytoplasm), GO:0016020 (membrane), and GO:0005623 (cell). The remaining seven GO terms were detected in both Pokkali and Nipponbare (fig. 4E). They are GO:0006629 (lipid metabolic process), GO:0015979 (photosynthesis), GO:0019748 (secondary metabolic process), GO:0009579 (thylakoid), GO:0005576 (extracellular region), GO:0030312 (external encapsulating structure), and GO:0005618 (cell wall). Interestingly, for each GO term, higher percentages of downregulated genes were detected in Nipponbare when compared with Pokkali (fig. 4E). Generally, under high salinity stress, genes with functions in response to stress or stimulus were down- or upregulated, and obvious differences were observed in the gene expression profilings between Nipponbare and Pokkali.
Expression Profile of Genes Encoding Transcription Factors
Among the differentially expressed genes between Nipponbare and Pokkali under normal and high salinity stress conditions, we were interested in these genes encoding transcription factors. We have identified 2,777 genes encoding 93 different transcription factor families in the rice genome (supplementary table S2, Supplementary Material online). Among them, 319 (11.5%) genes showed up- or downregulated expression under high salinity stress, and these genes were from either Nipponbare or Pokkali. Among these regulated genes, around one third of them showed similar expression patterns under NaCl stress between two genotypes and the remaining two thirds of them showed expression divergence between these two genotypes. The data suggested a high expression divergence of transcription factor encoding genes between Nipponbare and Pokkali under NaCl stress.
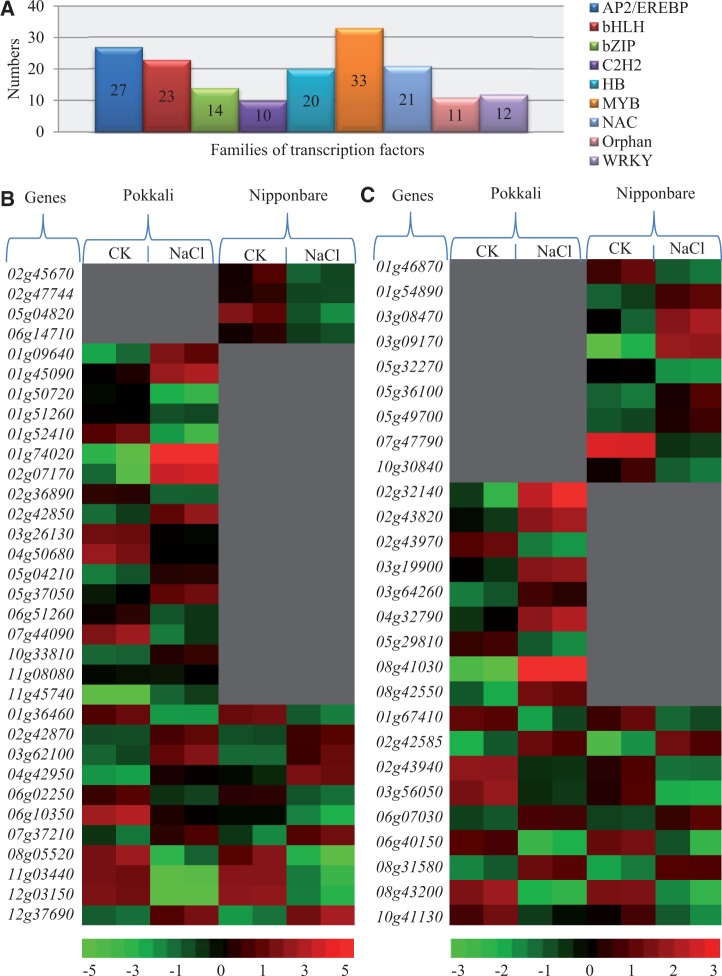
We then further analyzed the families with ten or more members in their responses to high salinity stress in detail. We identified a total of nine such families including Ap2/EREBP, bHLH (basic helix-loop-helix), bZIP (basic region/leucine zipper motif), C2H2 (Cysteine-2/Histidine-2), HB (homeobox), MYB (Myeloblast), NAC, Orphan, and WRKY (fig. 5A). The largest two families are MYB and AP2/EREBP. A total of 33 MYB and 27 AP2/EREBP family members were up- or downregulated by NaCl in either Nipponbare or Pokkali. For the 33 MYB family members, four genes were downregulated by NaCl only in Nipponbare and 18 genes were up- or downregulated by NaCl only in Pokkali (fig. 5B). The remaining 11 genes were regulated by NaCl in both Nipponbare and Pokkali (fig. 5B). For the 27 AP2/EREBP family members, nine genes were up- or downregulated by NaCl only in Nipponbare and other nine genes were up- or downregulated by NaCl only in Pokkali (fig. 5C). The remaining nine genes were regulated by NaCl in both Nipponbare and Pokkali (fig. 5C).
Fig. 5.—
Expression profiling of rice genes encoding transcription factors under normal and high salinity stress conditions. (A) Differentially expressed genes in top ten families of transcription factors. (B) and (C) Heat maps showed up- or downregulated genes encoding MYB and AP2/EREBP transcription factors, respectively. Normalized expression values were calculated by Agilent GeneSpring GX 11.5 software and were used for heat map analyses in figures 5 and 6. Red, black, and green colors indicated that normalized expression values were >0, =0, and <0, respectively, in the matrix. Gray color indicated that no statistical difference in their expression level has been detected in these genes under high salinity stress.
Expression Profile of Genes Encoding Na+/Other Cations and Abiotic Stress-Related Proteins
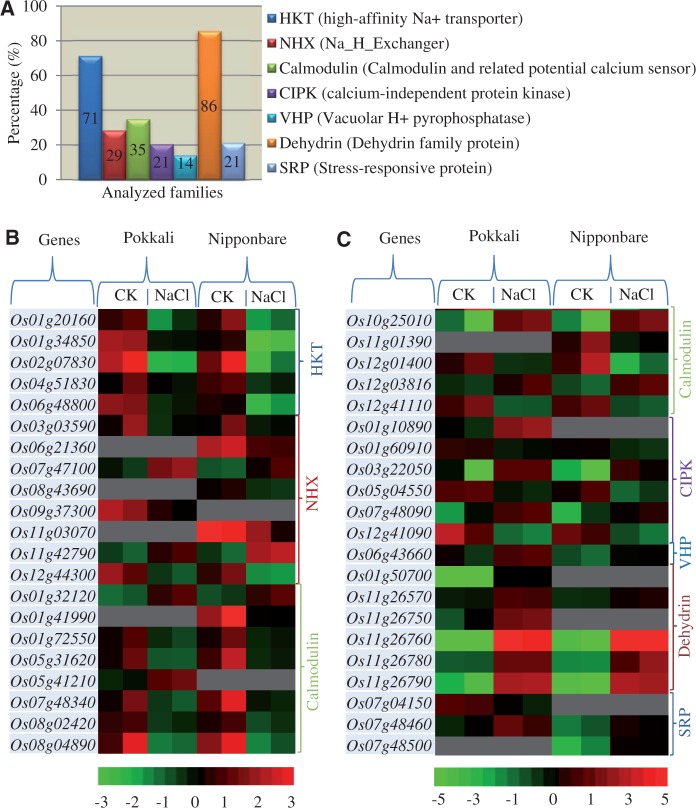
Because both Nipponbare and Pokkali showed obvious difference in their response to high salinity stress, we were interested in expression profiles of genes encoding stress-related proteins in these two genotypes. We have analyzed seven different families including HKT, NHX, calmodulin, CIPK, VHP, dehydrin, and SRP. We have identified a total of 129 genes encoding these seven families (supplementary table S3, Supplementary Material online) and 42 (32.6%) of them were regulated by NaCl in either Nipponbare or Pokkali. The rate is obviously higher than the average percentages of NaCl-regulated genes in Nipponbare and Pokkali. However, different families exhibited difference in their percentages of regulated genes, from 14% for VHP to 86% for dehydrin (fig. 6A). Generally, the percentages were significantly higher than that in other genes encoding transcription factors or other proteins. Figure 6B showed the detailed expression patterns of 42 up- or downregulated genes. For the HKT family, all five differentially expressed genes were downregulated by NaCl in both Nipponbare and Pokkali, and no significant difference was observed between these two genotypes. A similar situation was also observed for the VHP family, where only one upregulated gene was detected under NaCl treatment. For the remaining five gene families, expression divergence was observed between Nipponbare and Pokkali under high salinity stress. In these gene families, some genes were up- or downregulated by NaCl treatment only in either Nipponbare or Pokkali.
Fig. 6.—
Expression profiling of rice genes encoding Na+/other cation and abiotic stress-related proteins. (A) Differential expression profiling in seven families of genes encoding Na+/other cation and abiotic stress-related proteins. (B) and (C) Heat maps showed up- or downregulated genes. Red, black, and green colors indicate the same annotation as shown in figure 5. Gray color indicates that no statistical difference in their expression level has been detected in these genes under high salinity stress.
On the basis of our microarray analysis, we have selected a total of 30 candidate genes as listed in supplementary table S4, Supplementary Material online. These genes encode various transcription factors, Na+/other cations, and abiotic stress-related proteins. Their expression was differentially regulated by high salinity stress in either one or two tested varieties. They would be promising candidate genes for improving rice tolerance to high salinity stress by genetic manipulation.
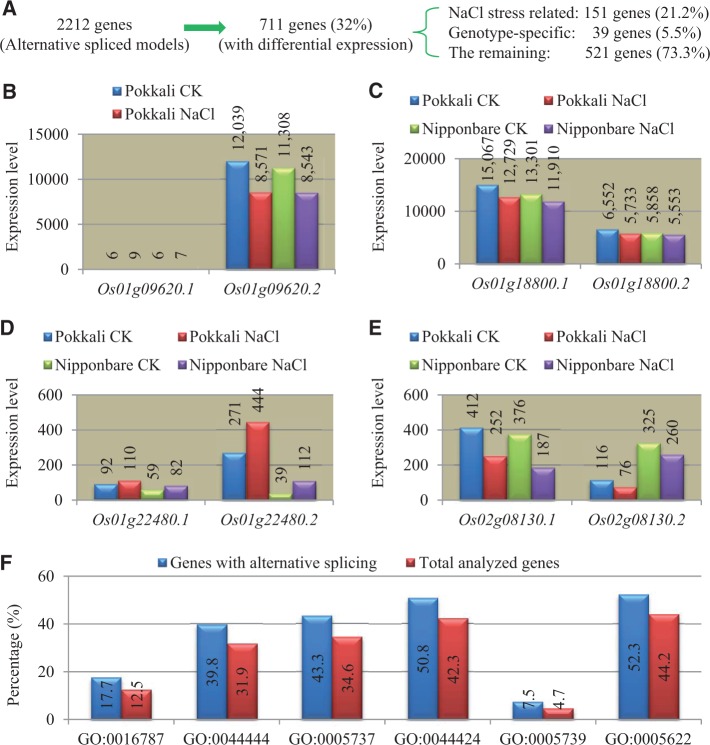
Expression Divergence of Genes with Alternative Spliced Models in Nipponbare and Pokkali
In the MSU database, a total of 6,457 genes/loci were annotated with alternative splicing models (http://rice.plantbiology.msu.edu/ [last accessed October 28, 2013]; Ouyang et al. 2007; Kawahara et al. 2013). To explore the expression divergence of genes with alternatively spliced models in Nipponbare and Pokkali under either normal or high salinity stress conditions, we randomly selected 2,282 out of 6,457 genes/loci annotated with alternatively spliced models. Two isoforms were analyzed for each gene. Among these genes/loci, 2,212 genes showed expression signals. A total of 711 loci (32%) were detected with differential expression between two alternatively spliced models (fig. 7A). Most of the expression divergence (521 genes, 73.3%) was observed in both Nipponbare and Pokkali under normal and high salinity stress conditions (fig. 7B and C). Some of the expression divergence (151 genes, 21.2%) occurred only under high salinity stress (fig. 7D). The other 39 divergent genes (5.5%) were genotype specific, that is, expression divergence was observed only in either Nipponbare or Pokkali (fig. 7E).
Fig. 7.—
Functional annotation and expression analysis of genes with alternative spliced models. (A) General outline of genes with alternative splicing models and their expression divergence. (B)—(E) Examples of genes with different types of expression divergence. (F) Overrepresented GO terms by GSEA in these genes with alternative spliced models. GO term annotation refers to the following: GO:0016787, hydrolase activity; GO:0044444, cytoplasmic part; GO:0005737, cytoplasm; GO:0044424, intracellular part; GO:0005739, mitochondrion; GO:0005622, intracellular.
To further investigate whether these genes with expression divergence between two alternatively spliced models are biased to certain functions, we submitted these genes for GSEA. In biological process (P) category, no over- or underrepresented GO term was detected under default parameters. In the molecular function (F) category, one GO term GO:0016787 (hydrolase activity) was detected with overrepresentation (fig. 7F). This may imply that hydrolase activity may be evolutionarily regulated by alternative splicing between two genotypes under either normal or NaCl stress conditions. A majority of overrepresented GO terms were observed in the cellular component (C) category. A total of five GO terms were detected with statistical overrepresentation (fig. 7F). They are GO:0044444 (cytoplasmic part), GO:0005737 (cytoplasm), GO:0044424 (intracellular part), GO:0005739 (mitochondrion), and GO:0005622 (intracellular).
Promoter Variation and Expression Divergence between Nipponbare and Pokkali
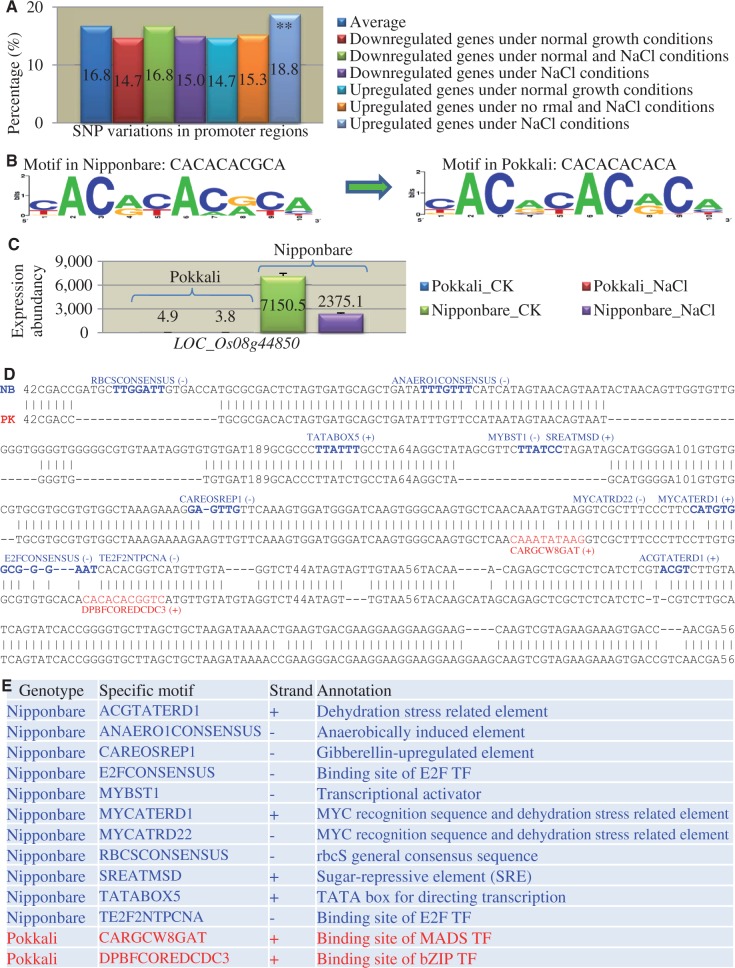
We have detected more than 5,000 differentially expressed genes under normal or NaCl stress conditions between Nipponbare and Pokkali, and they were classified into six groups as shown in figure 3A and B. To explore the molecular basis of their expression divergence, we surveyed the SNP variations in their 1-kb promoter regions upstream of start codon of genes. On average, 16.8% of rice genes showed SNP variations in their promoter regions (fig. 8A). However, among the six groups of differentially expressed genes, genes from five groups showed significant lower or similar SNP variations when compared with the average SNP variations (fig. 8A). Only one group showed significantly higher SNP variation (18.8%), and genes from this group exhibited at least two times higher expression only in Pokkali (upregulated) under NaCl stress (fig. 8A). The data suggested that SNP variations contributed to expression divergence mainly under certain stress conditions. To investigate how SNPs contribute to expression divergence, we further analyzed the difference in their overrepresented promoter motifs between these two genotypes. Promoter sequences from the group with 18.8% of SNP variations between these two genotypes were submitted to motif analysis (see Materials and Methods). We have detected three overrepresented promoter motifs and two of them were the same, and only one showed a difference between these two genotypes. In Nipponbare, most of the motif sequence is CACACACGCA and in Pokkali, the motif is CACACACACA (fig. 8B). In Nipponbare, the sequence encodes a promoter motif named DPBFCOREDCDC3, which is an abscisic acid response motif (Finkelstein and Lynch 2000). However, in Pokkali, no motif was detected due to the SNP variation from G to A (fig. 8B). The results suggest a contribution of SNPs to expression divergence.
Fig. 8.—
Genomic variations of promoter sequences and their motif analysis between Nipponbare and Pokkali. (A) Comparative analysis of promoter SNP variations among different types of regulated genes under high salinity stress. (B) Overrepresented promoter motifs between Nipponbare and Pokkali as revealed by SNPs. The BioProspector program (Liu et al. 2001) was used to detect overrepresented promoter motifs. (C) An example of a gene (LOC_Os08g44850) with expression divergence between Nipponbare and Pokkali. (D) Promoter sequencing and motif analysis of the gene LOC_Os08g44850. Motifs highlighted by blue fonts were Nipponbare (NB) specific, whereas these motifs with red fonts were Pokkali (PK) specific. (E) Species-specific motifs in Nipponbare and Pokkali and their annotation.
Because our data showed that SNP variations contribute to the expression divergence mainly for genes with two times higher Pokkali under NaCl (fig. 8A), we also analyzed the contribution of indels and SVs to expression divergence. As no whole-genome sequencing data are available for the Pokkali genome, we have randomly selected 24 genes with differential expression between Nipponbare and Pokkali under either normal or NaCl conditions for promoter sequencing analysis. Our data showed that 23 out of 24 genes showed variations in their promoter sequences. As a result, their promoters showed a difference in at least one motif. One of such genes is LOC_Os08g44850. This gene showed a very low expression level in Pokkali (fig. 8C). However, in Nipponbare, this gene showed high expression level and exhibited downregulated expression under NaCl stress (fig. 8C). Sequencing data revealed multiple indels and SVs in their promoter sequences besides SNPs (fig. 8D). As a result, 11 motifs lost and two motifs gained in Pokkali (fig. 8E). One of the lost motifs is TATABOX5, which might contribute to very low expression of this gene in Pokkali.
Effect of Duplication and Transposition on Gene Expression under High Salinity Stress
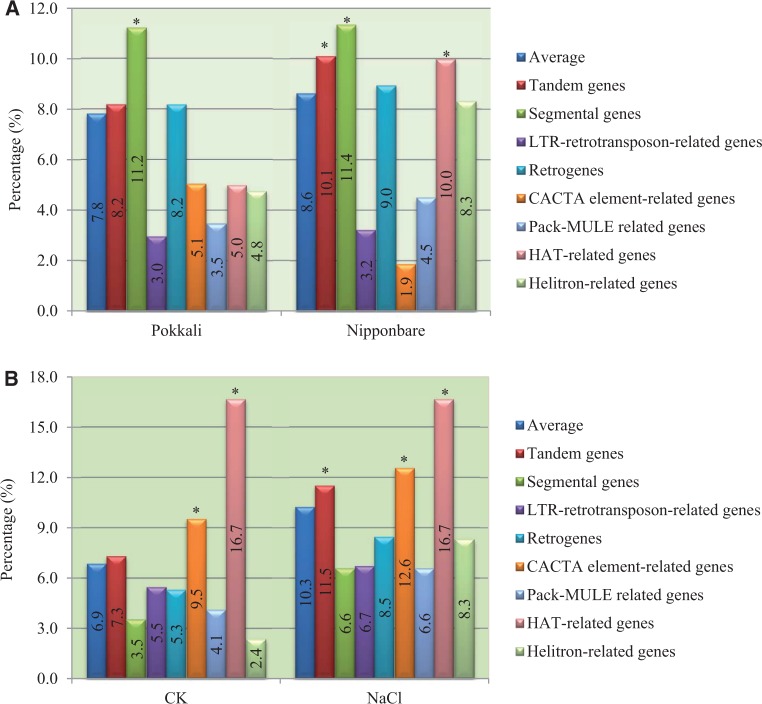
To explore the evolutionary basis of expression divergence within and between Nipponbare and Pokkali under normal and high salinity stress, we further analyzed the effect of both duplication and mobile elements on expression divergence. We have identified 5,111 tandemly and 6,244 segmentally duplicated genes, 8,502 LTR-retrotransposon-related, 2,912 retrogenes, 679 CACTA element-related, 3,707 Pack-MULE related, 60 hAT-related, and 81 Helitron-related genes in the rice genome (supplementary table S5, Supplementary Material online). We then analyzed their contribution to expression divergence under NaCl stress in both Nipponbare and Pokkali. In Pokkali, on average, we have detected 7.8% of genes that were regulated by NaCl stress. Among the analyzed two duplication models and six types of mobile elements, only segmental duplication significantly contributed to expression divergence and up to 11.2% of segmentally duplicated genes were up- or downregulated by high salinity stress (fig. 9A). However, in Nipponbare, higher expression divergence was observed in both tandemly and segmentally duplicated genes as well as hAT-related genes (fig. 9A).
Fig. 9.—
The effect of duplication/transposition on gene regulation and divergence. (A) The effect of duplication/transposition on gene regulation under high salinity stress. The average columns indicate the percentages of genes with up- or downregulated genes among total analyzed genes in either Pokkali or Nipponbare. (B) The effect of duplication/transposition on gene divergence between Nipponbare and Pokkali. The average columns indicate the percentages of genes with expression divergence between Nipponbare and Pokkali under either normal (CK) or high salinity stress conditions. The stars in (A) and (B) indicate statistical difference at P < 0.05.
We then surveyed the effects of duplication and transposition on gene expression divergence between Pokkali and Nipponbare. Under normal growth conditions, we have detected 6.9% of genes showing expression divergence between Pokkali and Nipponbare on average (fig. 9B). For CACTA and hAT-related genes, up to 9.5% and 16.7% of them showed expression divergence between these two genotypes. For duplicated genes and the remaining four types of mobile element-related genes, similar or lower percentages of them showed expression divergence under normal growth conditions (fig. 9B). However, a different situation was observed under NaCl stress conditions. On average, 10.3% of analyzed genes exhibited expression divergence under this stress (fig. 9B). We found that 11.5%, 12.6%, and 16.7% of genes expanded from tandem duplication, and CACTA and hAT mobile elements, respectively, were differentially expressed between Pokkali and Nipponbare under NaCl stress, which were significantly higher than the average (fig. 9B). Thus, different gene duplication/expansion mechanisms differentially contributed to the expression divergence between Nipponbare and Pokkali or under high salinity stress.
Discussion
Custom Microarray Analysis Revealed More Genes with Detectable Expression Signal in the Rice Genomes under Normal or Stress Conditions
How many genes are expressed in rice during seedling stage and what is the difference in gene expression profiling under certain stress? Large amounts of studies have been carried out to figure out these questions. However, no definite answer has been achieved. Because of the limited availability of expressed sequence tags (EST) and cDNA sequences at earlier stages, limited probes were printed for microarray analysis; thus, limited genes were analyzed (Kawasaki et al. 2001). Currently, widely used rice array chips were from Affymetrix (http://www.affymetrix.com, last accessed October 28, 2013). This array contains probes to query 51,279 transcripts representing two rice cultivars, with approximately 48,564 japonica transcripts and 1,260 transcripts representing the indica cultivar. Most of the rice microarray analyses were based on these chips. On the other hand, the rice genome was completely sequenced and annotated in its encoding genes (http://rice.plantbiology.msu.edu; Kawahara et al. 2013). Based on the various Affymetrix microarray-based expression analyses, only around 23,821 annotated genes were expressed (Jiang and Ramachandran 2010). This might be partially due to the fact that no probe has been printed in the Affymetrix chips for many annotated rice genes. Thus, currently available commercial microarray chips are not enough to cover all rice genes and to outline their expression profiling. In this study, we have designed 55,078 probes representing 38,517 non-TE and 14,278 TE genes using the Agilent 8 × 60 k format of microarray chips. Our data revealed 41,681 genes in Nipponbare and 41,295 genes in Pokkali with detectable expression signaling under either normal or high salinity stress conditions (fig. 2E). In a previous report, around 31,382 genes were detected with expression signal using microarray, massively parallel signature sequencing or by cDNA/EST (Jiang and Ramachandran 2010). By comparing these published data, our custom microarray analysis detected 16,230 (7,797 non-TE and 8,433 TE) genes in Nipponbare and 16,313 (7,863 non-E and 8,450 TE) genes in Pokkali, which were not probed or detected with expression signal in previous studies (Jiang and Ramachandran 2010). On the other hand, we only used 250 mM NaCl solution for high salinity stress. Thus, more expressed genes including differentially expressed genes should be detected if higher concentrations of NaCl (e.g., 300 mM) or a series of concentration gradients for salinity stress can be employed. Thus, more genes should have participated in rice growth and development as well as stress regulation during seedling stages. Furthermore, much more genes should be detected with expression signaling if more developmental stages and more abiotic or biotic stresses were investigated for gene expression analyses. Therefore, rice growth and development should be involved in a comprehensive gene regulation system with the participation of more than 40,000 genes.
Expression Divergence Significantly Contribute to Genetic Diversity in Rice
Genetic diversity has been assessed by morphological traits, physiological index, and molecular markers (Caldo et al. 1996; Jahn et al. 2011; Zhang et al. 2011). With the rapid progress on sequencing technologies, the whole genome sequencing data have been used to evaluate genetic diversity (Huang et al. 2012; Xu et al. 2012). In addition, expression divergence was also used to survey genetic diversity in animals and plants (Kliebenstein et al. 2006; Albert et al. 2012). However, in rice, little is known about genome-wide expression divergence of genes among different germplasms and its contribution to genetic diversity. We have identified more than 5,700 genes with differential expression in their transcription abundance between Nipponbare and Pokkali under normal conditions or high salinity stress (fig. 3). We have also identified differentially regulated genes by NaCl stress between these two genotypes (fig. 4). Compared with genetic variations revealed by SNPs and their contribution to the divergence of biological functions of corresponding genes, the number of genes with expression divergence between these two genotypes is obviously higher. These data suggested that expression divergence should also be regarded as an index to evaluate genetic diversity in rice. Furthermore, studies showed that upregulation or downregulation of certain genes in transgenic rice plants significantly altered their tolerance to salinity stress (Jiang and Ramachandran 2010; Nakashima et al. 2012), and some of these genes also showed expression divergence between Nipponbare and Pokkali under normal and salinity stress conditions. The data suggested that expression divergence might contribute to cultivar improvement in salinity tolerance during rice domestication. Our expression data might provide a reference for further improving rice salinity tolerance by selecting certain genes for genetic modification.
Molecular Basis of Gene Expression Divergence between Nipponbare and Pokkali
Generally, substantial differences in gene expression patterns between closely related species have been reported in several studies (Rifkin et al. 2003; Khaitovich et al. 2006; Tirosh et al. 2009, 2010; Rebeiz et al. 2011). Mechanisms underlying these differences have not been fully understood yet. A promoter is required to initiate transcription of a gene and expression divergence might occur in case of promoter mutation. Under normal growth conditions, promoter SNP variations showed no statistical difference between corresponding genes with expression divergence and no expression divergence (fig. 8). Similar results were also observed between genotypes Nipponbare and indica 93-11 (Zhang et al. 2008). Interestingly, our data showed that promoter SNP variations between Nipponbare and Pokkali were only detected in genes with higher level of expression in Pokkali under salinity stress (fig. 8A). SNP-mediated mutation in promoter motifs might be regarded as a reason for expression divergence (fig. 8B). Besides SNP, indels and SVs of promoter regions might also contribute to expression divergence. Out of the 24 differentially expressed genes between Nipponbare and Pokkali under normal and NaCl stress conditions, 23 of them were detected with promoter variations in either SNP/indel or SV. These variations significantly contributed to the change in promoter cis-regulatory elements, thus, resulting in expression divergence (fig. 8C and D). Previous study also showed that promoter indels significantly contributed to the expression divergence between Nipponbare and 93-11 (Zhang et al. 2008). On the other hand, our data showed that a few differentially expressed genes between Nipponbare and Pokkali also have the same promoter sequence. The data suggest the contribution of DNA methylation or other mechanisms to gene expression divergence between these two genotypes. Both genetic and epigenetic factors might affect gene expression divergence among different genotypes and their contribution rates might vary depending on different organisms. Up to 31.7% of genes with expression divergence showed no promoter variations between analyzed grain and sweet sorghum lines (Jiang, Ma, et al. 2013). Thus, in sweet and grain sorghum lines, DNA methylation might be regarded as an important factor to contribute to gene expression divergence (Jiang, Ma, et al. 2013). However, in Nipponbare and Pokkali, promoter variations from SNPs, indels, and SVs but not DNA methylation should be regarded as main factors to contribute to expression divergence.
Molecular Mechanism of Expression Divergence
Sequencing data of indica and japonica rice genomes showed that the majority of tandem and segmental duplications as well as transposition events of mobile elements occurred beyond the divergence between these two subspecies (Goff et al. 2002; Yu et al. 2002). These duplication or transposition events provided a basis for expression divergence. We were interested in whether these events contributed to expression divergence under normal or in response to high salinity stress. We have analyzed the effect of tandem and segmental duplications as well as six different mobile elements (LTR retrotransposon, non-LTR retrotransposon, CACTA element, MULE, hAT, and Helitron) on gene expression divergence. Our data showed that tandem and segmental duplication, CACTA and hAT elements significantly contributed to expression divergence under normal conditions or high salinity stress (fig. 9). Thus, duplication and mobile elements might partially contribute to species divergence through expression regulation. In this study, we only analyzed the expression divergence under normal and high salinity stress at seedling stage. Higher expression divergence should be revealed between these two genotypes by investigating their expression profiling under additional abiotic and biotic stresses and at multiple developmental stages.
Supplementary Material
Supplementary tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
This work was supported by the Singapore National Research Foundation under the Competitive Research Programme Funding Scheme (CRP award no. NRF-CRP7-2010-02).
Literature Cited
- Albert FW, et al. A comparison of brain gene expression levels in domesticated and wild animals. PLoS Genet. 2012;8:e1002962. doi: 10.1371/journal.pgen.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldo RA, Sebastian LS, Hernandez JE. Morphology-based genetic diversity analysis of ancestral lines of Philippine rice cultivars. Philipp J Crop Sci. 1996;21:86–92. [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu JK. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. [Google Scholar]
- Cotsaftis O, et al. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol Plant. 2011;4:25–41. doi: 10.1093/mp/ssq056. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gregorio GB, et al. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002;76:91–101. [Google Scholar]
- Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(database issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl R, Nacken WK, Krause A, Saedler H, Sommer H. Structural analysis of Tam3, a transposable element from Antirrhinum majus, reveals homologies to the Ac element from maize. Plant Mol Biol. 1991;16:369–371. doi: 10.1007/BF00020572. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009;14:660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, et al. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Horie T, et al. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, MacRae AF, Clegg MT. Molecular evolution of the Ac/Ds transposable-element family in pearl millet and other grasses. Genetics. 1995;139:1411–1419. doi: 10.1093/genetics/139.3.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MZ, Baset Mia MA, Islam MR, Akter A. Effect of different saline levels on growth and yield attributes of mutant rice. J Soil Nat. 2007;1:18–22. [Google Scholar]
- Ismail AM, Heuer S, Thomson MJ, Wissuwa M. Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol Biol. 2007;65:547–570. doi: 10.1007/s11103-007-9215-2. [DOI] [PubMed] [Google Scholar]
- Jahn CE, et al. Genetic variation in biomass traits among 20 diverse rice varieties. Plant Physiol. 2011;155:157–168. doi: 10.1104/pp.110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, González JM, Ramachandran S. Comparative genomic and transcriptomic analysis of tandemly and segmentally duplicated genes in rice. PLoS One. 2013;8:e63551. doi: 10.1371/journal.pone.0063551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Ma Z, Vanitha J, Ramachandran S. Genetic variation and expression diversity between grain and sweet sorghum lines. BMC Genomics. 2013;14:18. doi: 10.1186/1471-2164-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SY, Ramachandran S. Assigning biological functions to rice genes by genome annotation, expression analysis and mutagenesis. Biotechnol Lett. 2010;32:1753–1763. doi: 10.1007/s10529-010-0377-7. [DOI] [PubMed] [Google Scholar]
- Jiang SY, et al. Ds insertion mutagenesis as an efficient tool to produce diverse variations for rice breeding. Plant Mol Biol. 2007;65:385–402. doi: 10.1007/s11103-007-9233-0. [DOI] [PubMed] [Google Scholar]
- Joseph EA, Mohanan KV. A study on the effect of salinity stress on the growth and yield of some native rice cultivars of Kerala state of India. Agric For Fish. 2013;2:141–150. [Google Scholar]
- Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, et al. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempken F, Windhofer F. The hAT family: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma. 2001;110:1–9. doi: 10.1007/s004120000118. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, et al. Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics. 2006;172:1179–1189. doi: 10.1534/genetics.105.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Choi WY, Ko JC, Kim TS, Gregorio GB. Salinity tolerance of Japonica and Indica rice (Oryza sativa L.) at the seedling stage. Planta. 2003;216:1043–1046. doi: 10.1007/s00425-002-0958-3. [DOI] [PubMed] [Google Scholar]
- Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001;2001:127–138. [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Mäser P, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/s0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- McNally KL, et al. Genome-wide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, et al. Identification of active transposon dTok, a member of the hAT family, in rice. Plant Cell Physiol. 2006;47:1473–1483. doi: 10.1093/pcp/pcl012. [DOI] [PubMed] [Google Scholar]
- Mootha VK, et al. PGC-1alpharesponsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Ouyang S, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35(database issue):D846–D851. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. Molecular analysis of the En/Spm transposable element system of Zea mays. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, et al. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38(database issue):D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy R, Jiang SY, Kumar N, Venkatesh PN, Ramachandran S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008;49:865–879. doi: 10.1093/pcp/pcn061. [DOI] [PubMed] [Google Scholar]
- Ranty B, Aldon D, Galaud JP. Plant calmodulins and calmodulin-related proteins multifaceted relays to decode calcium signals. Plant Signal Behav. 2006;1:96–104. doi: 10.4161/psb.1.3.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci U S A. 2011;108:10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Ali GS, Celesnik H, Day IS. Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 2011;23:2010–2032. doi: 10.1105/tpc.111.084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosales MP, et al. Plant NHX cation/proton antiporters. Plant Signal Behav. 2009;4:265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P, Geros H. Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal Behav. 2009;4:718–726. doi: 10.4161/psb.4.8.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweredoski M, DeRose-Wilson L, Gaut BS. A comparative computational analysis of nonautonomous helitron elements between maize and rice. BMC Genomics. 2008;9:467. doi: 10.1186/1471-2164-9-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Sigal N, Assia Y, Barkai N. Chromatin regulators as capacitors of interspecies variations in gene expression. Mol Syst Biol. 2010;6:435. doi: 10.1038/msb.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, et al. Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol Biol. 2007;63:609–623. doi: 10.1007/s11103-006-9112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GD, et al. Genomic characterization of Rim2/Hipa elements reveals a CACTA-like transposon superfamily with unique features in the rice genome. Mol Genet Genomics. 2003;270:234–242. doi: 10.1007/s00438-003-0918-z. [DOI] [PubMed] [Google Scholar]
- Wang W, et al. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006;18:1791–1802. doi: 10.1105/tpc.106.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Guyot R, Yahiaoui N, Keller B. CACTA transposons in Triticeae: a diverse family of high-copy repetitive elements. Plant Physiol. 2003;132:52–63. doi: 10.1104/pp.102.015743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, et al. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33(web server issue):W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(Web Server issue):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012;12:140. doi: 10.1186/1471-2229-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, et al. GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 2009;149:171–180. doi: 10.1104/pp.108.128579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. Indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zeng L, Shannon MC, Grieve CM. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica. 2002;127:235–245. [Google Scholar]
- Zhang HY, et al. A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol Plant. 2008;1:720–731. doi: 10.1093/mp/ssn022. [DOI] [PubMed] [Google Scholar]
- Zhang G, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, et al. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One. 2011;6:e27565. doi: 10.1371/journal.pone.0027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.