Abstract
Miniature inverted-repeat transposable elements (MITEs) are a specific group of nonautonomous DNA transposons, and they are distributed in a wide range of hosts. However, the origin and evolutionary history of MITEs in eukaryotic genomes remain unclear. In this study, six MITEs were identified in the silkworm (Bombyx mori). Five elements are grouped into four known superfamilies of DNA transposons, and one represents a novel class of MITEs. Unexpectedly, six similar MITEs are also present in the triatomine bug (Rhodnius prolixus) that diverged from the common ancestor with the silkworm about 370 Ma. However, they show different lengths in two species, suggesting that they are different derivatives of progenitor transposons. Three direct progenitor transposons (Sola1, hobo/Ac/Tam [hAT], and Ginger2) are also identified in some other organisms, and several lines of evidence suggested that these autonomous elements might have been independently and horizontally transferred into their hosts. Furthermore, it is speculated that the twisted-wing parasites may be the candidate vectors for these horizontal transfers. The data presented in this study provide some new insights into the origin and evolutionary history of MITEs in the silkworm and triatomine bug.
Keywords: MITEs, origin, evolution, Bombyx mori, Rhodnius prolixus
Introduction
Transposons are discrete DNA fragments, and they are characterized by an inherent property of transposition and replication within their host genomes (Kidwell and Lisch 2001). An increasing number of studies have demonstrated that transposons are a major force to drive the evolution of genome complexity (Kazazian 2004).
Transposons can be classified into two classes (DNA and RNA transposons) based on their transposition intermediates. DNA transposons are a group of transposons that use a “cut-and-paste” mechanism to transpose (Wicker et al. 2007). They are characterized by terminal inverted repeats (TIRs) and target site duplication (TSD), and their autonomous copies encode a transposase that is responsible for their mobility. In the last decades, a specific group of nonautonomous elements that is derived from the autonomous DNA transposons has been found in plants (Bureau and Wessler 1992). These elements, named miniature inverted-repeat transposable elements (MITEs), are also flanked by TIRs and TSD. There are also other common features, including small size, A + T richness, high copy number, potential stable secondary structure, a lack of transposase, and size and sequence homogeneity (Feschotte et al. 2002; Han et al. 2010). The most important feature distinguishing them from typical defective elements is their high copy number and structural homogeneity (Feschotte and Pritham 2007). MITEs can be classified into different superfamilies based on their similar structure (such as TIRs or TSD). The distribution of MITEs in the genome is obviously associated with genes (Oki et al. 2008). These results suggested that MITEs might have great effects on gene regulation and genome evolution. For example, MITEs may upregulate (MITEs containing regulatory motifs) or downregulate (MITE-derived small RNAs) the expression levels of host genes where they reside (Yang et al. 2005; Oki et al. 2008; Kuang et al. 2009). However, the origin and evolutionary history of MITEs in insects are poorly understood.
The taxonomic distribution of DNA transposons suggests that they are almost present in the whole eukaryotic tree of life (Feschotte and Pritham 2007). However, transposons should be eliminated by drift and/or selection in their original lineages. Horizontal transfer (HT) between isolated species seems to be the most important means by which transposons can avoid inevitable stochastic loss (Schaack et al. 2010). Besides, HT of a transposon into a new genome is also regarded as an important force to drive genome variation and biological innovation. Furthermore, almost all transposons have the ability of HT (Schaack et al. 2010). The classic documented examples of transposon HT are the P and Tc1/mariner transposons of Drosophila (Daniels et al. 1990; Maruyama and Hartl 1991). Although 218 convincing cases of HT of transposons are described in invertebrates, vertebrates, and plants, the molecular mechanisms of most transposon HT have not been elucidated (Schaack et al. 2010). Recently, it has been demonstrated that the intimacy of parasitism might facilitate the HT of transposon (Yoshiyama et al. 2001; Laha et al. 2007; Gilbert et al. 2010).
Previously, we identified a novel cluster of the hobo/Ac/Tam (hAT) superfamily in Bombyx mori and Rhodnius prolixus and found that this novel element generated MITEs in B. mori rather than in R. prolixus (Zhang et al. 2013). It is likely that this element invaded into their hosts independently by HT (Zhang et al. 2013). Here, we report the characterization and classification of six MITEs identified in both B. mori and R. prolixus. Meanwhile, the direct or related progenitors of these MITEs were also found in some other organisms. Moreover, our results indicated that progenitors of these MITEs might have been independently and horizontally transferred into their hosts for a long time. Finally, a model is provided for the origin and evolutionary history of these MITEs in the silkworm and the triatomine bug.
Materials and Methods
Data Resources
Bombyx mori assembled genomic sequences, expressed sequence tag (EST) sequences, tRNAs, 5S rRNA, and 7SL RNA were downloaded from Silkworm Genome Database (SilkDB version 2, http://www.silkdb.org/silkdb/doc/download.html, last accessed October 29, 2013). Meanwhile, the full-length cDNA sequences of the silkworm were downloaded from the National Center for Biotechnology Information (NCBI) (accession numbers: AK377185-AK388575). Rhodnius prolixus genomic supercontig sequences, EST sequences, and transcript sequences were downloaded from VectorBase (Lawson et al. 2009). Both Manduca sexta and Me. moldrzyki whole genome shotgun (WGS) sequences were downloaded from NCBI (accession numbers: AIXA01000000 and AGDA00000000). Mengenilla moldrzyki EST sequences were downloaded from a data repository website (http://datadryad.org/resource/doi:10.5061/dryad.ts058, last accessed October 29, 2013). Heliconius melpomene genomic sequences were downloaded from Butterfly Genome Database (http://www.butterflygenome.org/, last accessed October 29, 2013). Leaf-cutter ant Atta cephalotes genome sequence and transcript sequences were downloaded from Hymenoptera Genome Database (http://hymenopteragenome.org/, last accessed October 29, 2013). Megachile rotundata and Hydra magnipapillata genomic sequences, and mRNA sequences of Meg. rotundata were obtained from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/, last accessed October 29, 2013). Transposase sequences of other known Sola, hAT, and Ginger transposons were obtained from Repbase (http://www.girinst.org/repbase/update/browse.php, last accessed October 29, 2013).
Identification and Copy Number Determination of MITEs
Six nonautonomous DNA transposable elements (bm_648, Bmori_800.383, bm_701, bm_1364, bm_295 and bm_248) were obtained from BmTEdb (Xu et al. 2013). Their copies were extracted with 500 bp flanking sequences using our Perl script. Then, these retrieved sequences were aligned using MUSCLE (Edgar 2004) to determine the exact boundary of these transposons. Then, we reconstructed their consensus sequences by multiple alignments of their copies in the silkworm genome using DAMBE (Xia and Xie 2001). To determine the distribution of these transposons, their consensus sequences were used as queries in BlastN searches against the NCBI genome databases of the species for which a complete or near complete genome is available. A transposon was considered to be present in a species if any copy was at least 80% identity at the nucleotide level to the consensus over at least 50% of its length. Consensus sequences for each transposon family identified in other species were also constructed based on multiple alignments of copies using DAMBE (Xia and Xie 2001). For families with only truncated copies or a few copies (<10) in the genome, the best hit was chosen as consensus sequence. For families with uncertainty in its internal regions, their reliable 5′ and 3′ terminal regions were used to construct consensus sequence (Gilbert et al. 2010).
To estimate copy number of each transposon family, consensus sequences or representatives were used to mask all genomes where these transposons families were found using BlastN. Blast hits longer than 100 bp and at least 80% identity at the nucleotide level to their consensus were used to calculate copy number (Gilbert et al. 2010). For MITEs, one copy was defined as full length according to the following criteria: >90% nucleotide identity to the consensus, covering >80% of the consensus and including complete TIRs (Han et al. 2010). As Novel_NA was not flanked by TIRs, copies that share at least 90% identity at the nucleotide level to the consensus and cover at least 90% of its length were considered as intact elements.
Identification of Autonomous DNA Transposons and Evolutionary Analysis
When we used all MITEs identified in the silkworm as queries to do BlastN search against NCBI nt database, autonomous candidates responsible for the spread of two families (Sola1_NA and hAT1_NA) were found. Then, their potential transposases were predicted using getorf in EMBOSS-6.3.1 package (Rice et al. 2000). To determine the distributions of these two autonomous families, their sequences were used as queries in BlastN searches against the NCBI genome databases of the species for which a complete or near complete genome is available. A transposon was considered to be present in a species if any copy was at least 80% identity at the nucleotide level to the consensus over at least 50% of its length.
For other four families, we identified their potential autonomous families using extensive analysis in the silkworm and the triatomine bug. Briefly, we searched the large DNA fragments that shared similar terminal sequences to the above four families. All DNA fragments ranging from 1,000 bp to 10 kb flanked by the termini of these four families were obtained. Then, their potential transposases were predicted using getorf in EMBOSS-6.3.1 package (Rice et al. 2000). These candidates with open reading frame (ORF) were used as queries in BlastN searches against the NCBI genome databases, and the same standard mentioned earlier was used.
To determine the relationships of autonomous Sola1, hAT1, and Ginger2 elements identified in this study and known elements from other organisms (downloaded from Repbase, http://www.girinst.org/repbase/update/browse.php, last accessed October 29, 2013), we aligned amino acid sequences of their transposases using MUSCLE (Edgar 2004). Gap sites and variable regions were excluded, and then MEGA 4 (Tamura et al. 2007) was used to build phylogenetic trees.
Sequence Analysis
The frequency distribution of sequence identities of MITEs in B. mori and R. prolixus was also determined. The percentage identity was based on the similarity between each copy and the respective consensus. Only copies longer than 100 bp were considered. Finally, we investigated insertion site preference of these six MITEs in B. mori and R. prolixus. To reflect their insertion preferences exactly, only copies flanked by intact TSDs were included in the analysis. Sequence logos were created by WebLogo (Crooks et al. 2004) based on 30 nt (15 upstream and 15 downstream) flanking the insertion site.
Sequences of three most conserved host genes, heat shock cognate 70 (Hsc70-4), Tubulin beta-3 (tub3), and elongation factor 1 alpha (EF1α), were used in the comparison with transposon Ka/Ks, with the purpose of testing HT hypothesis. Their accession numbers were listed in supplementary table S1, Supplementary Material online. Pairwise alignments of these genes were created using MUSCLE (Edgar 2004), and each pairwise identity was calculated by Bioedit (Hall 1999). Codon alignments of hAT1 and these host genes were constructed by PAL2NAL software (Suyama et al. 2006). Then, the Ka/Ks ratio was calculated using DnaSP v5 (Librado and Rozas 2009).
Polymerase Chain Reaction Analysis
Genomic DNA of the silkworm strain Dazao was obtained from State Key Laboratory of Silkworm Genome Biology (China). Genomic DNA of R. prolixus was kindly provided by Dr Ricardo N Araujo (Laboratório de Fisiologia de Insetos Hematófagos, Brasil). Polymerase chain reaction (PCR) primers for all MITEs identified in B. mori and R. prolixus were designed based on their flanking regions. The list of primers and expected product sizes were given in supplementary table S2, Supplementary Material online. PCRs were performed using the following thermal profiles: initial denaturation at 94 °C for 4 min and then 30–35 cycles (32 cycles for Sola1_NA_RP and hAT1_NA_RP, 35 cycles for hAT2_NA_RP, and 30 cycles for other families in B. mori and R. prolixus) of 94 °C for 40 s, 48–55 °C for 50 s (48 °C for hAT2_NA_RP, 50 °C for Novel_NA_BM and 55 °C for the rest families), and 72 °C for 1 min, PCR products were run in 1% agarose gels in 1× Tris acetate-EDTA (TAE) buffer and visualized under UV light.
Results and Discussion
Identification and Analysis of Six MITEs in B. mori and R. prolixus
Six families of nonautonomous transposon families were obtained from BmTEdb (Xu et al. 2013). These reference sequences were not classified in BmTEdb. However, we demonstrated that five families were grouped into Sola-like, hAT-like, Ginger-like, and Tc1/Mariner superfamilies based on their similarities to the characteristic features (TIRs and TSDs) of these superfamilies (table 1), and the last one may represent a novel class of transposons (please see the below results). We also demonstrated that these families may be MITE-like elements. (Each MITE identified in this study is named as X_NA_#$, where X represents a superfamily, NA represents nonautonomous elements, # is a letter representing the first letter of genus, and $ is a letter representing the first letter of species. Each autonomous element is named as X_ #$.) as they were highly homogeneous in sequence and lack encoding sequences. For example, the majority (about 65%) of members of Sola1_NA_BM were 300–340 bp in length (data not shown).
Table 1.
Characteristics of Sola1_NA, hAT1_NA, Ginger2_NA, Novel_NA, pogo_NA, hAT2_NA, and their Autonomous Elements
| Group | Length (bp) | TSD | TIR | Copy Number | Accession Number of Representatives |
|---|---|---|---|---|---|
| Species | |||||
| Sola1_NA | |||||
| B. mori | 334 | AWWT | GGACCGTTTATTTTGAAAGTGGTGTAAGTCCATTTT | 83 (41) | KF558364 |
| R. prolixus | 709 | AWDT | GGACCGTTTATTTTGAAAGTGGTGTAAG | 105 (40) | KF558358 |
| 1,489a | ND | ND | 1 | GL562232 10898 12386 − | |
| H. melpomene | 248 | AWDT | GGACCGTTTATTTTGAGAGTGGTGTAAGTCCATTTT | 263 (228) | CAEZ01006881 13054 13301 − |
| Danaus plexippus | 194 | ND | ND | 1 | AGBW01008816 6235 6428 + |
| M. sexta | 222 | ND | ND | 1 | AIXA01005774 19729 19950 − |
| A. cephalotes | 2,313a | ND | GACCATTTATTCTGAAAGT | 6 | ADTU01013967 5894 8206 + |
| Meg. rotundata | 2,778a | ND | GGACCGTTTATTTTGAAAGTGGTGCAAGTCCATTTT | 69 | AFJA01007769 17605 20382 − |
| hAT1_NA | |||||
| B. mori | 546 | 8 bp | CAGTGATTCCCAAA | 51 (32) | KF558365 |
| R. prolixus | 619 | 8 bp | CAGTGATTCCCAAA | 27 (25) | KF558359 |
| 2,889a | 8 bp | CAGTGATTCCCAAA | 2 | GL563087 1981027 1983915 − | |
| H. melpomene | 415 | 8 bp | CAGTGATTCCCAAA | 13 (4) | CAEZ01008298 7516 7940 − |
| M. sexta | 795 | 8 bp | CAGTGATTCCCAAA | 21 (15) | AIXA01005484 47113 47907 + |
| Hy. magnipapillata | 2,916a | 8 bp | CAGTGATTCCCAAA | 93 | NW_002143368 8030 10961 − |
| Me. moldrzyki | 1,573a | ND | ND | ND | AGDA01034126 1 1573 + |
| Ginger2_NA | |||||
| B. mori | 531 | WRTH | TGTTACGGTAAAAGTA | >120 (28) | KF558366 |
| 2,442a | NA | TGTTACGGTAGAAGTA | NA | BABH01026869 12853 15294 + | |
| R. prolixus | 651 | KHTM | TGTTACGGTAAAAGTA | 40 (25) | KF558360 |
| 3,767a | TATA | TGTTACGGTAAAAGTA | >1 | GL563081 872372 876138 − | |
| Me. moldrzyki | 599 | ND | TGTTACGGTAAAAGTA | 57 | AGDA01093000 3264 3854 − |
| Novel_NA | |||||
| B. mori | 892 | TT | — | 37 (11) | KF558367 |
| Danaus plexippus | 678 | ND | — | 2 | AGBW01012094 72750 73427 + |
| R. prolixus | 803 | TT | — | 881 (604) | KF558361 |
| pogo_NA | |||||
| B. mori | 270 | TA | CAGTAAAAGCTCTTTATTCGCG | >484 (15) | KF558368 |
| R. prolixus | 748 | TA | CAGTAAAAGCTCTTTATTCGCG | 297 (197) | KF558362 |
| M. sexta | 851 | TA | CAGTAGAAGCTCTTTATTCG | 1 | AIXA01008424 19396 20246 + |
| Danaus plexippus | 1,030 | TA | CAGTAAAATCTCTTTATTCGCG | 12 (3) | AGBW01000094 2627 3651 + |
| hAT2_NA | |||||
| B. mori | 310 | 8 bp | CGGTGTTTCCCAAC | 224 (29) | KF558369 |
| R. prolixus | 956 | 8 bp | CGGTGTTTCCCAAC | 126 (53) | KF558363 |
| H. melpomene | 522 | 8 bp | CGGTGTTTCCCAAC | 10 (1) | CAEZ01008877 20470 21038 + |
| M. sexta | 277 | 8 bp | CAGTGTTTCCCAAC | 78 (28) | AIXA01004312 63292 63566 + |
| Me. moldrzyki | 236 | ND | ND | 5 | AGDA01059403 8065 8300 − |
Note.—W, A/T; R, A/G; D, A/T/G; K, T/G; H, A/C/T; M, A/C. ND, not determined. The number in bracket is the number of full-length copies in each MITE family.
aThe potential autonomous elements.
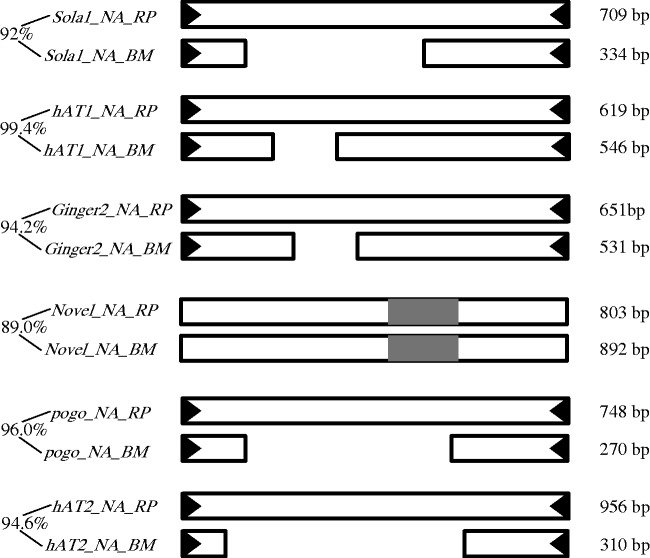
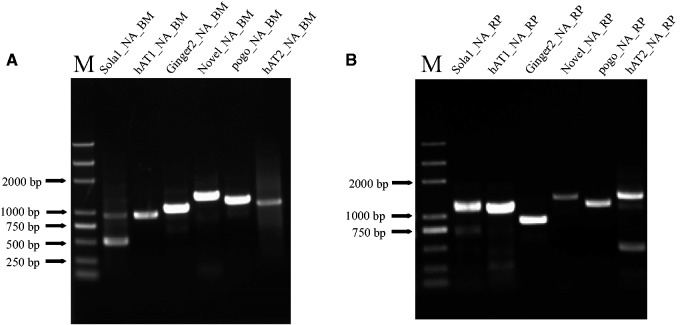
Unexpectedly, all these families are also identified in R. prolixus (supplementary table S3, Supplementary Material online) when we used the above silkworm transposons as queries to perform BlastN search against the R. prolixus genome. However, they showed different lengths in both species (fig. 1). This suggested that they were different derivatives of the same progenitor transposons in B. mori and R. prolixus. A pairwise comparison of these transposons in both species revealed an extremely high identity (89–99.4%) at the nucleotide level over the whole length (fig. 1). In R. prolixus, all these families also seemed to be MITEs because of their sequence and size homogeneity. The presence of these six MITEs in both species was confirmed by PCR (fig. 2).
Fig. 1.—
Structure and the homologous regions of six MITEs identified in the silkworm and triatomine bug. Black triangles are TIRs, and white rectangles are homologous regions of each transposon in both species. Gray regions represent the variable area of the Novel_NA family. The corresponding names and percentages of identity are shown on the left. The right numbers are the size of each consensus sequence.
Fig. 2.—
Experimental verification of the presence of six MITEs identified in the silkworm (A) and triatomine bug (B). PCR fragments of expected sizes were obtained from both species. “M” indicates marker, and the left numbers show their corresponding sizes.
To better understand their evolutionary history, we compared copy numbers of each family between B. mori and R. prolixus. Copy numbers of five families (Sola1_NA, hAT1_NA, Ginger2_NA, pogo_NA, and hAT2_NA) were from 27 to several hundreds, and these families had almost the same copy number in both species (table 1). In contrast, Novel_NA family had a very larger copy number in R. prolixus than B. mori (881 vs. 37). In addition, most copies of Novel_NA_RP (604/881) were the verified full-length elements, suggesting that this family might experience a burst of transposition in R. prolixus. A similar phenomenon was also observed in another family (pogo_NA_RP). Indeed, star shape trees of Novel_NA_RP and pogo_NA_RP also suggested that they might have experienced one round of burst amplification (supplementary fig. S1, Supplementary Material online).
To further clarify the evolutionary history of these families, the distributions of percentage identity scores between copies and corresponding consensus or representatives were estimated (supplementary fig. S2, Supplementary Material online). The results showed that copies of six families (except Sola1_NA) were more conserved in R. prolixus than B. mori. For example, R. prolixus Ginger2_NA family had an identity distribution with a peak at identity 99%. However, Ginger2_NA family in B. mori had a distribution with a peak at identity 93%, and it also included a long tail of more divergent copies. It appears that these families might have experienced bursts of transposition in the recent past in R. prolixus but in the more distant past in B. mori. However, it should be noted that these observations may not reflect the exact evolutionary histories of these transposons because they may have different nucleotide substitution rates in two different host species.
Finally, we analyzed insertion site preferences of these families. Sola1_NA in B. mori and R. prolixus showed insertion preferences for the AWWT (W: A/T) and AWDT (D: A/T/G) tetranucleotide, respectively (supplementary fig. S3, Supplementary Material online). This is a hallmark of the Sola1 transposons (Bao et al. 2009). HAT1_NA and hAT2_NA tended to integrate in sequences having conserved TA in positions + 4 and + 5 (supplementary fig. S3, Supplementary Material online). However, hAT1_NA also showed an additional preference for integration in sequence having a conserved A in position + 2 of their TSD in B. mori. Ginger2_NA family preferentially targeted 4-bp AT-rich sequences, TATA, an important characteristic of Ginger2 elements (Bao et al. 2010). Novel_NA family had a preferential insertion of T-rich sequences. Pogo_NA had strict insertion specificity for TA dinucleotide.
In summary, MITEs described here were strikingly similar to each other in B. mori and R. prolixus. They showed similar TIRs, high sequence identities, and similar targets for integration. All these data indicated that each family identified in both species originated from the same ancestral transposon and could be recognized as the same transposon evolving in two very different genomic backgrounds.
Annotation of Novel_NA in B. mori and R. prolixus
Classification of transposons is complex and difficult because of their complicated and diverse structures. Therefore, classification of a transposon needs several lines of evidence. Novel_NA identified in this study did not have TIRs and did not show similarity to any known transposons. However, the characteristics of nonautonomous nature, small size, and high copy number suggested that it might be a short interspersed element (SINE)-like or MITE-like element. Here, we demonstrated that Novel_NA was a MITE-like element based on three lines of evidence.
First, SINEs originate from tRNA, 5S rRNA, or 7S rRNA genes as a result of their 5′ sequences sharing homology with these genes (Frenkel et al. 2004). Moreover, SINEs also harbor an RNA pol III promoter that is necessary for their transposition. These are two major characteristics that distinguish SINEs from other nonautonomous transposable elements. However, Novel_NA elements lacked a RNA polymerase III (pol III) promoter and did not show homology with any tRNA, 5S rRNA, or 7S rRNA genes of the silkworm. This suggested that Novel_NA might not be a SINE-like element. Recently, large-scale full-length cDNA of the silkworm was determined (Suetsugu et al. 2013). A full-length Novel_NA, which was amplified by PCR from the silkworm genomic DNA (fig. 2A), was not correspondingly detected by using its sequence as a query in BlastN search against the above cDNA database. This suggested that the propagation of Novel_NA might not occur by reverse transcription.
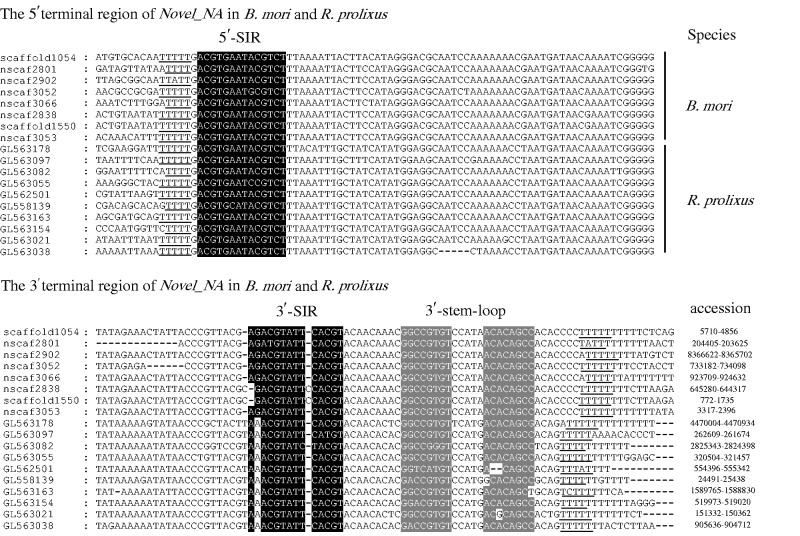
Second, many MITEs identified in insects tend to generate A + T biased TSD during their integration. For instance, HzMINE-2 elements from Helicoverpa zea and DINE-1 elements from Drosophila generated TT dinucleotide TSD during their integration (Yang and Barbash 2008; Coates et al. 2011). In particular, the HzMINE-2 and DINE-1 insertion sites were flanked by similar duplications (TTTTT or WTTTT). These nucleotide sequences provided the recognition sites where they integrated, and resulted in the creation of a TT TSD. Interestingly, Novel_NA was also characterized by the presence of TTTTT duplications (fig. 3). These findings suggested that Novel_NA might use the same integration mechanism as that of HzMINE-2 and DINE-1 elements.
Fig. 3.—
Alignment of 5′ and 3′ terminal regions from full-length Novel_NA in the silkworm and triatomine bug. Black color shows their subterminal inverted repeat (SIR) and gray indicates their 3′ stem loop. The underlines represent the potential target sites of Novel_NA. Accession numbers of Novel_NA transposons in their genomes are also shown.
Third, Novel_NA showed similar secondary structure to those of HzMINE-2 and DINE-1. They contain a 14 bp 5′-SIR (ACGTGAATACGTCT) that pairs with a reverse complementary 3′-SIR (AGACGTATTCACGT) and a 8 bp stem-loop structure downstream of the 3′-SIR (fig. 3). In addition, 5′ and 3′ boundaries of Novel_NA in R. prolixus are composed of TG and GT, and they are similar to those of HzMINE-2 and DINE-1 (TA and GT). This suggested that Novel_NA together with HzMINE-2 and DINE-1 might represent a novel class of MITEs in insects as a result of their unusual features distinguishing them from traditional MITEs. Moreover, their common features suggested that they could evolve from a common ancestor.
Autonomous Transposon Progenitors of MITEs
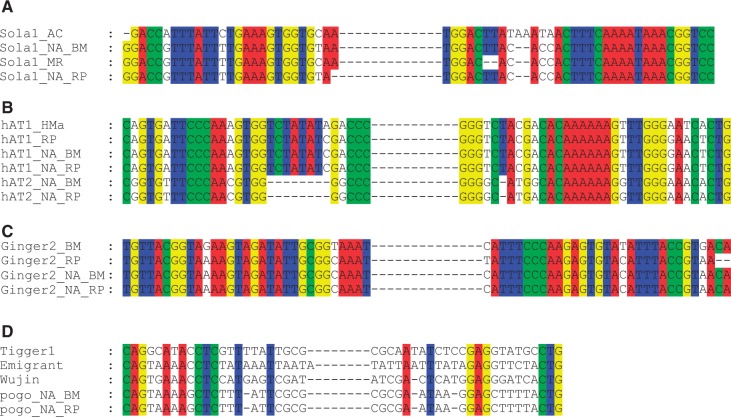
Genetic and structural features of MITEs imply that they originated from full-length cognate autonomous DNA transposons (Feschotte and Pritham 2007). To our surprise, potential autonomous elements responsible for the spread of three families (Sola1_NA, hAT1_NA, and Ginger2_NA) were found (table 1). For Sola1_NA, the overall size of these significant hits was 2,313, 1,489, and 2,778 bp in R. prolixus, A. cephalotes, and Meg. rotundata, respectively (table 1 and supplementary fig. S4A, Supplementary Material online). Such a size was not expected for a so-called miniature transposon. Interestingly, one intact ORF (630 aa), which did not have premature stop codon was clearly identified for transposon in Meg. rotundata. It suggested that it might be a source of functional protein and was still active in Meg. rotundata. However, it was difficult for us to determine the exact full-length transposase encoding by this transposon in A. cephalotes and R. prolixus due to stop codons or frameshifts (supplementary fig. S4A, Supplementary Material online). These elements in different hosts revealed high sequence identity (>85.8%), suggesting that they should belong to the same family (supplementary fig. S4A and table S3, Supplementary Material online). For hAT1_NA, its autonomous elements were 2,889, 2,916, and 1,573 bp in length in R. prolixus, Hy. magnipapillata and Me. moldrzyki, respectively (supplementary fig. S4B, Supplementary Material online). Their potential coding region could be determined. An alignment of this element from different hosts revealed a very high level of identity (>88.2%) over the whole length of the consensus sequences, suggesting these elements descended from the same active ancestral element (supplementary fig. S4B and table S3, Supplementary Material online). For Ginger2_NA, its autonomous element in B. mori and R. prolixus was 2,442 and 3,767 bp, respectively. However, we failed to identify a significant coding region for Ginger2_RP in the R. prolixus genome (supplementary fig. S4C, Supplementary Material online). By contrast, a significant coding region of Ginger2_BM was found when we used its nucleotide sequence to perform translated search against the Repbase (http://www.girinst.org/censor/index.php, last accessed October 29, 2013). Meanwhile, an alignment of these two elements showed that their similarity (93.2% pairwise nucleotide identity) is restricted to the terminal regions (supplementary fig. S4C and table S3, Supplementary Material online). However, their internal regions do not show any similarity. Therefore, we inferred that the internal region of Ginger2_RP was foreign sequences. Phylogenetic analyses of these elements with other known transposons indicated that they were grouped into the corresponding superfamily (supplementary fig. S5, Supplementary Material online). Multiple alignments of nonautonomous elements and their master elements suggested that they originated by internal deletions from master elements (fig. 4A– C).
Fig. 4.—
Homologies in 5′ and 3′ terminal regions between Sola1_NA (A), hAT1_NA (B), hAT2_NA (B), Ginger2_NA (C), pogo_NA (D), and their potential progenitor transposons. Alignment of the terminal sequences was done using MUSCLE. TIRs of the Tigger1, Wujin, and Emigrant families are from Feschotte and Mouchès (2000). “–” indicates a gap, and conserved sites are in white type on a green/blue/red/yellow background.
Although no direct autonomous element of pogo_NA has been found so far, TIRs of this family shared some similarity to Emigrant (fig. 4D), a MITE family identified in Arabidopsis thaliana (Casacuberta et al. 1998), and to a MITE family Wujin from yellow fever mosquito Aedes aegypti (Tu 1997). This indicated that pogo_NA and these MITEs could be grouped into the same MITE family. Especially, its TIRs also had some similarity to an autonomous pogo-like element, the human Tigger1 element (Robertson 1996). Therefore, our results supported the hypothesis that pogo-like elements had a strong tendency to generate MITEs (Feschotte and Mouches 2000). We also failed to find the autonomous elements for hAT2_NA family. However, TIR similarity (11/14) between hAT1_NA and hAT2_NA suggested that they may also belong to the same family (fig. 4B). Moreover, they tended to integrate in similar target sequences (supplementary fig. S3, Supplementary Material online). All these results indicated that hAT2_NA might use the transposition machinery of autonomous elements similar to that for hAT1_NA.
Evidence for HT
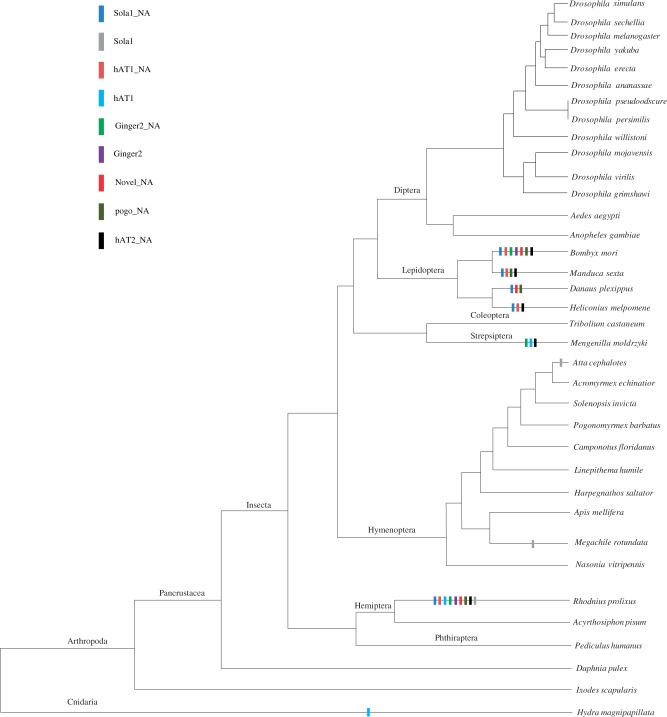
The screening of complete or near-complete genome sequences available in NCBI revealed that three highly similar autonomous elements (Sola1, hAT1, and Ginger2) were present in six invertebrate genomes, including the silkworm (B. mori), the triatomine bug (R. prolixus), the leaf-cutter ant (A. cephalotes), the alfalfa leafcutting bee (Meg. rotundata), the twisted-wing parasite (Me. moldrzyki), and the freshwater hydrozoan (Hy. magnipapillata) (table 1 and fig. 5).
Fig. 5.—
The taxonomic distribution of Sola1_NA, hAT1_NA, Ginger2_NA, Novel_NA, pogo_NA, hAT2_NA, and their autonomous elements among invertebrate genomes for which complete or near-complete genome sequences are available. Presence of these transposon families in each lineage are denoted by rectangles.
The autonomous Sola1 family, Sola1, existed in the triatomine bug, the leaf-cutter ant and the alfalfa leaf-cutting bee. In each of the species, a representative sequence was recovered because we were not able to construct their complete consensus sequences as a result of low copy number or fragmentation in their genomes. The nucleotide sequence identities between these transposons were unexpectedly high (>85.8 for all interspecific pairwise comparisons). This reflects that these transposons might have been horizontally transferred into these hosts because these three host species are distantly related (fig. 5). For instance, there was 93.9% sequence identity between the triatomine bug Sola1 (Hemiptera, Reduviidae) and the alfalfa leafcutting bee Sola1 (Hymenoptera, Megachilidae), and their hosts diverged about 370 Ma (Hedges et al. 2006). Surprisingly, Sola1 elements from closely related alfalfa leaf-cutting bee and leaf-cutter ant (Hymenoptera, Formicidae) showed higher level of nucleotide sequence divergence than Sola1 transposons from the triatomine bug and the alfalfa leaf-cutting bee (supplementary table S3 and fig. S4A, Supplementary Material online). High sequence identity of Sola1 transposons from divergent species and lower sequence identity of these elements from closely related species suggested horizontal transmission for Sola1 family.
The autonomous hAT family, hAT1, was present in the triatomine bug, the twisted-wing parasite, and the freshwater hydrozoan Hy. magnipapillata (table 1 and fig. 5). The hAT1 family in the Hy. magnipapillata genome was the only family whose consensus sequence could be reconstructed. However, this family in other two species appeared in partial copies or low copy number, and their representatives were chosen as respective consensuses. The nucleotide identity between the sequences from the triatomine bug and the twisted-wing parasite was extremely high (97%), suggesting TH (supplementary table S3 and fig. S4B, Supplementary Material online). Indeed, their hosts diverged from the common ancestor of Hemiptera and Strepsiptera about 370 Ma (Hedges et al. 2006). Similar patterns of sequence identity of hAT1 were observed between the fresh water hydrozoan and the twisted-wing parasite/the triatomine bug (88–92%). These species belong to different phyla (Cnidaria and Arthropoda) that diverged from their common ancestor >800 Ma (Hedges et al. 2006). Therefore, it was most likely that hAT1 elements might have been horizontally transferred into different host species.
The autonomous Ginger family, Ginger2, was only identified in the silkworm and the triatomine bug (table 1 and fig. 5). The member of this family appeared in partial or truncated copies in their respective hosts, which made it difficult to reconstruct reliable consensus sequences. The alignment of the Ginger2 sequences from the silkworm and the triatomine bug showed an extremely high level of nucleotide identity (93.2%) across a region of 1,217 bp (supplementary table S3 and fig. S4C, Supplementary Material online) despite the silkworm and the triatomine bug split at 370 Ma. This suggested that Ginger family might have been horizontally transferred into two host species.
There are generally three criteria used to detect HT events: 1) high sequence similarity of TEs from divergent taxa, 2) topological differences between TE and host phylogenies, and 3) discontinuous distribution of a transposon among closely related taxa (Schaack et al. 2010). Although the strikingly high level of sequence identity of the above three autonomous elements among such highly divergent invertebrates provided the strongest evidence to support the hypothesis that they had transferred horizontally into these lineages, such deduction only based on sequence identity may be problematic because we did not consider variable evolutionary rates that had been shown for some transposons (Malik et al. 1999). Therefore, making HT conclusion of these transposons should be cautious.
To obtain more evidence for HTs of these transposons, we investigated the phylogenetic distributions of the Sola1, hAT1, and Ginger2 families. Our results showed that they were patchy distributions in species (fig. 5). If these transposons had been vertically transferred from a common ancestor, it was difficult to explain the fact that there were as many as 11, 7, and 7 independent losses of the Sola1, hAT1, and Ginger2 families, respectively. For example, Sola1 family was identified only in A. cephalotes genome, it was absent in other six closely related sequenced ant genomes, including another leaf-cutter ant Acromyrmex echinatior, which shared a common ancestor with A. cephalotes about 8–12 Ma (Schultz and Brady 2008). Similar phenomenon was also observed for hAT1 family. It was present in the fresh water hydrozoan (Cnidaria) and insects (Arthropoda) but absent in Nematoda and Deuterostomes (data not shown). Additionally, it showed the discontinuous distribution across different insect species (fig. 5). Finally, the distribution of Ginger2 family was also patchy, because it was found only in two insect species (the silkworm and the triatomine bug).
A high level of sequence identity of these transposons among distantly related species may also result from intense purifying selection. To directly compare with the coding genes from host species, three most conserved genes (Hsc70-4, tub3, and EF1a) were examined. We retrieved full-length CDS sequences of these genes (except EF1a from Me. moldrzyki) from their hosts (supplementary table S1, Supplementary Material online). Ka/Ks, a ratio of the nonsynonymous and synonymous changes between two sequences, was calculated by DnaSP v5 (Librado and Rozas 2009). For Sola1 and Ginger2 families, we were not able to determine their exact coding sequences in at least two species, and they were not included in this analysis (supplementary fig. S4, Supplementary Material online). The frequency of nonsynonymous changes for the hAT1 families varied from 0.0295 to 0.1588, and the frequency of synonymous changes ranged from 0.0290 to 0.0574 (supplementary table S3, Supplementary Material online). Importantly, Ka/Ks for this family ranged from 0.2509 to 0.9831 for all interspecific comparisons, which is consistent with very low to no significant purifying selection. However, DnaSP v5 did not give an output for Ks for almost all pairwise comparisons of Hsc70-4 and tub3 (supplementary table S3, Supplementary Material online) possibly because synonymous changes were saturated. Meanwhile, EF1a comparisons were in accord with high selection pressure because the ratio Ka/Ks varied from 0.0067 to 0.0240. Therefore, these results suggested that high level of sequence identity of the hAT1 family did not result from intense purifying selection.
Taking the above evidence together, HT was the only reasonable explanation for such a high level of sequence identity among these transposons in highly divergent host species. This was consistent with previous studies in which the authors provided sufficient evidence supporting HTs of the transposons in the silkworm and the triatimine bug (Thomas et al. 2010; Zhang et al. 2013).
Mechanism for Transfer
Strepsiptera is an order of insect parasitoids that infect at least 35 families of insects belonging to seven orders, including Thysanura, Blattodea, Mantodea, Orthoptera, Heteroptera, Hymenoptera, and Diptera (Kathirithamby 1989). Meanwhile, some species of Strepsiptera are not completely host specific, and some of them might parasitize several genera and species (Kathirithamby 1989). During the process of parasitism, they obtain nutrients from a single host (Kathirithamby 2009). This order has two major groups: Stylopidia and Mengenillidia. Free living of female adult is one of the most important characters that distinguish Mengenillidae from other families (Kathirithamby 1989). Mengenilla moldrzyki is a newly discovered species and belongs to Mengenillidae group in the strepsipteran order (Pohl et al. 2012). However, its direct host was unknown.
Three families (hAT1, Ginger2_NA, and hAT2_NA) were identified in Me. moldrzyki (the twisted-wing parasite). These families shared an extremely high level of sequence identity with homologs in the silkworm, the triatomine bug, and the freshwater hydrozoan, which diverged from a common ancestor at least 300 Ma (table 1 and fig. 5). Several previous studies had demonstrated that the intimacy of parasite–host interaction might facilitate HTs of transposons (Yoshiyama et al. 2001; Laha et al. 2007; Gilbert et al. 2010). Although we could not directly connect the twisted-wing parasite with insects studied, these transposons identified in this insect parasitoid might also suggest that their transfers were likely facilitated by parasitism. Afterall, Me. moldrzyki is the only species of Strepsiptera whose complete genome has been sequenced (Niehuis et al. 2012). Meanwhile, there were only 33 insect species whose complete or near-complete genomes were available (fig. 5). This means that our present view of the phylogenetic distribution of these transposons may be biased by the sampling of insect genome projects. Therefore, to have more insights into the origin and evolutionary history of these transposons, it is better to focus on a systematic analysis of the phylogenetic distribution of these transposons in future. To the best of our knowledge, this was the first time to identify transposons in the order Strepsiptera and to report the phenomenon of HT of transposons in these entomophagous parasitoids. Moreover, the published complete genome of Me. moldrzyki will also provide a good opportunity for studying the effect of parasite–host interaction on the evolution of transposons and speciation.
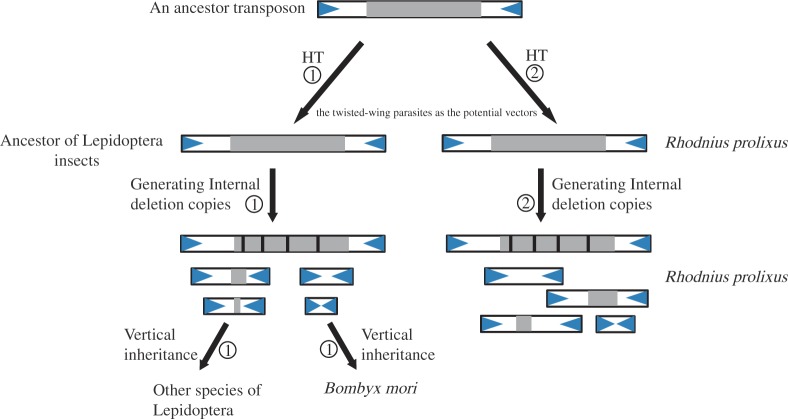
A Model for the Origin and Evolutionary History of MITEs in the Silkworm and the Triatomine Bug
Using MITEs identified in the silkworm as queries, five families were also found in recent published genomes of three other Lepidpoteran insects (Danaus plexippus, H. melpomene, and M. sexta) (Zhan et al. 2011; Heliconius Genome Consortium, 2012) (table 1 and fig. 5). This suggested that these MITEs in the silkworm were vertically inherited. Thus, it is most likely that the progenitor elements of these MITEs might have been horizontally introduced into the ancestor of lepidopteran insects and MITEs were generated within the ancestor genome. Then the silkworm obtained MITEs from its ancestor through vertical inheritance (fig. 6). Meanwhile, the progenitor elements of the MITEs have been horizontally invaded into the triatomine bug genome and then generated the MITEs within its genome (fig. 6). Furthermore, we speculate that the twisted-wing parasites may be the candidate vectors for these HTs (fig. 6). In summary, our results provide some new insights into the origin and evolutionary history of the MITEs in B. mori and R. prolixus.
Fig. 6.—
A model for the origin and evolution of MITEs in the silkworm and triatomine bug. Gray regions indicate the transposase of autonomous elements, and blue triangles are TIRs. The potential processes involved in the origin and evolution of MITEs in the silkworm and in the triatomine bug. 1. An ancestor transposon was first introduced into the ancestor of Lepidopteran insects by HT. Then, many internal deletion copies (including MITEs) of this autonomous element are generated. Meanwhile, this autonomous copy might be stochastically lost or fossilized in the ancestor genome. Finally, the silkworm MITEs are obtained from its ancestor through vertical inheritance. 2. An ancestor transposon was horizontally transferred into the triatomine bug genome. Then, kinds of newly internal deletion versions of this autonomous element were generated in this species. It is speculated that the twisted-wing parasites may be the candidate vectors for these transfers.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Dr Ricardo Nascimento Araujo (Laboratório de Fisiologia de Insetos Hematófagos, Brasil) and Dr Adriana Gámez (Instituto Venezolano de Investigaciones Científicas, Venezuela) for providing the materials of R. prolixus. The authors also thank Dr Cedric Feschotte for his advice in writing paper. This work was supported by the Hi-Tech R&D Program (863) of China (2013AA102507).
Literature Cited
- Bao W, Jurka MG, Kapitonov VV, Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol Biol Evol. 2009;26:983–993. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Kapitonov VV, Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau TE, Wessler SR. Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Casacuberta JM, Puigdomènech P, Monfort A. Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterisation of the Emigrant family of elements. Plant J. 1998;16:79–85. doi: 10.1046/j.1365-313x.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- Coates BS, Kroemer JA, Sumerford DV, Hellmich RL. A novel class of miniature inverted repeat transposable elements (MITEs) that contain hitchhiking (GTCY) n microsatellites. Insect Mol Biol. 2011;20:15–27. doi: 10.1111/j.1365-2583.2010.01046.x. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990;124:339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Mouchès C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel FE, Chaley MB, Korotkov EV, Skryabin KG. Evolution of tRNA-like sequences and genome variability. Gene. 2004;335:57–71. doi: 10.1016/j.gene.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Han MJ, et al. Burst expansion, distribution and diversification of MITEs in the silkworm genome. BMC Genomics. 2010;11:520. doi: 10.1186/1471-2164-11-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathirithamby J. Review of the order Strepsiptera. Syst Ent. 1989;14:41–92. [Google Scholar]
- Kathirithamby J. Host-parasitoid associations in Strepsiptera. Annu Rev Ent. 2009;54:227–249. doi: 10.1146/annurev.ento.54.110807.090525. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Kuang H, et al. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res. 2009;19:42–56. doi: 10.1101/gr.078196.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha T, et al. The bandit, a new DNA transposon from a hookworm—possible horizontal genetic transfer between host and parasite. PLoS Negl Trop Dis. 2007;1:e35. doi: 10.1371/journal.pntd.0000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Hartl DL. Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J Mol Evol. 1991;33:514–524. doi: 10.1007/BF02102804. [DOI] [PubMed] [Google Scholar]
- Niehuis O, et al. Genomic and morphological evidence converge to resolve the enigma of Strepsiptera. Curr Biol. 2012;22:1309–1313. doi: 10.1016/j.cub.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Oki N, et al. A genome-wide view of miniature inverted-repeat transposable elements (MITEs) in rice, Oryza sativa ssp. japonica. Genes Genet Syst. 2008;83:321–329. doi: 10.1266/ggs.83.321. [DOI] [PubMed] [Google Scholar]
- Pohl H, et al. A new species of Mengenilla (Insecta, Strepsiptera) from Tunisia. ZooKeys. 2012;198:79–101. doi: 10.3897/zookeys.198.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robertson HM. Members of the pogo superfamily of DNA-mediated transposons in the human genome. Mol Gen Genet. 1996;252:761–766. doi: 10.1007/BF02173985. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci U S A. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu Y, et al. Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3. 2013;3:1481–1492. doi: 10.1534/g3.113.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomas J, Schaack S, Pritham EJ. Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol Evol. 2010;2:656–664. doi: 10.1093/gbe/evq050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z. Three novel families of miniature inverted-repeat transposable elements are associated with genes of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1997;94:7475–7480. doi: 10.1073/pnas.94.14.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xu HE, et al. BmTEdb: a collective database of transposable elements in the silkworm genome. Database. 2013 doi: 10.1093/database/bat055. 2013:bat055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, et al. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–1568. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HP, Barbash DA. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 2008;9:R39. doi: 10.1186/gb-2008-9-2-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama M, et al. Possible horizontal transfer of a transposable element from host to parasitoid. Mol Biol Evol. 2001;18:1952–1958. doi: 10.1093/oxfordjournals.molbev.a003735. [DOI] [PubMed] [Google Scholar]
- Zhang HH, et al. A novel hAT element in Bombyx mori and Rhodnius prolixus: its relationship with miniature inverted repeat transposable elements (MITEs) and horizontal transfer. Insect Mol Biol. 2013;22:584–596. doi: 10.1111/imb.12047. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147:1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.