Abstract
Plants share a common history of successive whole-genome duplication (WGD) events retaining genomic patterns of duplicate gene copies (ohnologs) organized in conserved syntenic blocks. Duplication was often proposed to affect the origin of novel traits during evolution. However, genetic evidence linking WGD to pathway diversification is scarce. We show that WGD and tandem duplication (TD) accelerated genetic versatility of plant secondary metabolism, exemplified with the glucosinolate (GS) pathway in the mustard family. GS biosynthesis is a well-studied trait, employing at least 52 biosynthetic and regulatory genes in the model plant Arabidopsis. In a phylogenomics approach, we identified 67 GS loci in Aethionema arabicum of the tribe Aethionemae, sister group to all mustard family members. All but one of the Arabidopsis GS gene families evolved orthologs in Aethionema and all but one of the orthologous sequence pairs exhibit synteny. The 45% fraction of duplicates among all protein-coding genes in Arabidopsis was increased to 95% and 97% for Arabidopsis and Aethionema GS pathway inventory, respectively. Compared with the 22% average for all protein-coding genes in Arabidopsis, 52% and 56% of Aethionema and Arabidopsis GS loci align to ohnolog copies dating back to the last common WGD event. Although 15% of all Arabidopsis genes are organized in tandem arrays, 45% and 48% of GS loci in Arabidopsis and Aethionema descend from TD, respectively. We describe a sequential combination of TD and WGD events driving gene family extension, thereby expanding the evolutionary playground for functional diversification and thus potential novelty and success.
Keywords: comparative genomics, systems biology, whole-genome duplication, functional diversification, Brassicaceae
Introduction
Gene duplication has played an important evolutionary role in angiosperm adaptation and success, for example, by contributing to regulatory and enzymatic pathways involved in generating more than 200,000 diverse biochemical plant secondary metabolites found in the Angiosperm lineage (Hartmann 2007). Functional diversification refers to processes of gene duplication followed by sub- or neofunctionalization of the enzymes encoded by duplicate copies (Ohno 1970; Roth et al. 2007; Wang et al. 2011c), mediating specificities to extended classes of substrates or catalysis of novel reactions (Stehle et al. 2008). Fast expansion of gene copy number occurs in various ways. In this study, we focus on whole-genome duplication (WGD), tandem duplication (TD), and gene transposition duplication (GTD). For example, approximately 45% of the Arabidopsis nuclear protein-coding genes have been affected by such processes (Bowers et al. 2003; Rizzon et al. 2006; Huang et al. 2012). In this study, we investigated the impact of gene duplication to the diversification of plant secondary metabolites exemplified with glucosinolate (GS) biosynthesis. GS biosynthesis is a well-studied key trait shared by all Brassicales including the mustard family (Brassicaceae) crown group (Schranz et al. 2011) and its sister lineage Aethionemae. Comparative genomics analysis unraveled a history of successive paleopolyploidy events commonly shared by almost all Angiosperms (Bowers et al. 2003). The Arabidopsis lineage underwent at least five polyploidy events in the history of life, two preceding and three following Angiosperms radiation (Bowers et al. 2003; Jiao et al. 2011). The most recent WGD is commonly referred to as At-α and occurred approximately 30–60 Ma in the ancestor of all Brassicaceae, including the sister group Aethionemae (Edger PP, Pires Jc, submitted for publication). As a result, pairwise syntenic regions are scattered throughout the genome (genomic blocks), defined as copies of consecutive ohnologs derived from At-α (Bowers et al. 2003). It is known that polyploidy is succeeded by a genome-wide process of biased fractionation, preferentially targeting one subgenome to retain clusters of dose-sensitive genes organized in functional modules (Thomas et al. 2006). Furthermore, several studies have established a potential link of polyploidy to natural variation due to differential expression of ohnolog copies (Wang et al. 2011c), seed and flower origin and diversification (De Bodt et al. 2005; Irish and Litt 2005; Jiao et al. 2011), morphological complexity (Freeling and Thomas 2006), and survival of plant lineages at the Cretaceous-Tertiary extinction event (Fawcett et al. 2009). In this study, we provide solid evidence for the link of WGD to pathway expansion of a distinct key trait relevant for herbivore defense and hence highly connected to fitness. Interestingly, polyploidy also affects other kinds of duplication, creating networks of factors with mutual influence. Recent studies have shown an interaction between polyploidy and the fractionation rate of tandem duplicate copies in both Arabidopsis and Brassica rapa (having undergone an additional genome triplication). Hence, we analyzed short-sequence duplications to utilize the evolutionary significance of different duplication classes.
TD of short sequences can be caused by unequal crossing-over or template slippage during DNA repair, producing tandem arrays (TARs) of homologous genes in close genomic vicinity (Kane et al. 2010). Depending on the number of allowed gene spacers, TAR genes include about 10–15% of the Arabidopsis thaliana genome (0 and 10 spacers, respectively) (Rizzon et al. 2006). Comparison of TARs in Arabidopsis and rice revealed enrichment of genes encoding membrane proteins and function in biotic and abiotic stress (Rizzon et al. 2006). Notably, the impact of TD to trait evolution has been elucidated in multiple taxa, including disease resistance in Solanaceae (Parniske et al. 1999) and Brassicaceae (Leister 2004). Likewise, TD played a role in the evolution of signal transduction, for example, the expansion and functional diversification of the F-box type transcriptional activator gene family in Fabaceae (Bellieny-Rabelo et al. 2013). Moreover, TD is an important factor for increasing versatility of defense response in Brassicaceae. In GS biosynthesis, subfunctionalization of TAR genes is evident for 2-oxoglutarate-dependent dioxygenases (AOP) (Kliebenstein et al. 2001), flavin-monooxygenases (FMOGSOX) (Li et al. 2008) and methylthioalkylmalate synthases (MAM) (Kroymann et al. 2003; Heidel et al. 2006; Textor et al. 2007). In this study, we integrate previous findings to dissect the influence of polyploidy with TD and GTD in the last 30–60 Ma of GS pathway expansion since Aethionema and Arabidopsis lineage divergence.
Duplicate gene copies can move to a new genomic location. The observed frequency of gene movements explains the observed erosion of synteny between plant genomes during evolution (Wicker et al. 2010), defining the limits of synteny-based approaches for ortholog detection. Gene movements are often caused by transposition. GTD events occur when a single nontransposon gene relocates to a new position, and segregants contain duplicates (Freeling 2009). Although transposable elements (TEs) account for approximately 10% of the Arabidopsis genome (Huang et al. 2012) and show nonrandom association to syntenic blocks (Hughes et al. 2003), 14% of all protein-coding genes in Arabidopsis transposed at least once during Rosid evolution (Freeling et al. 2008; Woodhouse et al. 2011). Importantly, a novel genomic context of the transposed copy potentially influences rates of gene expression (Wang et al. 2013) and might thereby contribute to the phenotypic consequences of the duplication event (Kliebenstein 2008). Accordingly, TE activity was shown to foster variation of NBS-resistance proteins in grape (Malacarne et al. 2012) as well as natural growth variation and expansion of ERF family transcriptional regulators in Arabidopsis (Nakano et al. 2006; Vlad et al. 2010). In contrast, evolutionary dynamics of GTD events affecting genetic versatility of plant secondary metabolism has not yet been investigated.
GS comprise a class of secondary plant metabolites derived from amino acids and sugars, part of a two-component chemical defense against herbivory in Brassicales (Rodman 1998; Windsor et al. 2005; Beekwilder et al. 2008). Myrosinase enzymes are the other component of the defense system and confer GS hydrolysis activity. They are released from the vacuole upon tissue damage, producing a plethora of GS degradation products such as nitriles, isothiocyanates, thiocyanates, and ephithioalkanes with various bioactivities (Rask et al. 2000; Bones and Rossiter 2006). GS are of particular interest for human health because they can inhibit carcinogen activation (Hecht 2000; Nakajima et al. 2001) and carcinogenesis by triggering cell cycle arrest and stimulating apoptosis (Wittstock et al. 2003; Hayes et al. 2008). The observed variation in GS biochemistry across Brassicales is due to the differences in biochemistry among their amino acid precursors (Fahey et al. 2001; Windsor et al. 2005) and allows GS grouping to four distinct classes. Oxidative deamination of Phe and Tyr initiates biosynthesis of indolic GS (I); Trp is the substrate for indolic GS production (II); Ala, Val, Leu, and Ile are precursors for biosynthesis of aliphatic GS (III) (Mithen et al. 2010). Although aromatic and aliphatic GS have been detected in other eudicot families including Phytolaccaceae, Euphorbiaceae, and Pittosporaceae (Rodman et al. 1996; Fahey et al. 2001), indolic GS are Brassicales specific. Met-derived GS form a fourth class of GS (IV), referring to a subset of aliphatic GS specific to the Brassicales crown group, including the sister group Aethionemae. The utilization of Trp- and Met-derived amino acids for GS production may be tied to pathway expansion caused by ancient WGD events (Schranz et al. 2011).
The genus Aethionema of the tribe Aethionemae is an ideal group for comparative genomics of polyploidy and GS pathway evolution. First, it shares the composite GS chemotype observed in the larger and more diverse Brassicaceae crown group (Schranz et al. 2012). Second, phylogenetic analysis highly support the tribe Aethionemae as the earliest diverged clade and extant sister to the crown group Brassicaceae (Couvreur et al. 2010) with an estimated split of the two lineages approximately 30–60 Ma. However, a high degree of interspecies synteny (see Results) is maintained. Third, the most recent WGD event identified in the lineage of Arabidopsis (referred to as At-α) predated the divergence of Arabidopsis and Aethionema. Furthermore, it was not succeeded by an additional species-specific genome polyploidation, preventing additional fractionation of synteny (Bowers et al. 2003; Haudry et al. 2013) (Edger PP, Pires Jc, submitted for publication). In contrast, B. rapa underwent an additional genome triplication event (Wang et al. 2011b), complicating efforts to analyze the potential impact of At-α on the evolution of the GS pathway inventory.
Materials and Methods
Aethionema arabicum Genome Assembly and Set of Annotated Genes
Sequence assembly and annotation of the Aethionema arabicum genome was obtained from Haudry et al. (2013).
RNA Isolation and Sequencing
Aethionema arabicum RNA was isolated from fresh apical meristematic tissue or very young leaves using an RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Samples were kept on liquid nitrogen before RNA isolation. The optional step of heating the lysis solution to 65 °C was used to maximize RNA yield. RNA was eluted into a final volume of 100 µl RNase-free water. Total mass of RNA and quality was estimated using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). Samples were deemed acceptable if RIN (RNA integrity number) scores were greater than 8.0. A minimum of 20 µg of total RNA was required for library building and sequencing. RNA-seq (Wang et al. 2009) paired-end libraries with average fragment lengths of 250 bp were constructed, and each library was sequenced on a single lane of an Illumina GAIIX sequencer flow cell (Illumina, San Diego, CA) to generate a minimum of 3 gigabases of 75 bp, paired-end sequences.
Database of A. thaliana Genes Retaining an Ohnologous Copy Dating Back to the At-α WGD Event
First, we generated a spreadsheet with information on all 33,323 A. thaliana nuclear genes annotated in the TAIR database v10, including 1) Arabidopsis gene identifiers (AGIs), 2) locus type, 3) locus name, and 4) short description of encoded function. Second, we integrated optional affiliation to 5) syntenic block and 6) ohnolog copy dating back to At-α WGD event as described previously (Freeling and Thomas 2006). The corresponding authors did not account for every gene in their analysis and inferred the genomic location of ohnolog blocks dating back to At-α using the TIGR Arabidopsis genome annotation v5 from 2005. Third, we added an additional column (i), to indicate coverage of the gene in Feeling’s study (yes/not considered/not present in TIGR5).
Database for GS Biosynthetic Gene Identification in A. thaliana and Aet. arabicum
Files containing the coding sequences representing the complete set of GS biosynthetic and regulatory (AtGS) genes in A. thaliana (Sonderby et al. 2010) were acquired from the TAIR database v10 (www.arabidopsis.org, last accessed November 9, 2013). We highlighted the AGIs in the spreadsheet covering all nuclear genes in Arabidopsis (see Materials and Methods).
Interpolation of Novel Putative GS Biosynthetic Genes and Retained At-α Ohnolog Pairs in A. thaliana
We utilized similarity among ohnologous gene copies. We employed the spreadsheet containing information on genomic location of AtGS genes and α-blocks with optional retained duplicates therein. We visually screened for all ohnolog copies of AtGS genes not sharing annotation as AtGS genes themselves. Differential expression of ohnolog pairs was tested using the botany array resource (http://bar.utoronto.ca/welcome.htm, last accessed November 8, 2013). We included all ohnolog copies for our analysis to create an extended AtGS gene set. For the spreadsheet, see supplementary table S1, Supplementary Material online. Previous approaches of ohnologous gene pair identification did not consider every protein-coding gene (Bowers et al. 2003; Thomas et al. 2006). To minimize resulting errors for analysis of AtGS loci, we performed a BlastP screen without a cut-off e value, querying all AtGS genes with nonretained At-α ohnologs against all other Arabidopsis genes with nonretained At-α ohnologs. Highest-scoring sequence pairs sharing genomic location within converse copies of the same α-block were tested for synteny and positives defined as additional pairs of retained At-α ohnolog copies (marked as “our addition/OA” in table 1 and supplementary table S1, Supplementary Material online).
Table 1.
Retained At-α Ohnolog Duplicate Gene Pairs in Arabidopsis and Aethionema GS Pathway Inventory
| Protein Namea | AGI | α-Block | Evident SSDb | AabIDc | Syntelog | % Identity | Col-0 → Aabd |
|---|---|---|---|---|---|---|---|
| Core-structure formation | |||||||
| UGT74C1 | AT2G31790 | A02N051 | TD | Aab37175 | Yes | 79.44 | 6 → 10 |
| [UGT like] | AT1G05670 | A02N051 | TD | Aab31930 | Yes | 81.11 | 6 → 10 |
| FMO-GSOX-2 | AT1G62540 | A03N117 | TD | Aab10869 | Yes | 76.6 | 11 → 8 |
| [FMO like] | AT1G12130 | A03N117 | TD | Aab13543 | Yes | 65.09 | 11 → 8 |
| CYP79F2 | AT1G16400 | A05N062 | TD | — | — | — | 8 → 9 |
| CYP79C1 | AT1G79370 | A05N062 | GTD | Aab34143 | Yes | 76.8 | 8 → 9 |
| SOT16 | AT1G74100 | A05N186 | TD | Aab14278 | Yes | 91.07 | 3 → 3 |
| SOT17 | AT1G18590 | A05N186 | Aab19675 | Yes | 86.05 | 3 → 3 | |
| SUR1 | AT2G20610 | A10N194 | Aab30136 | Yes | 89.15 | 3 → 2 | |
| [SUR like] | AT4G28420 | A10N194 | TD | Aab31155 | Yes | 57.93 | 3 → 2 |
| CYP79B2 | AT4G39950 | A10N257 | GTD | Aab17805 | Yes | 81.38 | 8 → 9 |
| CYP79B3 | AT2G22330 | A10N257 | GTD | Aab19477 | Yes | 81.4 | 8 → 9 |
| GGP1 | AT4G30530 | A10N314 | TD | Aab24374 | Yes | 87.6 | 5 → 5 |
| [GGP like] | AT2G23960 | A10N314 | TD | Aab11021 | Yes | 61.38 | 5 → 5 |
| GSTF11 | AT3G03190 | A12N102 | Aab14996 | Yes | 77.1 | 4 → 4 | |
| [GSTF12] | AT5G17220 | A12N102 | Aab14791 | Yes | 77.84 | 4 → 4 | |
| Cosubstrate pathways | |||||||
| [AAO3] | AT2G27150 | A02NOA1 | GTD | Aab27016 | 77.67 | 2 → 2 | |
| AAO4 | AT1G04580 | A02NOA1 | Aab24896 | Yes | 79.58 | 2 → 2 | |
| APK1 | AT2G14750 | A10NOA2 | Aab32150 | Yes | 83.75 | 2 → 2 | |
| APK2 | AT4G39940 | A10NOA2 | GTD | Aab17804 | Yes | 86.05 | 2 → 2 |
| Side-chain elongation | |||||||
| BCAT4 | AT3G19710 | A08N074 | Aab21007 | Yes | 75 | 6 → 6 | |
| [BCAT7] | AT1G50090 | A08N074 | TD | Aab22548 | Yes | 76.9 | 6 → 6 |
| IPMI1 | AT3G58990 | A11N226 | Aab13092 | Yes | 83 | 3 → 3 | |
| [IPMI like] | AT2G43090 | A11N226 | TD | Aab19619 | Yes | 85.99 | 3 → 3 |
| BCAT3 | AT3G49680 | A19N002 | Aab33782 | Yes | 76.02 | 6 → 6 | |
| [BCAT5] | AT5G65780 | A19N002 | TD | Aab23605 | Yes | 75.3 | 6 → 6 |
| BAT5 | AT4G12030 | A20N095 | Aab32285 | Yes | 76.21 | 2 → 2 | |
| [BAT like] | AT4G22840 | A20N095 | Aab23321 | Yes | 91.82 | 2 → 2 | |
| TF regulation | |||||||
| OBP2 | AT1G07640 | A02N142 | Aab18330 | Yes | 80.28 | 2 → 2 | |
| [OBP like] | AT2G28810 | A02N142 | Aab24559 | Yes | 70.03 | 2 → 2 | |
| MYB122 | AT1G74080 | A05N185 | Aab14276 | Yes | 57.56 | 6 → 4 | |
| MYB51 | AT1G18570 | A05N185 | Aab19683 | Yes | 59.89 | 6 → 4 | |
| IQD1 | AT3G09710 | A14N046 | Aab18852 | Yes | 65.59 | 2 → 2 | |
| [IQD2] | AT5G03040 | A14N046 | Aab18368 | Yes | 77.39 | 2 → 2 | |
| MYB28 | AT5G61420 | A26N034 | Aab12163 | Yes | 67.39 | 6 → 4 | |
| MYB29 | AT5G07690 | A26N034 | TD | Aab33585 | Yes | 65.13 | 6 → 4 |
| 36% TD (13/36) | 29% TD (10/35) | Ø 76.58 | |||||
| 14% GTD (5/36) | 14% GTD (5/35) | ||||||
aSquared brackets indicate ohnolog copies of GS biosynthetic genes without GO!-annotation to GS biosynthetic process.
bTD refers to members of TARs and GTD refers to a history of transposition in Arabidopsis.
cPredicted Aethionema CDS.
dChange of gene family locus count in Arabidospsis → Aethionema order.
Database of A. thaliana Tandem Arrayed Genes
A database of A. thaliana coding sequences organized in TARs was generated for the TAIR annotation v10 as described previously (Rizzon et al. 2006), using a low-stringency approach with a number of N = 10 allowed gene pacers. Information was updated to TAIR10 and included to the spreadsheet covering all Arabidopsis nuclear genes (supplementary table S1, Supplementary Material online).
Database of A. thaliana GS Genes Affected by GTD
A database of the epoch-independent positional history of all Arabidopsis genes was generated as described previously for TAIR9 (Woodhouse et al. 2011). We updated all putative GTD copies to TAIR10. Woodhouse et al. scored gene duplicates as transposed based on a function of synteny across taxa in the direction A. thaliana → A. lyrata → Carica papaya → Populus trichocarpa → Vitis vinifera. For analysis of Brassicaceae genome evolution, methodical restrictions apply due to the low resolution within that clade, covered by only two tribes. Thus, we screened the genomic context of AtGS genes within a narrow window of 3 kb for flanking TE-like sequences, using the GEvo function from the CoGe comparative genomics platform (http://genomevolution.org/CoGe/GEvo.pl, last accessed November 8, 2013) (Lyons and Freeling 2008). Graphical highlights of TE-like sequences have been customized by choosing “show other features” in the “results visualization” screen. By that means, we confirmed AtGS genes that transposed at least once during lineage evolution as defined by Woodhouse et al. and identified further GTDs missed by that approach due to lack of synteny data (i.e., GTDs predating Vitis speciation and recent GTDs of Brassicaceae-specific genes; marked by asterisks in table 2). Information on GTD events was added to an additional column in supplementary table S1, Supplementary Material online.
Table 2.
GTDs in Arabidopsis and Aethionema GS Pathway Inventory
| Protein Name | AGIa | α-Block | AabIDb | Syntelog | % Identity | Lineage Specific? | Col-0 → Aabc |
|---|---|---|---|---|---|---|---|
| GS genes with retained α-ohnolog | |||||||
| [AAO3] | AT2G27150 | A02NOA1 | Aab27016 | Yes | 77.67 | No | 2 → 2 |
| CYP79C1 | AT1G79370 | A05N062 | Aab34143 | Yes | 76.8 | No | 8 → 9 |
| APK2 | AT4G39940 | A10NOA2 | Aab17804 | Yes | 86.05 | No | 2 → 2 |
| CYP79B2 | AT4G39950 | A10N257 | Aab17805 | Yes | 81.38 | No | 8 → 9 |
| CYP79B3 | AT2G22330 | A10N257 | Aab19477 | Yes | 81.4 | No | 8 → 9 |
| GS genes with tandem duplicate copy | |||||||
| AOP1 | AT4G03070 | A01 | Aab37231 | Yes | 70.03 | No | 2 → 1 |
| AOP3 | AT4G03050 | A01 | — | — | — | Arabidopsis | 2 → 1 |
| CYP79F1 | AT1G16410 | A05 | Aab27579 | Yes | 72.79 | Aethionema | 8 → 9 |
| CYP79C2 | AT1G58260 | A03 | Aab17711 | Yes | 71.85 | Aethionema | 8 → 9 |
| A11 | Aab22600 | No | 61.8 | Aethionema | 8 → 9 | ||
| CYP83A1 | AT4G13770 | A15 | Aab32506 | Yes | 69.67 | No | 2 → 3 |
| — | Aab30975 | No | 82.8 | Aethionema | 2 → 3 | ||
| CYP81F2 | AT5G57220 | A22 | — | — | — | Arabidopsis | 2 → 1 |
| GS genes without α-ohnolog or tandem duplicate copy | |||||||
| UGT74B1 | AT1G24100* | A05 | Aab07826 | Yes | 70.35 | No | 6 → 10 |
| A05 | Aab07827 | Yes | 80.65 | Aethionema | 6 → 10 | ||
| IMD3 | AT1G31180 | A06 | — | — | — | Arabidopsis | 2 → 1 |
| CYP83B1 | AT4G31500 | A10 | Aab12019 | No | 92.73 | No | 2 → 3 |
| GSL-OH | AT2G25450 | A10 | — | — | — | Arabidopsis | 1 → 0 |
| IMD1 | AT5G14200 | A12 | Aab14760 | Yes | 89.5 | No | 2 → 1 |
| CYP79A2 | AT5G05260* | A14 | Aab36760 | Yes | 73.37 | No | 8 → 9 |
| CHY1 | AT5G65940* | A19 | Aab05851 | Yes | 80.75 | No | 2 → 2 |
| FMO-GSOX-1 | AT1G65860* | A25 | Aab30109 | Yes (minimum) | 58.45 | No | 11 → 8 |
| 24% TD (4/17) | 18% TD (3/17) | Ø 76.78% | |||||
| 29% retained α-ohnolog (5/17) | 29% retained α-ohnolog (5/17) | ||||||
aAsterisks mark GTDs inferred by flanking TE-like sequences using GEVo.
bPredicted Aethionema CDS.
cChange of gene family locus count in Arabidospsis → Aethionema order.
Analysis of Putative GS Genes Not Affected by TD, GTD, or At-α Ohnolog Retention in Arabidopsis
We performed additional analysis of AtGS loci beyond the aforementioned types of duplication by considering more ancient WGD events. Information on Arabidopsis genome-wide distribution of ohnolog duplicate pairs dating back to the At-ß and At-γ WGD events (Bowers et al. 2003) were added to an additional column in supplementary table S1, Supplementary Material online. GS genes were referenced accordingly. Remnants did not show significant similarities to any other locus in Arabidopsis by definition, and evolutionary stability was confirmed using the Arabidopsis transpositional history database (http://nature.berkeley.edu/freelinglab, last accessed November 8, 2013) and data on AtGS syntelogs in B. rapa (Wang et al. 2011a).
Orthologous Gene Identification of Arabidopsis GS Biosynthetic Genes in Aet. Arabicum
We considered multiple lines of evidence for identification of orthologs between A. thaliana GS loci and Aet. arabicum. We defined orthologous pairs of A. thaliana and Aet. arabicum GS loci as reciprocal best hits (RBHs) within a given region of gene colinearity (synteny). First, we screened for regions in the Aet. arabicum genome displaying synteny to genomic regions in A. thaliana harboring GS loci, using the “Synfind” function with standard parameters from the CoGe comparative genomics system (www.genomeevolution.org, last accessed November 8, 2013) (Tang and Lyons 2012). Second, we determined RBHs between A. thaliana GS genes and Aet. arabicum genes within the syntenic regions from (i), using BlastP with a minimum query coverage of N = 0.5 and a cut-off e-value of 1E−10. Third, we queried all putative Aet. arabicum GS loci against the Aet. arabicum genome in a BlastP screen with a cut-off e-value of 1E−30. We screened for subject sequences not sharing the query sequence scaffold and identified syntenic regions in A. thaliana. If a GS biosynthetic gene was present in syntenic A. thaliana region (BlastP with a cut-off e value of 1E−30), we defined the aligned Aet. arabicum subject sequence as ortholog to the A. thaliana query sequence.
Tandem Arrayed Gene Copy Identification of Putative GS Biosynthetic Genes in Aet. arabicum
We queried all putative Aet. arabicum GS loci against the Aet. arabicum genome in a BlastP screen with a cut-off e-value of 1E−30. For identification of TDs, GS query sequences were grouped with the subset of respective subject sequences located within a window of N = 10 allowed gene spacers to form Aet. arabicum superfamilies of putative TAR genes. TDs were visualized using the MAFFT package (http://mafft.cbrc.jp/alignment/software/, last accessed November 8, 2013) (Katoh et al. 2002). We further confirmed Aet. arabicum GS genes expression by querying the RNA-seq data. Transcriptome data were mined for expression of GS genes using TBlastX with a cut-off e-value of 1E−10 (data not shown).
Identification of Lineage-Specific GS GTDs Comparing Putative GS Biosynthetic Genes in Aet. arabicum and A. thaliana
We queried all putative Aet. arabicum GS loci against the Aet. arabicum genome in a BlastP screen with a cut-off e-value of 1E−30. For identification of GTDs following divergence of these lineages, we screened for subject sequences not sharing the query sequence scaffold and identified syntenic regions in A. thaliana. If GS biosynthetic gene is absent in syntenic A. thaliana region (BlastP with a cut-off e-value of 1E−30), we defined the aligned Aet. arabicum subject sequence as lineage-specific GTD copy.
Phylogenetic and Similarity Analysis
A number of Arabidopsis flavin-monooxygenases involved in GS biosynthesis (FMO GS-OX) are encoded in clusters consisting of retained ohnolog copies as well as both tandem- and gene transposition duplicates. To visualize the evolution of FMO-like sequences in Brassicales, Carica. papaya, and Tarenaya hasslerania (Cheng et al. 2013), FMO orthologs from these species were obtained using the CoGe comparative genomics system. A phylogenetic tree was constructed using the maximum-likelihood method with PhyML 3.1 software (Guindon et al. 2010), employing the Le/Gascuel (LG) model for amino acid substitution. Protein sequence similarity analysis were performed using the Needle program from the EMBOSS software package (http://emboss.sourceforge.net/, last accessed November 8, 2013) (Rice et al. 2000).
Genome Data Visualization and Statistics
Fisher’s exact test for count data was performed using the R package for statistical computing (www.r-project.org, last accessed November 8, 2013). Circular visualization of genome data was performed using the circos package (www.circos.ca, last accessed November 8, 2013) (Krzywinski et al. 2009) and graphically edited with the GIMP-package (www.gimp.org, last accessed November 8, 2013).
Results
The Influence of the At-α WGD Event to GS Pathway Evolution in Arabidopsis
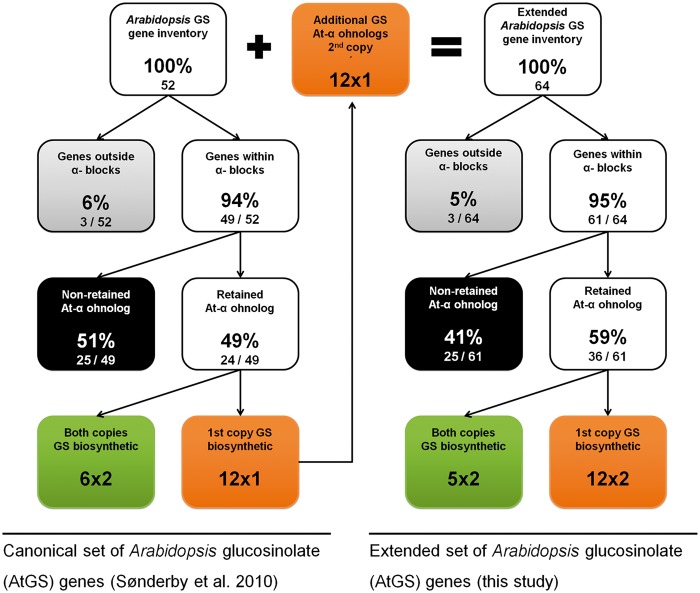
We first updated the genomic location of all ohnolog blocks dating back to the At-α WGD event (α-blocks thereafter) in A. thaliana from the TIGR5 to the TAIR10 annotation, leading to minor changes in the list published by Thomas et al. (2006) (supplementary table S1, Supplementary Material online). As a first step to understanding the dynamics of GS pathway evolution, we divided the 52 to-date known AtGS genes into three groups: first, genes with a retained At-α ohnolog copy (table 1). Second, genes with lost At-α ohnolog copy, but a genomic location covered by α-blocks (table 3). Third, genes located outside the genomic borders of α-blocks (table 4). For the original set of AtGS genes published by Sonderby et al. (2010), we found an increased retention rate of 49% (24/49) for retained ohnolog copies dating back to the At-α WGD event (fig. 1), compared with a 22% average observed for all Arabidopsis protein-coding genes (fig. 2A and B). These 24 canonical AtGS genes group to six ohnolog pairs with annotation to GS metabolism and 12 loci lacking annotation of one ohnolog copy to GS biosynthesis (figs. 1 and 3). Notably, the 12 ohnolog pairs sharing GS annotation and the six ohnolog pairs lacking GS annotation of one member (forming18 AtGS ohnolog copy pairs in total) either display high degrees of pairwise similarity and/or show similar tendencies in gene expression following treatment with methyljasmonic acid, an organic volatile important for plant defense signaling (Cheong and Choi 2003) (tables 5 and 6). Therefore, we inferred functional redundancy of ohnolog copies due to structural homology. We propose a significant contribution to GS metabolism and consistently include all 12 ohnolog copies lacking GS annotation to our analysis, forming 12 pairs of two ohnolog copies each. We thereby created an extended set of 64 putative AtGS genes (figs. 1 and 3). Among genes located within α-block boundaries, we found an At-α ohnolog retention rate of 59% (36/61) for the extended AtGS set (fig. 1), which is more than double of the observed 22% average rate for ohnolog retention among all Arabidopsis protein-coding loci harbored within the boundaries of α-blocks (fig. 2B).
Table 3.
Genes with Nonretained At-α Ohnolog Duplicate Gene Copy in Arabidopsis and Aethionema GS Pathway Inventory
| Protein Name | AGI | α-Block | Evident SSDa | AabIDb | Syntelog | % Identity | Col-0 → Aabc |
|---|---|---|---|---|---|---|---|
| Core-structure formation | |||||||
| AOP1 | AT4G03070 | A01 | TD/GTD | Aab37231 | Yes | 70.03 | 2 → 1 |
| AOP3 | AT4G03050 | A01 | TD/GTD | — | — | — | 2 → 1 |
| UGT74-like | Aab specific | A02 | TD | Aab37178 | Yes | 82.05 | 6 → 10 |
| UGT74-like | Aab specific | A02 | TD | Aab37179 | Yes | 77.63 | 6 → 10 |
| UGT74-like | Aab specific | A02 | TD | Aab37180 | Yes | 78.33 | 6 → 10 |
| GSTF10 | AT2G30870 | A02 | TD | Aab28612 | Yes | 91.59 | 4 → 4 |
| FMO-GSOX-3 | AT1G62560 | A03 | TD | Aab10867 | Yes | 71.9 | 11 → 8 |
| FMO-GSOX-4 | AT1G62570 | A03 | TD | Aab10866 | Yes | 55.2 | 11 → 8 |
| FMO-GSOX-5 | AT1G12140 | A03 | TD | Aab13546 | Yes | 71.9 | 11 → 8 |
| CYP79C2 | AT1G58260 | A03 | TD | Aab17711 | Yes | 71.85 | 8 → 9 |
| Aab specific | A11 | Aab22600 | No | 61.8 | 8 → 9 | ||
| CYP79F1 | AT1G16410 | A05 | TD | Aab27579 | Yes | 72.79 | 8 → 9 |
| SOT18 | AT1G74090 | A05 | TD | Aab14277 | Yes | 83.9 | 3 → 3 |
| GSTU20 | AT1G78370 | A05 | TD | Aab7000 | Yes | 67.29 | 5 → 6 |
| Aab specific | Aab6995 | Yes | 48.86 | 5 → 6 | |||
| UGT74B1 | AT1G24100 | A05 | GTD | Aab07826 | Yes | 70.35 | 6 → 10 |
| Aab specific | Aab07827 | Yes | 80.65 | 6 → 10 | |||
| CYP83B1 | AT4G31500 | A10 | GTD | Aab12019 | No | 92.73 | 2 → 3 |
| GSL-OH | AT2G25450 | A10 | GTD | — | — | — | 1 → 0 |
| CYP79A2 | AT5G05260 | A14 | GTD | Aab36760 | Yes | 73.37 | 8 → 9 |
| CYP83A1 | AT4G13770 | A15 | GTD | Aab32506 | Yes | 69.67 | 2 → 3 |
| Aab specific | Aab30975 | No | 82.8 | 2 → 3 | |||
| CYP81F2 | AT5G57220 | A22 | TD/GTD | — | — | — | 2 → 1 |
| FMO-GSOX-1 | AT1G65860 | A25 | GTD | Aab30109 | Yes | 58.45 | 11 → 8 |
| Cosubstrate pathways | |||||||
| CHY1 | AT5G65940 | A19 | GTD | Aab05851 | Yes | 80.75 | 2 → 2 |
| GSH1 | AT4G23100 | A20 | NA | Aab22781 | Yes | 91.81 | 2 → 2 |
| BZO1 | AT1G65880 | A25 | TD | Aab31601 | Yes | 70.04 | 2 → 4 |
| TD | Aab31602 | Yes | 69.4 | 2 → 4 | |||
| Side-chain elongation | |||||||
| IMD3 | AT1G31180 | A06 | GTD | — | — | — | 2 → 1 |
| IPMI2 | AT2G43100 | A11 | TD | Aab19630 | Yes | 78.71 | 3 → 3 |
| IMD1 | AT5G14200 | A12 | GTD | Aab14760 | Yes | 89.5 | 2 → 1 |
| IIL1 | AT4G13430 | A15 | NA | Aab18132 | Yes | 93.9 | 1 → 1 |
| TF regulation | |||||||
| MYB76 | AT5G07700 | A26 | TD | — | — | — | 6 → 4 |
| 60% TD (15/25) | 57% TD (16/28) | Ø 76.46% | |||||
| 48% GTD (12/25) | 39% GTD (11/28) | ||||||
Note.—NA, not applicable.
aTD (Tandem Dupicate) refers to members of tandem arrays and GTD (Gene Transposition Duplication) refers to a history of transposition in Arabidopsis.
bPredicted Aethionema CDS
cChange of gene family locus count in Arabidospsis → Aethionema order
Table 4.
Genes Not Covered by α-Blocks in Arabidopsis and Aethionema GS Pathway Inventory
| Protein Namea | AGI | α-Block | Evident SSDb | AabIDc | Syntelog | % Identity | Col-0 → Aabd |
|---|---|---|---|---|---|---|---|
| Side-chain elongation | |||||||
| MAM1 | AT5G23010 | — | TD | Aab12229 | Yes | 72.31 | 2 → 4 |
| Aab12230 | Yes | 71.5 | 2 → 4 | ||||
| MAM-L | AT5G23020 | — | TD | Aab12225 | Yes | 70.67 | 2 → 4 |
| Aab12226 | Yes | 68.36 | 2 → 4 | ||||
| TF regulation | |||||||
| MYB34 | AT5G60890 | — | NA | — | — | — | 6 → 4 |
| 66% TD (2/3) | 100% TD (4/4) | Ø 70.71% | |||||
| 0% GTD | 0% GTD | ||||||
aSquared brackets indicate ohnolog copies of GS biosynthetic genes without GO!-annotation to GS biosynthetic process.
bTD refers to members of TARs and GTD refers to a history of transposition in Arabidopsis.
cPredicted Aethionema CDS.
dChange of gene family locus count in Arabidospsis → Aethionema order.
Fig. 1.—
Distribution of GS pathway inventory relative to At-α WGD event. AtGS genes are shown before (left) and after (right) interpolation of ohnolog duplicate copies. We hypothesize functional redundancy of 12 additional ohnologs to canonical GS biosynthetic genes.
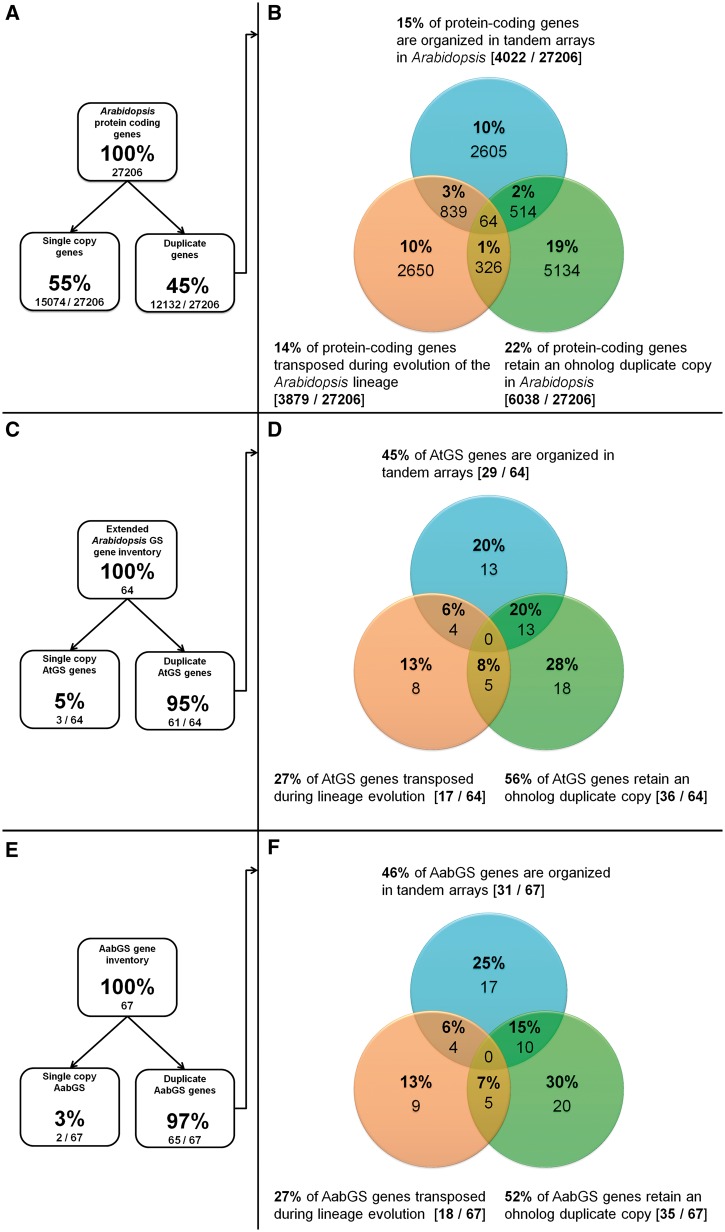
Fig. 2.—
Duplicate distribution among (A, B) Arabidopsis protein-coding genes compared with (C, D) AtGS and (E, F) Aethionema GS loci. Shown are retained ohnologs (green), tandem duplicates (blue), and gene transposition duplicates (orange). GS metabolic versatility resulted from a combination of increased ohnolog retention and TD rates.
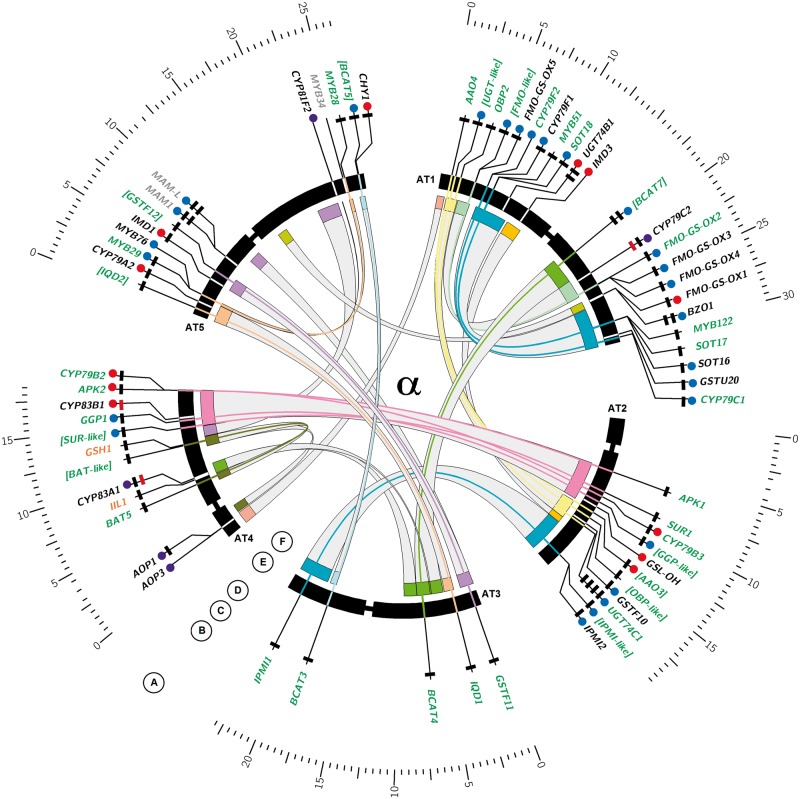
Fig. 3.—
Ideogram of Arabidopsis thaliana chromosomes with GS biosynthetic genes. Circos plot visualizing the evolutionary contribution of different duplication types to GS pathway inventory in Arabidopsis and Aethionema. (A) Inner chromosome scale (Mb). (B) Arabidopsis thaliana GS biosynthetic genes. Gray text indicates genomic location outside ohnolog blocks. Black text indicates genomic location within ohnolog blocks but nonretained ohnolog copy. Green text indicates retained pairs of ohnolog copies with missing GO annotation to GS biosynthetic process shown in edged brackets. Orange text indicates single copy genes without clear paralogs in both species. (C) Blue circles indicate genes organized in TARs (i). Red circles indicate genes with transpositional history (ii). Purple circles indicate loci sharing (i) and (ii). (D) Number of rectangles indicates number of homologs present in the Aethionema arabicum draft genome (0–4). Color of rectangles indicates presence (black) or absence (red) of synteny between A. thaliana and Aet. arabicum in the genomic context of the target gene. (E) Arabidopsis thaliana chromosomes with labels showing GS biosynthetic genes. Bands for genes retained in ohnolog pairs are connected with colors of corresponding ohnolog blocks, as defined by Bowers et al. (2003). (F) Genomic location of ohnolog block copies harboring GS biosnthetic genes in A. thaliana, connected by gray bands. All ranges are in scale.
Table 5.
Intraspecies Protein Similarities for At-α Ohnolog Pairs Sharing GS Annotation, Shown with Differential Expression in Arabidopsis Following MeJA Treatment
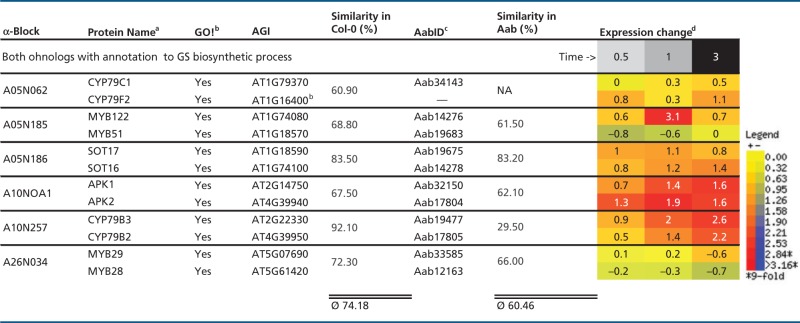 |
Note.—MeJA, methyljasmonic acid; NA, not applicable.
aSquared brackets indicate ohnolog copies of GS biosynthetic genes without GO! annotation to GS biosynthetic process.
bGO! column indicates if gene is part of canonical GS pathway inventory set (Sonderby et al. 2010).
cPredicted Aethionema CDS.
dWhole WT plant averages of log-transferred expression change in Arabidopsis.
Table 6.
Intraspecies Protein Similarities for At-α Ohnolog Pairs Not Sharing GS Annotation, Shown with Differential Expression in Arabidopsis Following MeJA Treatment
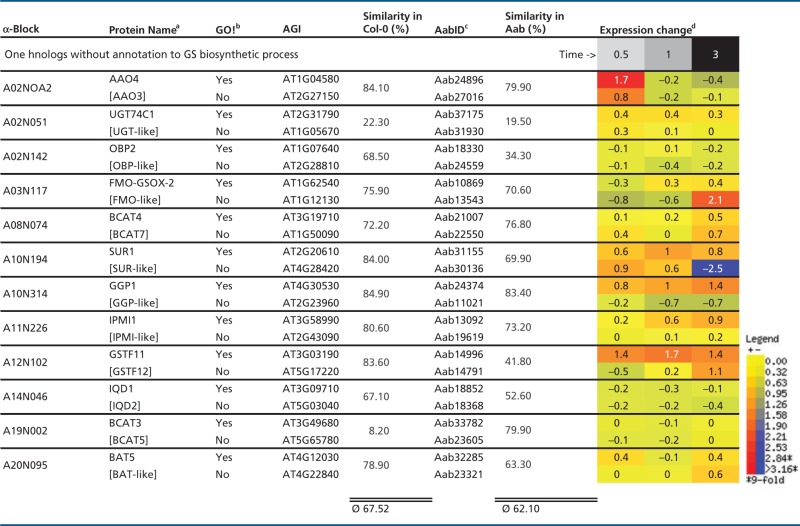 |
Note.—MeJA, methyljasmonic acid.
aSquared brackets indicate ohnolog copies of GS biosynthetic genes without GO! annotation to GS biosynthetic process.
bGO! column indicates if gene is part of canonical GS pathway inventory set (Sonderby et al. 2010).
cPredicted Aethionema CDS.
dWhole WT plant averages of log-transferred expression change in Arabidopsis.
Quantification of TD Influence to GS Pathway Evolution in Arabidopsis
In the next step, we quantified the impact of TD to GS pathway versatility in Arabidopsis. Minor changes were made in the list of Arabidopsis TAR genes by Rizzon et al. (2006) due to the gene updates to TAIR10 (supplementary table S1, Supplementary Material online). We mined the 1,497 Arabidopsis TARs comprising 4,034 duplicate gene copies for AtGS genes. Forty-five percent (29/64) of AtGS genes are members of TARs, compared with a genome-wide average of 15% (figs. 2B and D). Contribution of GTDs to GS pathway evolution in Arabidopsis
We quantified the influence of GTD to GS pathway evolution in Arabidopsis. Initially, a list of 4,575 genes with putative origin due to a GTD was proposed for TAIR9 (Woodhouse et al. 2011). Our update to TAIR10 retained 4,539 loci clearly referenced to transposition events (supplementary table S1, Supplementary Material online), illustrating a 14% average for GTD genes among protein-coding loci in Arabidopsis (fig. 2B). Among those, we confirmed all 13 references to AtGS loci (table 2), using the GEvo function from the CoGe platform (see Materials and Methods). We thereby discovered that four additional AtGS genes (marked by asterisks in table 2) lost all syntenic anchor genes in genomic proximity (±1,000 kb) but are surrounded by TE-like sequences. Thus they may have transposed following the At-α WGD event (marked by asterisks in table 2). These additional genes may be Brassicaceae specific but lost secondarily in A. lyrata. This might explain their absence in the Arabidopsis gene transpositional history database (that mainly scores pre-α GTD events due to the lack of further Brassicaceae synteny data necessary for scoring of post-α GTDs). Hence, the total fraction of GTD copies among AtGS genes sums up to 27% (18/67) (fig. 2D).
Analysis of GS Genes Not Affected by TD, Ohnolog Retention, or Transposition
In addition, we performed a more in-depth analysis of the three AtGS genes lacking TD, retained At-α ohnolog copy or evidence for transposition during evolution of the Arabidopsis lineage (table 7). Among those, MYB34 is the only locus retaining an ohnologous copy dating back to the At-ß WGD event, leaving two putative nonduplicate genes in AtGS pathway inventory: GSH1 and IIL1, functioning in GS cosubstrate pathways and side-chain elongation, respectively (table 3). To confirm the observed evolutionary stability of these genes, we identified syntelogs in V. vinifera (IIL1) and B. rapa (GSH1), respectively. Syntelogs in Vitis proof that IIL1 did not transpose since the birth of the Rosids. Therefore, this gene represents a very ancient unigene. In case of duplication before Vitis lineage evolution, all copies were lost subsequently before radiation of the Rosid clade. In contrast, GSH1 may be Brassicaceae-specific unigene that likewise lost all duplicates with above-threshold similarity.
Table 7.
Putative Single-Copy Genes in Aethionema and Arabidopsis GS Pathway Inventory
| AGI | Name | α-Block | Retained At-ß/-γ Ohnolog | Most Ancient Syntelog | Closest Paralog | BlastP e Value | Name | α-Block | Retained At-ß/-γ Ohnolog |
|---|---|---|---|---|---|---|---|---|---|
| AT4G13430 | IIL1 | A15 | No | Vitis vinifera | AT4G26970 | 1.00E−16 | ACO2 | A22N121 | No |
| AT4G23100 | GSH1 | A20 | No | Brassica rapa | AT1G19220a | 0.19 | ARF11 | A05 | No |
| AT5G60890b | MYB34 | — | B20N001 | V. vinifera | AT1G74080 | 4.00E−62 | MYB122 | A05N185 | B20N004 |
aGene transpositional duplicate copy.
bAbsent in Aethionema.
GS Biosynthetic Gene Identification from Draft Aet. arabicum Genome
On the basis of the Aet. arabicum genome v1.0 and 37,839 annotated genes (Haudry et al. 2013), we identified homologs of A. thaliana loci coding for GS biosynthetic and regulatory genes. Combining reciprocal best BlastP hits with LAST screens for large scale gene colinearity/synteny (100 kb–1.2 Mb) (employed by the Synfind algorithm, see Materials and Methods), we found putative Aet. arabicum orthologs covering 57 of the 64 proposed AtGS genes with an observed nucleotide sequence identity of 45–94% (tables 1, 3, and 4). Among those, seven loci gave rise to an additional 10 further paralogs due to TD and GTD in Aethionema (see later), thereby extending the copy number of six multigene families to a total of 67 putative AabGS genes. The mRNA sequencing data for Aet. arabicum supported the evidence that all 67 putative AabGS genes were expressed (data not shown).
GS Gene Families with Expanded Copy Number in Aethionema
Among the 10 novel paralogs, eight were identified as descendants from TD events. Intriguingly, the Arabidopsis methylthiomalate synthase array MAM1/MAM-L underwent a further duplication in Aethionema, retaining four MAM-like loci (supplementary fig. S1, Supplementary Material online). Likewise, we observed a further TD of the Arabidopsis benzoate-CoA ligase array BZO1/BZO-like in Aethionema, adding two paralogs to the set of putative AabGS genes (table 8). Notably, both clusters encode functions in GS side-chain elongation (MAM) or cosubstrate pathways (BZO), indicating the connection of TD to metabolic versatility in both Arabidopsis and Aethionema. Furthermore, TD extended the gene inventory of GS core-structure modification in two cases. First, we detected one additional duplicate of the Arabidopsis tau-type gluthation-s-transferase array GSTU19-23 in Aethionema (table 8). Second, we identified extension of the UGT-like superfamily that is present with five members in Arabidopsis and organized in two TARs of distant genomic location (fig. 3B). Intriguingly, both regions represent the sister copies of α-block a02 (fig. 3F). Furthermore, both UGT-like TARs comprise neighboring pairs of the At-α ohnologs duplicates A02N051 (UGT74C1 or AT2G31790/UGT-like or AT1G05670) and A02N053 (UGT74D1 or AT2G31750/UGT74E2 or AT1G05680) (fig. 3B, table 8), indicating a pre-At-α TD event generating both precursors of the above-mentioned UGT-like ohnolog pairs. In Aethionema, we find a further TD-driven extension of this superfamily, adding three more copies to reach a total number of 8 UGT-like sequences (table 8). Therefore, the diversity of UGT-like sequences in Brassicaceae is expanded by the combination of WGD with pre- and post-At-α TD events.
Table 8.
Tandem Duplicate Genes in Arabidopsis and Aethionema GS Pathway Inventory
| Protein Namea,b | AGI | α-Blockc | AabIDd | Syntelog | % Identitye | Lineage Specific? | Col-0 → Aabf |
|---|---|---|---|---|---|---|---|
| Tandem duplicates of syntenic anchor genes retaining an At-α ohnolog | |||||||
| UGT74C1 | AT2G31790 | A02N051 | Aab37175 | Yes | 79.44 | No | 6 → 10 |
| Aab37178 | Yes | 82.05 | Aethionema | 6 → 10 | |||
| Aab37179 | Yes | 77.63 | Aethionema | 6 → 10 | |||
| Aab37180 | Yes | 78.95 | Aethionema | 6 → 10 | |||
| UGT74D1_oa | AT2G31750 | A02N053 | Aab37181 | Yes | 78.33 | (no GS gene) | 6 → 10 |
| [UGT-like] | AT1G05670 | A02N051 | Aab31930 | Yes | 81.11 | No | 6 → 10 |
| UGT-like_oa | AT1G05675 | A02 | Aab31932 | Yes | 71.4 | (no GS gene) | 6 → 10 |
| UGT74E2_oa | AT1G05680 | A02N053 | Aab31933 | Yes | 59.8 | (no GS gene) | 6 → 10 |
| FMO-GSOX-2 | AT1G62540 | A03N117 | Aab10869 | Yes | 76.6 | No | 11 → 8 |
| FMO-GSOX-3 | AT1G62560 | A03 | Aab10867 | Yes | 71.9 | No | 11 → 8 |
| FMO-GSOX-4 | AT1G62570 | A03 | Aab10866 | Yes | 55.2 | No | 11 → 8 |
| FMO-like_oa | AT1G62580 | A03 | — | — | — | (no GS gene) | 10 → 7 |
| FMO-like_oa | AT1G62600 | A03 | — | — | — | (no GS gene) | 10 → 7 |
| FMO-like_oa | AT1G62620 | A03 | — | — | — | (no GS gene) | 10 → 7 |
| [FMO-like] | AT1G12130 | A03N117 | Aab13543 | Yes | 65.09 | No | 11 → 8 |
| FMO-GSOX-5 | AT1G12140 | A03 | Aab13546 | Yes | 71.9 | No | 11 → 8 |
| FMO-like_oa | AT1G12200 | A03 | Aab13549 | Yes | 66.66 | (no GS gene) | 10 → 7 |
| CYP79F2 | AT1G16400 | A05N062 | — | — | — | Arabidopsis | 8 → 9 |
| CYP79F1 | AT1G16410 | A05 | Aab27579 | Yes | 72.79 | Arabidopsis | 8 → 9 |
| SOT16 | AT1G74100 | A05N186 | Aab14278 | Yes | 91.07 | No | 3 → 3 |
| SOT18 | AT1G74090 | A05 | Aab14277 | Yes | 83.9 | No | 3 → 3 |
| GSTU20 | AT1G78370 | A05 | Aab07000 | Yes | 67.29 | No | 5 → 6 |
| GSTU23_oa | AT1G78320 | A05 | Aab06994 | Yes | 81.74 | (no GS gene) | 5 → 6 |
| GSTU22_oa | AT1G78340 | A05 | Aab06997 | Yes | 71.1 | (no GS gene) | 5 → 6 |
| GSTU21_oa | AT1G78360 | A05 | Aab06998 | Yes | 76.71 | (no GS gene) | 5 → 6 |
| GSTU19_oa | AT1G78380 | A05N104 | Aab06999 | Yes | 83.41 | (no GS gene) | 5 → 6 |
| [BCAT7] | AT1G50090 | A08N074 | Aab22548 | Yes | 69 | No | 6 → 6 |
| BCAT-like_oa | AT1G50110 | A08 | Aab22550 | Yes | 78.12 | (no GS gene) | 6 → 6 |
| [SUR-like] | AT4G28420 | A10N194 | Aab31155 | Yes | 47.42 | No | 3 → 2 |
| A10 | Aab31154 | Yes | Aethionema | ||||
| SUR-like_oa | AT4G28410 | A10 | Aab31153 | Yes | 63.96 | (no GS gene) | 3 → 2 |
| GGP1 | AT4G30530 | A10N314 | Aab24374 | Yes | 87.6 | No | 5 → 5 |
| GGP-like_oa | AT4G30540 | A10 | Aab24373 | Yes | 75 | (no GS gene) | 5 → 5 |
| GGP3_oa | AT4G30550 | A10 | Aab24372 | Yes | 82 | (no GS gene) | 5 → 5 |
| [GGP-like] | AT2G23960 | A10N314 | Aab11021 | Yes | 69.67 | No | 5 → 5 |
| GGP-like _oa | AT2G23970 | A10 | Aab11018 | Yes | 83.6 | (no GS gene) | 5 → 5 |
| [IPMI-like] | AT2G43090 | A11N226 | Aab19619 | Yes | 85.99 | No | 3 → 3 |
| IPMI2 | AT2G43100 | A11 | Aab19630 | Yes | 78.71 | No | 3 → 3 |
| [BCAT5] | AT5G65780 | A19N002 | Aab23605 | Yes | 75.3 | No | 6 → 6 |
| LINC4_oa | AT5G65770 | A19 | Aab23607 | Yes | 70.08 | (no GS gene) | 6 → 6 |
| MYB29 | AT5G07690 | A26N034 | Aab33585 | Yes | 65.13 | Arabidopsis | 6 → 4 |
| MYB76 | AT5G07700 | A26 | — | — | — | Arabidopsis | 6 → 4 |
| Tandem duplicates of genes inside the boundaries of α-blocks with nonretained At-αohnolog | |||||||
| AOP1 | AT4G03070 | A01 | Aab37231 | Yes | 70.03 | Arabidopsis | 2 → 1 |
| AOP3 | AT4G03050 | A01 | — | — | — | Arabidopsis | 2 → 1 |
| GSTF10 | AT2G30870 | A02 | Aab28612 | Yes | 91.59 | No | 4 → 4 |
| GSTF9_oa | AT2G30860 | A02 | Aab28613 | Yes | 89.76 | (no GS gene) | 4 → 4 |
| CYP79C2 | AT1G58260 | A03 | Aab17711 | Yes | 71.85 | No | 8 → 9 |
| CYP-like_oa | AT1G58265 | A03 | Aab17712 | Yes | 60.71 | (no GS gene) | 8 → 9 |
| UGT74B1 | AT1G24100 | A05 | Aab07827 | Yes | 80.65 | Aethionema | 6 → 10 |
| A05 | Aab07826 | Yes | 70.35 | Aethionema | 6 → 10 | ||
| CYP81F2 | AT5G57220 | A22 | — | — | — | Arabidopsis | 2 → 1 |
| CYP71B10 _oa | AT5G57260 | A22 | Aab25774 | Yes | 73.21 | (no GS gene) | 2 → 1 |
| BZO1 | AT1G65880 | A25 | Aab31601 | Yes | 70.04 | No | 2 → 4 |
| Aab31602 | Yes | 69.4 | Aethionema | 2 → 4 | |||
| BZO-like_oa | AT1G65890 | A25 | Aab31603 | Yes | 68.85 | (no GS gene) | 2 → 4 |
| Aab31604 | Yes | 67.83 | (no GS gene) | 2 → 4 | |||
| Tandem duplicates of genes outside the boundaries of α-blocks | |||||||
| MAM1 | AT5G23010 | — | Aab12229 | Yes | 72.31 | No | 2 → 4 |
| Aab12230 | Yes | 71.5 | Aethionema | 2 → 4 | |||
| MAM-L | AT5G23020 | — | Aab12225 | Yes | 70.67 | No | 2 → 4 |
| Aab12226 | Yes | 68.36 | Aethionema | 2 → 4 | |||
| AtGS genes: 45% TD (29/64) | AabGS genes: 46% TD (31/67) | Ø 73.43% | |||||
aSquared brackets indicate ohnolog copies of GS biosynthetic genes without GO! annotation to GS biosynthetic process.
bThe “_oa” suffix indicates tandem duplicate copies without GO! annotation to GS biosynthetic process. These genes have not been considered for the tandem duplicate count of GS loci in both organisms.
cUnderlined items refer to offspring of one pre-α TD event.
dPredicted Aethionema CDS.
eIn case of Aethionema-specific TAR expansion, the corresponding Arabidopsis sequence for identity comparison was determined based on both genomic location and homology criteria.
fChange of gene family locus count in Arabidospsis → Aethionema order.
GTD accounts for the copy number expansions of two putative AabGS loci. Both cases involve CYP-like genes that play a role in GS core-structure formation (Sonderby et al. 2010). In Aethionema, the TAR formed by 1) CYP79C2 (At1G85260) and 2) the CYP-like locus AT1G58265 transposed an additional copy of the TAR to a different genomic location (supplementary fig. S2, Supplementary Material online). Likewise, we identified an additional GTD of CYP83A1 in Aethionema (table 3). CYP83A1 metabolizes oximes in GS biosynthesis, is not redundant to CYP83B1, and interestingly also possesses a history of GTD events in Arabidopsis (Naur et al. 2003).
Arabidopsis GS Loci without Orthologs in Aethionema
In seven cases, RBH- and synteny-based evidence was not sufficient to clearly assign orthologs to AtGS loci in the Aet. arabicum draft genome (tables 1, 3, and 4), leading to the contraction of six multigene families and loss of one single-copy gene.
Four of those loci, namely AOP3, CYP79F2, IMD3, and MYB76, are likewise absent in the B. rapa genome and may therefore be specific to A. thaliana and more closely related species (Wang et al. 2011a). AOP1/AOP3 andCYP79F1/2 represent two neighboring TARs with evident subfunctionalization in Arabidopsis (Kliebenstein et al. 2001; Prasad et al. 2012). Although AOP3 functions in GS side-chain elongation in Arabidopsis (Kliebenstein et al. 2001), CYP79F2 encodes an enzyme involved in core structure formation of long-chain aliphatic GS. Furthermore, overexpression of the MYB76 transcription factor correlates with increased levels of both long-chained and short-chained aliphatic GS in Arabidopsis (Gigolashvili et al. 2008). However, experiments with Arabidopsis myb76 T-DNA insertion lines to date failed to show any significant change in GS chemotype, making a strict requirement of MYB76 for GS biosynthesis unlikely (Gigolashvili et al. 2008). Moreover, IMD3 encodes a predicted enzyme with proposed functional redundancy to (as well as strong coexpression with) IMD1, a protein that was shown to be involved in GS accumulation in Arabidopsis (Hirai et al. 2007; Wentzell et al. 2007; Gigolashvili et al. 2009; Sawada et al. 2009; He et al. 2010). Therefore, absence of IMD1 (IMD3) in B. rapa (Aet. arabicum) supports the hypothesized capability of mutual phenotype rescue among IMD1/3 double knock-outs in Brassicaceae, eventually preventing significant alterations of GS chemotype due to fractionation of IMD-like genes in Aethionema.
The other three of the seven AtGS loci that lack a clear ortholog in Aethionema are not found in the B. rapa genome: MYB34, CYP81F2, and GSL-OH (tables 3 and 4). Therefore, they represent Aethionema lineage-specific gene losses. MYB34 was shown to control indolic GS biosynthesis in Arabidopsis (Celenza et al. 2005). Interestingly, overexpression of MYB34 in Arabidopsis partially rescued the altered GS chemotype caused by MYB51 knockout (Gigolashvili et al. 2007). Because of functional redundancy of MYB51/34, the loss of MYB34 likely does not cause significant changes in GS chemotype in Aethionema lineages. In contrast, CYP81F2 and GSL-OH encode functions associated with secondary modification of the GS core structures (Sonderby et al. 2010). CYP81F2 has been shown to control the indole GS modifier1 quantitative trait loci in Arabidopsis, catalyzing the conversion of indole-3-yl-methyl GS to 4-hydroxy-indole-3-yl-methyl GS (Pfalz et al. 2009). Notably, these metabolites play a significant role in MAMP-triggered immunity in Arabidopsis (Bednarek et al. 2009). Among various known cytochrome p450s active in indolic GS biosynthesis, CYP81F2 is the only locus impairing callose deposition after detection of the nonself-infection in Arabidopsis (Clay et al. 2009). On the basis of these findings, we concluded a CYP81F2-specific onset of sub/neofunctionalization from GS biosynthesis toward plant innate immunity after divergence of the Arabidopsis and Aethionema lineages, thereby mitigating fatal consequences of the absent ortholog for GS chemotype in Aethionema.
Moreover, we determined the absence of the 2-oxoacid-dependend oxygenase activity GSL-OH in Aethionema (table 3). GSL-OH is necessary for biosynthesis of 2-hydroxybut-3-enyl GS in Arabidopsis (Wentzell et al. 2007; Hansen et al. 2008) and present in the B. rapa genome (Wang et al. 2011a). Noteworthy, GSL-OH was the only multigene family member within the extended AtGS set whose loss in Aethionema was not accompanied by copy number expansion of one or more paralogs (table 3). Accordingly, we concluded an Aethionema-specific loss of this locus after divergence from the Arabidopsis lineage, creating measurable differences in GS chemotype among Arabidopsis and Aethionema. Consistently, we could not detect traces of 2-hydroxybut-3-enyl GSs in Aet. arabicum root-, leaf-, and seed extract using uHPLC (data not shown).
GS Gene Families with Lower Copy Number in Aethionema
Considering the Aethionemae-specific loss of GSL-OH, six putative GS-annotated multigene families display a lower copy number in Aethionema (AOPx, CYP79x, CYP81x, GSLx, IMDx, and MYBx) (tables 1, 3, 4, and 8). In sum, their total gene count increased from 20 genes in Aethionema to 27 observed in Arabidopsis, thereby mediating a 35% increase (tables 1, 4, and 8).
Although IMD3 and GSL-OH possess GTD copies in Arabidopsis, these loci are absent in Aethionema. In contrast, AOP1/2, IMD1/3, MYB29/76, and CYP81F2 with its CYP-like neighbor AT1G58265 comprise TARs in Arabidopsis but are likewise absent in Aethionema (table 8) and B. rapa (Wang et al. 2011a).
Therefore, the underlying TD events may be Arabidopsis specific. Thus, TD facilitated GS pathway expansion in the Arabidopsis lineage after split from the tribe Aethionemae.
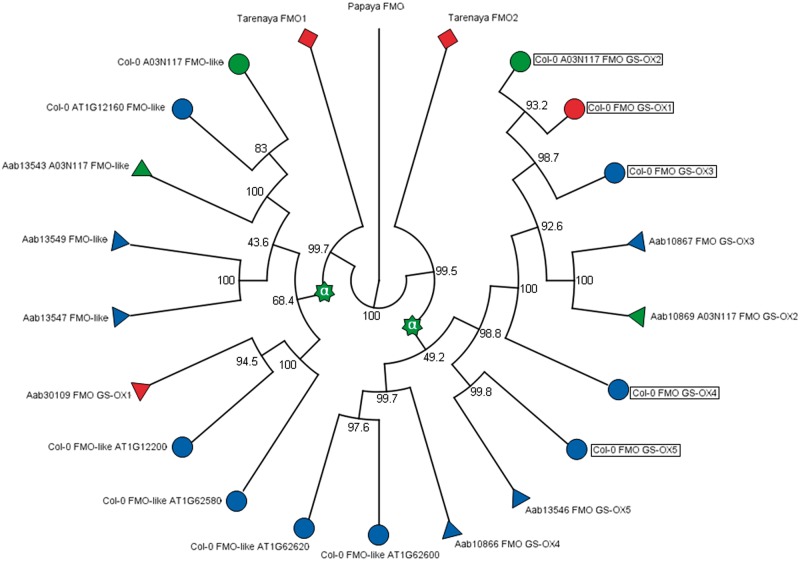
Furthermore, we found evidence for Arabidopsis-specific TD of three neighboring FMO-like loci (FMO GS-OX2-4) (figs. 3 and 4), leading to lower copy number in Aethionema. These genes were lost in B. rapa (Wang et al. 2011a), illustrating a degree of plasticity across Brassicaceae. FMO-like loci comprise a multigene family with five members annotated to GS biosynthesis in Arabidopsis mapping to three distant genomic locations (fig. 3B). Among those, two regions are embedded in ohnolog copies of α-block A02 (fig. 3E) and contain the retained α-pair of AT1G62540 (FMO GS-OX2) and AT1G12130 (FMO-like) (fig. 3B). The latter is not annotated to GS biosynthesis in Arabidopsis. However, AT1G12130 is member of a FMO-like 4-gene TAR with its 3′ neighbor FMO GS-OX5 involved in aliphatic GS biosynthesis (Li et al. 2008). The third genomic region in Arabidopsis harboring a FMO-like sequence with encoded function in GS metabolism is defined by AT1G65860 (FMO GS-OX1), representing a transposed duplicate gene copy (fig. 4). Interestingly, FMO GS-OX1-4 share broad substrate specificity and catalyze the conversion from methylthioalkyl GS to the related methylsulfinylalkyl GS independent of chain length. In contrast, FMO GS-OX5 shows substrate specificity for 8-methylthiooctyl GS (Hansen et al. 2007; Li et al. 2008). This example is similar to the case of UGT-like loci (see earlier) and again illustrates the combination of ohnolog retention with tandem- and GTD leading to increased GS pathway versatility in Brassicaceae (fig. 4).
Fig. 4.—
Phylogenetic relationships among FMO proteins. Col-0, Aab, and papaya refer to Arabidopsis thaliana (circles), Aethionema arabicum (triangles), and Carica papaya (colorless), respectively. Boxes indicate annotation to GS metabolic activity in Arabidopsis. Tarenaya hasslerania (diamonds) represents the closest-related outgroup of Brassicaceae, including the sister clade Aethionemae. Stars: At-α WGD event. Blue: proteins encoded by members of TARs. Red: protein encoded by GTD locus. The At-α WGD leads to duplication of a FMO locus, resulting in two clades comprising all FMO-like sequences of both Aethionema and Arabidopsis. Thus, FMO versatility has been promoted by a combination of increased degree of ohnolog retention and TD events.
Deduction of Total Duplicate Frequencies in Aethionema and Arabidopsis GS Pathway Inventory and Comparison to Arabidopsis Genome-Wide Average
We found the fraction of retained ohnolog duplicate gene pairs among Arabidopsis (56%) and Aethionema (52%) GS biosynthetic and regulatory genes significantly increased compared with the genome-wide average in Arabidopsis (22%) (fig. 2, table 9).
Table 9.
Statistical Testa on Duplicate Fractions in Arabidopsis and Aethionema GS Pathway Inventory Compared with Genome-Wide Average in Arabidopsis
| Arabidopsis Genome | Arabidopsis GS Genesb | Aethionema GS Genes | |
|---|---|---|---|
| Protein-coding genes | 27,206 | 64 | 67 |
| Retained At-α ohnologs | 6,038/22% | 36/56% | 35/52% |
| P value | 3.87E−09 | 1.26E−07 | |
| Tandem duplicates | 4,022/15% | 29/45% | 32/48% |
| P value | 5.71E−09 | 9.14E−10 | |
| GTDs | 3,879/14% | 17/27% | 18/27% |
| P value | 0.01066 | 0.07462 | |
| Sum duplicates | 12,132/45% | 61/95% | 65/97% |
| P value | 2.20E−16 | 2.20E−16 |
aFisher's exact test on count data.
bExtended set (fig. 1).
Moreover, 46% (31/67) of AabGS genes are organized in TARs (fig. 2F), compared with a 22% average for all protein-coding genes in Arabidopsis, thereby significantly surpassing the TAR coverage rate of 45% (29/64) observed for AtGS loci (fig. 2B and D, table 9).
For duplication by gene transposition, we detected 27% (17/67) of affected AabGS loci (fig. 2). In summary, we found no significant enrichment of GTD events among GS pathway inventory in both species (table 9).
Discussion
The Aethionemae/Brassicaceae crown-group/sister-group lineages split about 30–60 Ma shortly after the last common WGD event and independently evolved ever since (Couvreur et al. 2010; Mithen et al. 2010; Schranz et al. 2011). Notably, the radiation process evident for the Brassicaceae lineage created about 3,700 species (Couvreur et al. 2010). In contrast, the species-poor Aethionema lineage (Schranz et al. 2012) is well established as most ancient Brassicaceae extant sister and may therefore possess a more “ancient” genome organization when compared with Arabidopsis. This facilitates the recognition and quantification of common factors underlying rapid innovation of complex traits shared by both species. We exploit novel genomics resources for evolutionary analysis of the complete GS pathway inventory in both A. thaliana and Aet. arabicum to utilize the impact of different kinds of duplication classes to diversification of plant secondary metabolites. In a comparative genomics approach, we employ the phylogenetic relation of Aet. arabicum and A. thaliana to identify key factors driving GS pathway divergence. In this context, we establish GSs genetics/genomics as a scaffold to incorporate further phenotypic data for better understanding the impact of duplication to rapid evolution of novel key traits. In Arabidopsis, several GS genes retained duplicate gene copies dating back to the last WGD event but lacking annotation to GS metabolic processes (fig. 1). Illustrating high degrees of protein similarities among these ohnolog copy pairs and/or similar responses in gene regulation following GS pathway induction (tables 5 and 6), we identified 12 novel putative Arabidopsis genes associated to GS biosynthesis (figs. 1 and 3). Given the fact that these loci remained unknown despite their putative relevance for an experimentally very well-studied trait-like GS biosynthesis, we highlight the importance of considering ohnolog copies when analyzing a plethora of other highly diverged multigene pathways (i.e., terpenoid biosynthesis). We thereby provided an easy-to-follow framework on how to use existing data on WGD in Arabidopsis to better understand the networks of functional redundancy, especially involving genes that are targeted for knock-out experiments in functional studies.
Evolutionary analysis of homologous GS loci in Arabidopsis and Aethionema found a majority (all but two) comprising duplicate groups organized in multigene families (figs. 2 and 3). This underlined the dominant role of duplication for creation and expansion of biochemical diversity in plant (secondary) metabolism.
Clear orthologs of seven Arabidopsis GS genes are absent in the Aethionema draft genome (due to three Aethionema-specific GS gene losses and four Arabidopsis-specific TDs). Evolution of 10 additional Aethionema paralogs (two due to gene transposition and eight due to TD events, fig. 3) lead to an almost 100% conserved GS pathway inventory across the crown group/sister group system. This sheds light upon the relevance of genome plasticity for key trait maintenance despite of scattered gene losses. To test this hypothesis, we indicate the requirement of further research on additional multigene pathways in a deeper phylogenetic resolution. Identification of Aethionema GS gene homologs allowed confirming the increased frequency of duplicates in lineages that diverged more than 30–60 Ma. The absence of lineage-specific polyploidy events in either species facilitated the comparative analysis of genes duplicated due to the common ancient WGD events (particularly At-α) as well as lineage-specific gene tandem and transposition duplications. Partitioning the duplicate genes set in GS pathway inventory revealed significant enrichments of retained At-α ohnologs and tandem duplicates (but not GTD events) in both species compared to the average observed for protein-coding genes in Arabidopsis (table 2). We therefore conclude that WGD and TD facilitated the early and continued evolution of GS biosynthesis in the mustard family. To our knowledge, this is the first study providing distinct indications on a genetics level for the connection of WGD to the emergence of key traits in planta.
Various duplicates of different GS gene families code for proteins encoding functions in consecutive steps of GS biosynthesis (Kliebenstein et al. 2001; Hansen et al. 2007). Among GS biosynthetic and regulatory genes, pairs of retained At-α ohnolog duplicates in distant genomic location further expand to TARs (fig. 3). In Arabidopsis, the S-oxygenase activity FMO is provided by a pair of retained ohnologs on distant arms on chromosome 1 (figs. 3 and 4). Both copies evolved further tandem duplicates with different substrate specificities (Li et al. 2008). Different groups of substrates are products of SOT-type sulfotransferases provided by another retained ohnolog pair on At1 with additional TD copies sharing annotation to GS production (Bowers et al. 2003; Piotrowski et al. 2004) (fig. 3). The reaction delivering substrates for GS SOT-type sulfotransferases is catalyzed by UGT-type proteins, likewise encoded by a pair of retained ohnologs that evolved multiple tandem and gene transposition duplicates in both Aethionema and Arabidopsis (fig. 3). It is thus inferred that subfunctionalization of both TD and retained At-α ohnolog pairs caused functional diversification of GS biosynthetic and regulatory elements. Showing mutual influence of ohnolog retention and TD rate across a crown group–sister group system, we describe a complex network of gene duplication fostering the expansion of a composite trait, thereby contributing to the means of mutation and selection to create evolutionary innovation in a limited timeframe. Evidence of the model of evolution by gene duplication can be found in comparative GS pathway analysis. Thus, GS may provide a framework for investigating the expansion of complex traits.
Supplementary Material
Supplementary figures S1 and S2 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
I am grateful for the input and support of Erik van den Bergh, Nicole van Dam, Mike Freeling, Tom Mitchell-Olds, Benjamin Schweßinger, Cyril Zipfel, and Martin Parniske. Thanks to Chiara Vercesi for her assistance with graphical editing of the figures. Likewise, I want to acknowledge the contributions of two anonymous reviewers. This work was supported by a Netherlands Organization for Scientific Research (NWO) Ecogenomics grant to M.E.S.
Literature Cited
- Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- Beekwilder J, et al. The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS One. 2008;3:e2068. doi: 10.1371/journal.pone.0002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellieny-Rabelo D, Oliveira AE, Venancio TM. Impact of whole-genome and tandem duplications in the expansion and functional diversification of the F-box family in legumes (Fabaceae) PLoS One. 2013;8:e55127. doi: 10.1371/journal.pone.0055127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bones AM, Rossiter JT. The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry. 2006;67:1053–1067. doi: 10.1016/j.phytochem.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Celenza JL, et al. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell. 2013;25:2813–2830. doi: 10.1105/tpc.113.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J-J, Choi YD. Methyl jasmonate as a vital substance in plants. Trends Genet. 2003;19:409–413. doi: 10.1016/S0168-9525(03)00138-0. [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur TL, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci U S A. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Freeling M, et al. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Muller C, Flugge UI. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, et al. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, et al. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell. 2009;21:1813–1829. doi: 10.1105/tpc.109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 2007;50:902–910. doi: 10.1111/j.1365-313X.2007.03101.x. [DOI] [PubMed] [Google Scholar]
- Hansen BG, et al. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008;148:2096–2108. doi: 10.1104/pp.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry. 2007;68:2831–2846. doi: 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Haudry A, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45:891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Kelleher MO, Eggleston IM. The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur J Nutr. 2008;47(2 Suppl):73–88. doi: 10.1007/s00394-008-2009-8. [DOI] [PubMed] [Google Scholar]
- He Y, Chen B, Pang Q, Strul JM, Chen S. Functional specification of Arabidopsis isopropylmalate isomerases in glucosinolate and leucine biosynthesis. Plant Cell Physiol. 2010;51:1480–1487. doi: 10.1093/pcp/pcq113. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Clauss MJ, Kroymann J, Savolainen O, Mitchell-Olds T. Natural variation in MAM within and between populations of Arabidopsis lyrata determines glucosinolate phenotype. Genetics. 2006;173:1629–1636. doi: 10.1534/genetics.106.056986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci U S A. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R, Ekollu V, Rose JR. Non-random association of transposable elements with duplicated genomic blocks in Arabidopsis thaliana. Mol Phylogenet Evol. 2003;29:410–416. doi: 10.1016/s1055-7903(03)00262-8. [DOI] [PubMed] [Google Scholar]
- Irish VF, Litt A. Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev. 2005;15:454–460. doi: 10.1016/j.gde.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kane J, Freeling M, Lyons E. The evolution of a high copy gene array in Arabidopsis. J Mol Evol. 2010;70:531–544. doi: 10.1007/s00239-010-9350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One. 2008;3:e1838. doi: 10.1371/journal.pone.0001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci U S A. 2003;100(2 Suppl):14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA. Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 2008;148:1721–1733. doi: 10.1104/pp.108.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Malacarne G, et al. Deconstruction of the (paleo)polyploid grapevine genome based on the analysis of transposition events involving NBS resistance genes. PLoS One. 2012;7:e29762. doi: 10.1371/journal.pone.0029762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R, Bennett R, Marquez J. Glucosinolate biochemical diversity and innovation in the Brassicales. Phytochemistry. 2010;71:2074–2086. doi: 10.1016/j.phytochem.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Yoshida R, Shimada N, Yamazaki H, Yokoi T. Inhibition and inactivation of human cytochrome P450 isoforms by phenethyl isothiocyanate. Drug Metab Dispos. 2001;29:1110–1113. [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, et al. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003;133:63–72. doi: 10.1104/pp.102.019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer Publishing Group; 1970. p. 160. [Google Scholar]
- Parniske M, et al. Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol Plant Microbe Interact. 1999;12:93–102. doi: 10.1094/MPMI.1999.12.2.93. [DOI] [PubMed] [Google Scholar]
- Pfalz M, Vogel H, Kroymann J. The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell. 2009;21:985–999. doi: 10.1105/tpc.108.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M, et al. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem. 2004;279:50717–50725. doi: 10.1074/jbc.M407681200. [DOI] [PubMed] [Google Scholar]
- Prasad KV, et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, et al. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:e115. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman J. Parallel evolution of glucosinolate biosynthesis inferred from congruent nuclear and plastid gene phylogenies. Am J Bot. 1998;85:997. [PubMed] [Google Scholar]
- Rodman JE, Karol KG, Price RA, Sytsma KJ. Molecules, morphology, and Dahlgren's expanded order capparales. Syst Bot. 1996;21:289–307. [Google Scholar]
- Roth C, et al. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J Exp Zool B Mol Dev Evol. 2007;308:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Sawada Y, et al. Omics-based approaches to methionine side chain elongation in Arabidopsis: characterization of the genes encoding methylthioalkylmalate isomerase and methylthioalkylmalate dehydrogenase. Plant Cell Physiol. 2009;50:1181–1190. doi: 10.1093/pcp/pcp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Edger PP, Pires JC, van Dam NM, Wheat CW. Comparative genomics in the Brassicales: ancient genome duplications, glucosinolate diversification and pierinae herbivore radiation. In: Edwards D, Batley J, Parkin I, Kole C, editors. Genetics, genomics and breeding in crop plants. Boca Raton (FL): CRC press; 2011. pp. 206–218. [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr Opin Plant Biol. 2012;15:147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Sonderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Stehle F, Brandt W, Schmidt J, Milkowski C, Strack D. Activities of Arabidopsis sinapoylglucose: malate sinapoyltransferase shed light on functional diversification of serine carboxypeptidase-like acyltransferases. Phytochemistry. 2008;69:1826–1831. doi: 10.1016/j.phytochem.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Tang H, Lyons E. Unleashing the genome of Brassica rapa. Front Plant Sci. 2012;3:172. doi: 10.3389/fpls.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, de Kraker JW, Hause B, Gershenzon J, Tokuhisa JG. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007;144:60–71. doi: 10.1104/pp.106.091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D, Rappaport F, Simon M, Loudet O. Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000945. doi: 10.1371/journal.pgen.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Glucosinolate biosynthetic genes in Brassica rapa. Gene. 2011a;487:135–142. doi: 10.1016/j.gene.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 2013;9:e1003255. doi: 10.1371/journal.pgen.1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011b;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS One. 2011c;6:e28150. doi: 10.1371/journal.pone.0028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Buchmann JP, Keller B. Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 2010;20:1229–1237. doi: 10.1101/gr.107284.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor AJ, et al. Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae) Phytochemistry. 2005;66:1321–1333. doi: 10.1016/j.phytochem.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Wittstock U, Kliebenstein DJ, Lambrix V, Reichelt M, Gershenzon J. Glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores. In: Romeo JT, editor. Integrative phytochemistry: from ethnobotany to molecular ecology. Amsterdam (The Netherlands): Elsevier; 2003. pp. 101–126. [Google Scholar]
- Woodhouse MR, Tang H, Freeling M. Different gene families in Arabidopsis thaliana transposed in different epochs and at different frequencies throughout the rosids. Plant Cell. 2011;23:4241–4253. doi: 10.1105/tpc.111.093567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.