Abstract
Over the past 40 years, the incidence and prevalence of respiratory diseases have increased significantly throughout the world, damaging economic productivity and challenging health care systems. Current diagnoses of different respiratory diseases generally involve invasive sampling methods such as induced sputum or bronchoalveolar lavage that are uncomfortable, or even painful, for the patient. In this paper, we present a platform incorporating fiber-optic bundles and antibody based microarrays to perform multiplexed protein profiling of a panel of six salivary biomarkers for asthma and cystic fibrosis (CF) diagnosis. The platform utilizes an optical fiber bundle containing approximately 50,000 individual 4.5 μm diameter fibers that are chemically etched to create microwells in which modified microspheres decorated with monoclonal capture antibodies can be deposited. Based on a sandwich immunoassay format, the array quantifies human vascular endothelial growth factor (VEGF), interferon gamma-induced protein 10 (IP-10), interleukin 8 (IL-8), epidermal growth factor (EGF), matrix metalloproteinase 9 (MMP-9), and interleukin 1 beta (IL-1β) salivary biomarkers in the sub-picomolar range. Saliva supernatants collected from 291 individuals (164 asthmatics, 71 CF patients, and 56 healthy controls (HC)) were analyzed on the platform to profile each group of patients using this six-analyte suite. It was found that four of the six proteins were observed to be significantly elevated (p<0.01) in asthma and CF patients compared with HC. These results demonstrate the potential to use the multiplexed protein array platform for respiratory disease diagnosis.
Keywords: Saliva analysis, protein microarray, asthma, cystic fibrosis, fiber-optic bundle, multiplexed fluorescent detection, microsphere-based microarrays
Introduction
Saliva has potential to be a preferred diagnostic fluid compared with traditional samples such as serum, urine, or tissue biopsy, as its collection is noninvasive and requires minimal training.1-4 The traditional samples used in the study and monitoring of respiratory diseases such as asthma and cystic fibrosis (CF) are induced sputum5,8 or bronchoalveolar lavage (BAL)9,12 fluids. However, the collection of these fluids involves invasive sampling procedures, which are uncomfortable and preclude the possibility of frequent sampling in the same individual.13,14 Due to the direct anatomic relationship between the oral cavity and respiratory system, saliva has been proposed as a noninvasive alternative biospecimen for respiratory diseases.15,18 Furthermore, saliva is an ideal specimen for diagnosis or monitoring of diseases that require repeated sampling, considering its high daily secretion volume (up to 1.5 L).2 It is also a preferred diagnostic biospecimen compared to urine and blood, due to the convenience and comfort of sampling.19
Asthma is a chronic disease characterized by recurrent attacks, inflammation, and airflow limitation.20-23 It can be atopic (triggered by environmental allergens) or non-atopic and varies in severity and frequency between patients.24,25 Asthma affects more than 300 million people worldwide and causes 250,000 deaths annually—most of the deaths are avoidable. Asthma is also the leading cause of chronic disease in children.26 CF is a chronic obstructive pulmonary disease that affects both the lungs and the digestive system and may result in premature death.27,28 CF is an inherited autosomal recessive disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene located on the long arm of human chromosome 7.29 As of 2009, there were more than 30,000 CF patients in the United States, with 1,000 new cases reported annually.30 Asthma and CF are heterogeneous diseases with diverse pathologies and phenotypes. Currently physicians’ diagnostic tools for these conditions are limited to conventional pulmonary function tests.31 However, specific biomarkers from saliva specimens may enable physicians to tailor a detailed diagnosis and point-of-care treatment plan to individual patients.
Quantitative protein profiling has become a crucial tool for the discovery of new biomarkers, including respiratory diseases.32 Multiplexed proteomics platforms commonly combine different analytical techniques for pre-fractionation, multidimensional separation, and detection. Single-plex immunoassays such as enzyme linked immunosorbent assay (ELISA) are of course the mainstay of quantitative protein analysis, but for the past 15 years they have been supplemented with the use of protein microarrays.33,35 Major advantages of antibody-based microarrays include the ability to analyze multiple proteins simultaneously with reduced reagent consumption, high throughput, cost effectiveness, and simplified labor. In particular, multiplexed protein microarrays based on established microsphere-based technology have grown rapidly, with multiple platforms that have their roots in two main array classes: suspension arrays, such as the Luminex® system36 and randomly ordered arrays, such as the Illumina BeadChip® system, although to date the latter has been used exclusively for nucleic acids.37
Suspension arrays have been broadly used to simultaneously analyze hundreds of proteins in clinical applications with demonstrated reproducibility, good dynamic range, and excellent acquisition rates.38-40 However, they appear to be more sensitive than randomly ordered arrays to altered levels of undesired circulating proteins and inhibitors, because all reactions take place with the microspheres floating freely in the sample.41 In addition, readout devices for suspension arrays are usually expensive, whereas randomly ordered arrays have an inherent ability to be adapted to point-of-care diagnosis systems due to their ease of adaptation to cheap, portable, and battery-powered imaging systems.42,44 For these reasons, we previously created as a proof of concept a multiplexed protein microarray based on fiber-optic bundles and microsphere-based antibody arrays for the detection of salivary cytokines.45
Here we report the optimized clinical application of the optical fiber array platform, implemented by refining our candidate biomarker list to a six-analyte panel that is informative for asthma and CF diagnosis. The encoding, coupling, and assay protocols were optimized for better sensitivity. Quality assessment and data handling were improved to preserve both sensitivity and specificity. The limits of detection (LODs) were improved 13 to ~1500 times compared to our previous report.45 A total of 291 saliva samples collected from asthmatics and CF patients, as well as healthy control individuals (HC), have been tested. The large number of samples ensures the results are statistically representative. Analysis of the results demonstrates the potential to use saliva as a valuable specimen for both research and diagnosis of asthma and CF.
Experimental Section
Materials
Fiber-optic bundles containing approximately 50,000 individual optical fibers (with diameters of 4.5 μm) were purchased from SCHOTT North America, Inc. (Elmsford, NY). Carboxyl functionalized polystyrene microspheres (containing 5.5% divinylbenzene and 5% methacrylic acid) with an average diameter of 4.5 μm were ordered from Bangs Laboratories, Inc. (Fishers, IN). Phosphate buffered saline (PBS, 10× concentrated), molecular biology grade water, tetrahydrofuran (THF), coumarin 30 (C30), methanol, sodium dodecyl sulfate solution (SDS, 20%), and Tween 20 (T20) were purchased from Sigma-Aldrich (St. Louis, MO). Tris[4,4,4-trifluoro-1-(2-thienyl)-1,3-butanediono]europium (III) (Eu-TTA) and sodium hydroxide solution (5N/certified) were obtained from Fisher Scientific (Pittsburgh, PA). PBS Protein-free blocking buffer (PBSPF), PBS StartingBlock T20 blocking buffer (PBSS), TBS StartingBlock blocking buffer (TBSS), Blocker bovine serum albumin (BSA) (10% w/v in PBS), 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC), BupH 2-(N-morpholino)ethanesulfonic acid (MES) buffered saline, and Sulfo-N-hydroxysulfosuccinimide (sulfo-NHS) were ordered from Thermo Scientific (Rockford, IL). Streptavidin R-phycoerythrin conjugate (SARPE, premium grade) was purchased from Invitrogen (Grand Island, NY).
Fiber-optic bundles were imaged with an epi-fluorescence microscope (Olympus America Inc., Center Valley, PA). The microscope is equipped with a 75W xenon lamp, iKon-L CCD camera (Andor, South Windsor, CT), and fluorescence filter sets (Chroma, Bellos Falls, VT) for Eu-TTA (excitation 365/10, dichroic 525DCLP, emission 620/60), C30 (excitation 350/50, dichroic 400DCLP, emission 460/50), and SARPE (excitation 546/10, dichroic 560DCLP, emission 580/30).
All antibodies and recombinant human proteins used in this work were purchased from R&D Systems (Minneapolis, MN). Monoclonal mouse antibodies were used as capture antibodies for human VEGF (clone no. 26503), IP-10 (clone no. 33036), IL-8 (clone no. 6217), EGF (clone no. 10827), MMP 9 (clone no. 36020), and IL-1β (clone no. 2805). Mouse IgG isotype control antibody (clone no. 11711) was chosen as a negative control, as recommended by the manufacturer. Purified recombinant human proteins and biotinylated polyclonal goat antibodies were used as standards for microarray characterizations and detection antibodies, respectively.
Microsphere Encoding
The microspheres were encoded with Eu-TTA and C30 dyes as follows (Figure S1a): 60 μL of microsphere stock solution (containing approximately 6 mg, 1.17 × 109 solid microspheres) was washed by centrifugation/resuspension with 600 μL of 1× PBS three times and then with 600 μL of THF three times. The microspheres were suspended in 600 μL of THF containing different concentrations of dyes and incubated on a M3 shaker (IKA, Wilmington, NC) at 3,000 rpm for 24 h at room temperature (RT), protected from light. Afterwards, the microspheres were washed with 600 μL of methanol six times and then with 600 μL of 1× PBS containing 0.01% T20 (PBST) six times. The microspheres were finally re-suspended in 600 μL of PBST, stored at 4 °C and protected from light before antibody coupling.
Microsphere Coupling with Capture Antibodies
The encoded microspheres were coupled with different capture antibodies as follows (Figure S1b): 200 μL of encoded microsphere solution (containing approximately 2 mg microspheres) was washed with 500 μL of MES buffer (0.1 M MES, 0.9 % NaCl, 0.01% SDS, pH 5.7) three times. The microspheres were then activated in 1 mL of MES buffer containing 15.8 mg EDC and 24.4 mg sulfo-NHS for 4 h at RT in the dark. After activation, the microspheres were washed with 500 μL of PBS containing 0.01 % SDS (PBSSDS) three times. The microspheres were then incubated in 500 μL of PBSSDS buffer containing 60 μg of capture antibody for 4 h at RT in the dark. After incubation, the microspheres were washed three times with 500 μL of TBS StartingBlock buffer (TBSS), and blocked in 1 mL of TBSS buffer for 1 h at RT in the dark. After washing with 500 μL of TBSS buffer three times, the microspheres were stored in 100 μL of TBSS buffer at 4 °C, protected from light. In our experience, the coupled microspheres are stable for more than five months without detectable loss of response signals.
Microarray Fabrication
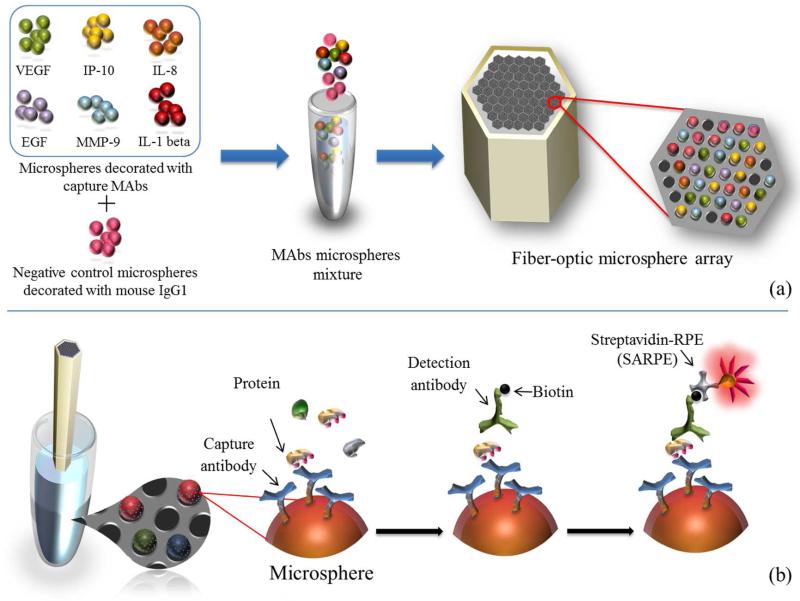
The workflow of the microarray fabrication is depicted in Figure 1a. Fiber-optic bundles were cut to ~5 cm pieces and both ends were polished sequentially using a fiber polisher (Allied High Tech, Ranch Dominguez, CA), using 30, 15, 9, 6, 3, 1, 0.5, and 0.05 μm-sized diamond lapping films (Allied High Tech). One end of the fiber was etched in 0.025 N hydrochloric acid (HCl) for 150 s to form microwells with a depth of approximately 3.5 μm. The etched end was rinsed for 1 min, sonicated in deionized water for 1 min and subsequently blocked in 400 μL of PBSPF buffer for 1 h. Seven types of antibody modified microsphere solutions (~100 μL of each solution) were mixed and concentrated to 200 μL by removing the suspension buffer. A 1 μL droplet of the microsphere mixture (containing approximately 2 × 106 solid microspheres) was loaded onto the etched end of the fiber bundle and kept in the dark for 15 min; the volume of the microsphere solution decreased by evaporation and the microspheres were loaded into the etched microwells. Almost all of the microspheres will remain in the microwells through the entire assay procedure. The process was repeated a second time to increase the loading efficiency (i.e., the percentage of microwells loaded with one microsphere over the total number of microwells in the array). The typical loading efficiency is between 10% and 45% (1000–4500 microspheres in the area of view) in our experiments. To remove excess microspheres, the proximal end of the loaded fiber bundle was wiped by a swab saturated with the sample which going to be analyzed.
Figure 1.
Workflow of the multiplexed protein microarray. (a) Microarray assembly process: seven different types of microspheres (including one negative control) were mixed and loaded into the microwells on the etched fiber-optic bundle. (b) Saliva analysis: the assembled microarray was incubated sequentially with the saliva sample, a mixture of different biotinylated detection antibodies, and SARPE dye.
Protein Microarray Assay
The workflow of the assay is depicted in Figure 1b. Protein quantification was based on a sandwich assay format where the capture antibody and detection antibody were simultaneously bound to different epitopes on the antigen (target protein). The freshly assembled protein microarray was immediately incubated in 200 μL of two-fold diluted saliva sample (100 μL saliva diluted with equal volume of PBSS buffer) for 2 h at 600 rpm. After the incubation, the fiber was dipped in a microcentrifuge tube containing 200 μL of washing buffer (1× PBS containing 0.1% BSA and 0.1% T20) and washed by shaking at 600 rpm for 2 min. The washing step was repeated three times. The microarray was then incubated in 100 μL of biotinylated detection antibody mixture (5 μg/mL each of all six detection antibodies in PBSS buffer) for 30 min and then washed three times with 200 μL of washing buffer. The microarray was finally incubated in 200 μL of 20 μg/mL streptavidin conjugated fluorophore (SARPE) in 1× PBS for 10 min and then washed with 200 μL of washing buffer five times. The microarray was protected from light throughout the analysis. The fiber bundle was dried with compressed air and the polished end without microspheres was cleaned with a swab saturated in absolute ethanol.
Microarray Imaging and Data Analysis
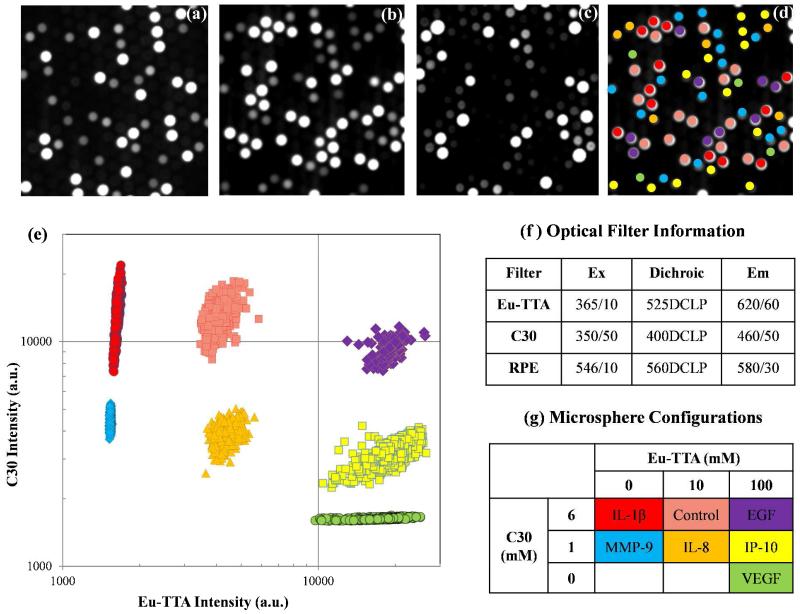
The fiber-optic bundle was placed onto a home built fiber holder and imaged on an upright epi-fluorescence microscope equipped with a 16-bit CCD camera. The fiber was imaged at the distal end sequentially in the Eu-TTA, C30, and SARPE channels. Images of a representative portion of a fiber bundle from each channel are shown in Figure 2a-c. Images were analyzed using a custom designed algorithm in MATLAB R2009b (MathWorks, Inc., Natick, MA); the decoding results are shown in Figure 2d. After the microspheres were categorized, the tri-mean of signals from all microspheres of each type were calculated (a minimum of 25 microspheres of each type was required) and adjusted by subtracting the signal from the control microspheres. The detailed analysis process is described in the Supplemental Information.
Figure 2.
A set of fluorescent images and decoding results for seven microsphere types. (a) EuTTA encoding image, (b) C30 encoding image, (c) SARPE signal image, and (d) decoding results of a small portion of one fiber-optic bundle; (e) decoding results of seven microsphere types, (f) optical filter information, and (g) the microsphere configurations.
Microarray Characterization
For calibrations, recombinant protein standards at different concentrations were mixed and tested by the microarray, and the normalized responses were plotted using a 4-parameter logistic regression (4-PL) model (eq. 1):46
| (eq. 1) |
where D is the estimated response at infinite concentration of the analyte, A is the estimated response at zero concentration of the analyte, x is the concentration of the analyte, B represents the slope of the curve at the inflection point, and C (IC50) is the midrange concentration. Data were analyzed using Sigma Plot (Systat Software Inc.).
Saliva Sample Collection and Preparation
The saliva samples were collected at Boston Medical Center and Children’s Hospital Boston from 291 study participants and kept at −80 °C until they were analyzed at Tufts University. The detailed protocols for sample collection and processing have been described previously.45 Previous studies on protein stability of saliva suggest that our collection and storage protocols would have a minimal impact on the analyte concentrations.47,49 For saliva analysis, frozen saliva supernatants were warmed to RT and diluted with an equal volume of PBSS buffer to decrease the sample viscosity and potential matrix effects.
Results and discussion
Salivary Protein Biomarker Selection
Numerous proteins play a key role in the chronic inflammation of pulmonary disorders50-52 and developing and refining a working protein panel for diagnosis is not straightforward. In our previous work, we screened 72 salivary cytokine mediators using commercial kits, and refined the candidate list to 10 proteins that appeared have a potential association with asthma severity.45 For the present clinical study, we built a six-plex panel including four of these 10 salivary cytokines, as well as MMP-9 and IL-1β. MMP-9 is one of the major proteinases involved in airway inflammation processes,53 while IL-1β is one of the most important mediators of the inflammatory response.54
Microsphere Encoding
The encoding protocol has been thoroughly optimized to achieve higher accuracy of decoding. The microspheres were suspended in THF to make the polymer matrix swell, and different proportions of Eu-TTA and C30 were loaded into the swollen microspheres, providing each microsphere type with a unique identifier. After incubation, the THF was replaced by water, which de-swelled the microspheres so that the encoding dyes were physically trapped in the polymer matrix. In our current protocol, we used three concentrations of both Eu-TTA and C30. By varying the combination of these two dyes, we can prepare eight different encodings to identify different types of microspheres (only seven types were used in this study, Figure 2g).
We have made several additional improvements to the encoding protocol. First, the incubation time was increased to 24 h for more homogeneous trapping of encoding dyes. Second, T20 was added to the wash buffer to enable better suspension of the microspheres, which helped to remove any excess dyes on the microspheres’ surface. Furthermore, the coumarin dye used in the previous study (i.e., 7-amino-4-methylcoumarin)45 was replaced with a much brighter analog, C30. The decoding process also employed a stricter classification rule, and any hard-to-categorize microspheres were excluded from the analysis. The detailed decoding process is described in the Supplemental Information. With the current protocols, an overall encoding accuracy above 99% is achieved (Figure 2e). Encoded microspheres can be stored in the dark at 4 °C for more than five months without degradation of the encoding accuracy.
Microspheres Coupled with Capture Antibodies
The capture antibodies were coupled to the carboxyl microspheres through the amine groups on the lysine residues using a carbodiimide-mediated process. According to the manufacturer of the microspheres, 3 μg of antibody is required to form a monolayer on 1 mg of microspheres with an average diameter of 4.5 μm; this antibody amount is henceforth named 1×. The manufacturers state that the optimal antibody amount in the coupling solution may be up to 10×.55 To find the optimal amount for our particular antibodies, different batches of microspheres were coupled with antibody amounts of 1.5 μg (0.5×), 3 μg (1×), 6 μg (2×), 15 μg (5×), and 30 μg (10×). These microspheres were tested using the multiplexed sandwich assay, and the microspheres coupled with 30 μg of antibody resulted in the highest signal responses (see Figure S3 in the Supporting Information). This observation also suggests that the signal responses of the microspheres could be adjusted by controlling the amount of antibodies added to the coupling solution. This strategy would be useful for multiplexed detection of different proteins spanning a wide range of concentrations. The reproducibility of the coupling protocol was evaluated by multiplexed detection of 10 ng/mL MMP-9 using different batches of microspheres coupled by the exact same protocol. The results showed no statistical variance between different batches prepared by different researchers on different days (see Table S1 in the Supporting Information).
To minimize the effect of nonspecific binding during the immunoassay, negative control microspheres were included in every assay, and the measured signals for the active microspheres were calculated by subtracting the signals from the negative controls. The negative control microspheres were fabricated by coupling with mouse IgG isotype control antibody. This particular antibody is specifically designed as a negative control for direct ELISAs and has been confirmed to have no cross-reactivity with more than 40 common human proteins.56 The response on the control microspheres was defined as a baseline; thus, the response for each microsphere type was calculated by subtracting the signal intensity from that of the control microspheres.
Image Capturing and Data Analysis
The image capturing process and analysis algorithm were designed and optimized to enhance the reproducibility of the multiplexed analysis. We used a large format (2048 × 2048 pixels) 16-bit CCD camera for image acquisition.57 The 16-bit detection range (0 to 65,536) of the camera helps us achieve a broad dynamic range as well as high sensitivity in detecting analytes at very low concentrations. The larger format CCD chip helps to accommodate the required numbers of all microsphere types in the image.
In the subsequent data analysis, a custom-designed unsupervised analysis algorithm written in MATLAB was used to generate analysis reports automatically. To have statistical confidence in the data, we require that a minimum of 25 microspheres for each analyte must be identified in each experiment; otherwise the entire set of data will be excluded from analysis. To calculate the average signal intensity on each microsphere type, the conventional mean statistic is replaced by the more robust trimean.58 This protocol protects the population center against outliers, which are usually caused by mis-categorizing the encoding signals and can change the mean value significantly. The trimean is the weighted average of the distribution median and its two quartiles as defined below, where Q1, Q2, and Q3 denote the 25%, 50% and 75% points in a distribution, respectively (eq. 2).
| (eq. 2) |
Array Characterization
The performance of a multiplexed protein microarray strongly relies on limiting interference problems. The presence of interfering substances may either generate a large background signal or inhibit specific analyte binding to its complementary capture antibody, thereby decreasing both assay sensitivity and dynamic range.59 Based on these considerations, six independent blank tests were performed using PBSS buffer to evaluate the background signal generated during the assay. Negligible responses were observed from all six types of microspheres and no statistical difference was observed between different experiments (see Table 1). Moreover, the potential cross reactivities between antibodies and non-complementary proteins were also evaluated by testing the protein microarray with single protein standards. The normalized responses caused by cross-reactivity were less than 3% compared with those from complementary proteins (see Figure S4 in the Supporting Information). These results confirmed that the array-based assay exhibits minimal non-specific interactions between different antibodies and targeted proteins. This lack of cross-reactivity represents a major improvement over many commercially available multiplex arrays.41,59,60
Table 1.
Results for blank and reproducibility tests. The results for six independent blank tests (average and standard deviation (SD)) are listed in the top section of the table. One human saliva sample was tested independently three times; the intensities from these three detections are listed in the bottom table.
| Analyte | VEGF | IP-10 | IL-8 | EGF | MMP9-1 | IL-1β |
|---|---|---|---|---|---|---|
| Blank Test (n = 6) |
96 | 78 | 194 | 7.2 | −84 | −66 |
| 141 | 70 | 125 | 12.3 | −59 | −40 | |
| 132 | 47 | 173 | 28.6 | −83 | −42 | |
| 132 | 88 | 148 | 21.1 | −50 | −35 | |
| 138 | 63 | 206 | 13.6 | −40 | −35 | |
| 109 | 69 | 136.3 | 7.7 | −58 | −59 | |
| Average (a.u.) | 125 | 68 | 164 | 15 | −62 | −46 |
| SD (a.u.) | 18 | 14 | 33 | 8 | 18 | 13 |
|
| ||||||
| Saliva Supernatant (n = 3) |
4947 | 2474 | 6189 | 2338 | 356 | 1099 |
| 5491 | 2503 | 6996 | 2565 | 285 | 1081 | |
| 4614 | 2735 | 6432 | 3180 | 414 | 1034 | |
| Average (a.u.) | 5017.3 | 2570 | 6539 | 2694 | 352 | 10712 |
| SD (a.u.) | 442.5 | 143 | 414 | 436 | 65 | 33 |
| CV (%) | 9 | 6 | 6 | 16 | 18 | 3 |
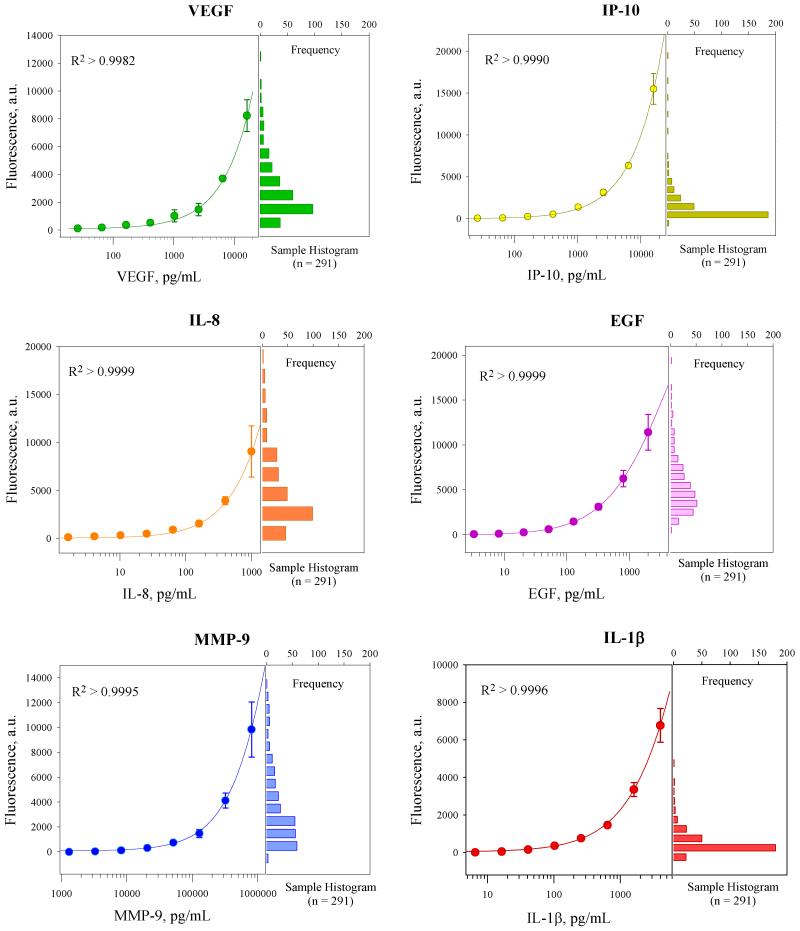
Multiplexed calibration curves for the optimized microarray were obtained by testing a series of protein standards prepared with recombinant proteins in PBSS buffer. Standards with a serial dilution of 2.5 at eight different concentrations were tested, which covered a three-log concentration range. Averages of the normalized responses of three independent detections at each concentration level are shown in Figure 3. The responses were modeled using 4-PL model and R2 values higher than 0.9982 were obtained for all six proteins, suggesting that multiplexed quantitative analysis of unknown samples can be performed using this platform.
Figure 3.
Six-plex multiplexed calibration curves built by protein standards in PBSS buffer and fit using 4-PL regression model. Histograms of responses from real human saliva samples are also featured on the right, suggesting that responses from most samples fall within the assay’s dynamic range.
The LODs for different proteins were calculated as the target protein concentration that corresponds to the mean fluorescence intensity of the blank measurements plus three times the standard deviation (10 times for the quantification limit, LOQ).61 The theoretical LODs for the six proteins ranged from 3 to 26 pg/mL for VEGF, IP-10, EGF, IL-8, IL-1β, and 1311 pg/mL for MMP-9 (details are listed in Table 2). The LODs were as good as or better than those from other microsphere-based multiplexed array systems or commercial kits.45,62,64 The LOQs for different proteins were calculated from their calibration curves and the highest concentrations extend even above the three orders of magnitude covered by the standards.
Table 2.
Characteristic parameters of the targeted proteins and their standard curves.
| Analyte | UniProtKB/ Swiss-Prot |
Molecular Weight (kDa) |
LOD (pg/mL) |
LOD (pM) |
Working Range (pg/mL) |
|---|---|---|---|---|---|
| VEGF | NP_001165097.1a | 19.2 | 6 | 0.3 | 202 – 16000b |
| IP-10 | P02778.2 | 8.7 | 26 | 3.0 | 99 – 16000b |
| IL-8 | P10145.1 | 8.0 | 4 | 0.6 | 24 – 1000b |
| EGF | P01133.2 | 6.0 | 3 | 0.5 | 6 – 10000b |
| MMP-9 | P14780.3 | 77 | 1311 | 17 | 1311 – 800000b |
| IL-1β | NP_000567.1a | 17 | 5 | 0.3 | 378 – 4000b |
NCBI Reference Sequence
the measurement range extends beyond the concentration of the standards.
Reproducibility Test using Saliva Samples
The assay reproducibility was also tested and characterized by the coefficient of variation (CV in %) for a saliva sample measured in three independent tests. Saliva samples collected from patients with respiratory diseases are sometimes very viscous and difficult to analyze by suspension microarrays. To decrease the viscosity of the sample, saliva supernatants were diluted with an equal volume of PBSS buffer before analysis. Previous studies in our lab have shown that matrix effects for twofold dilution are negligible (recovery rate of 80-120%).48 The average CV obtained for VEGF, IP-10, IL-8, and IL-1β ranged from 3% to 9%, whereas 16% and 18% were obtained for EGF and MMP-9, respectively (see Table 1). These results suggest that the assay is highly stable for measuring saliva samples. The CVs obtained are comparable to results from other studies using multiplexed microsphere based platforms for the same proteins (10 – 14%).65,66 In addition, significant differences in signal intensities were observed from saliva samples compared with blank tests (see Table 1), which demonstrated the potential suitability of this multiplexed platform for protein biomarker quantification in saliva samples.
Saliva Testing Results Analysis
A total of 291 human saliva samples were tested, which includes 164 samples collected from asthmatic patients, 71 from CF patients, and 56 HC. Concentrations of six protein biomarkers from each group were statistically analyzed and distinct patterns were observed in patients with asthma and CF compared to the HC group (Table 3). Protein concentrations in the different groups were compared using Student’s t-test. Patients with CF showed elevated levels of VEGF, IP-10, IL-8, and EGF compared with HC, while patients with asthma showed elevated levels of VEGF, IL-8, and EGF compared with HC. The p-test showed that all elevations were statistically very significant (p<0.01). The elevated levels of these inflammatory biomarkers suggest inflammation in the respiratory system, which agrees with our hypothesis. The elevated levels of these proteins also demonstrate the potential use of these biomarkers in respiratory diseases research and diagnosis.
Table 3.
Statistical results of the saliva samples from each group.
| Protein | Group | n | Mean | SD | p value |
|---|---|---|---|---|---|
|
VEGF (pg/mL) |
Asthma | 164 | 10764 | 9511 | 7.5 × 10−4* |
| CF | 71 | 9700 | 6784 | 1.1 × l0−3* | |
| HC | 56 | 6286 | 3940 | ||
|
| |||||
|
IP-10 (pg/mL) |
Asthma | 164 | 1532 | 2900 | 0.087 |
| CF | 71 | 4893 | 7332 | 8.1 × 10−5 | |
| HC | 56 | 838 | 1424 | ||
|
| |||||
|
IL-8 (pg/mL) |
Asthma | 164 | 1253 | 1239 | 2.6 × 10−3 |
| CF | 71 | 1542 | 1131 | 1.9 × 10−6 | |
| HC | 56 | 735 | 468 | ||
|
| |||||
|
EGF (pg/mL) |
Asthma | 164 | 1584 | 1452 | 8.4 × 10 −3 |
| CF | 71 | 1556 | 1039 | 4.8 × 10−3 | |
| HC | 56 | 1025 | 1028 | ||
|
| |||||
|
MMP-9 (ng/mL) |
Asthma | 164 | 544 | 516 | 0.65 |
| CF | 71 | 462 | 439 | 0.12 | |
| HC | 56 | 578 | 379 | ||
|
| |||||
|
IL-1β (pg/mL) |
Asthma | 164 | 395 | 708 | 0.28 |
| CF | 71 | 357 | 439 | 0.37 | |
| HC | 56 | 290 | 266 | ||
compared with data from HC group.
While obtaining the average concentrations of six protein concentrations for the three groups suggests their value, the real value of performing these protein assays ultimately depends on their utility for individual patients. Toward this end, the potential clinical applications of these results for diagnosing disease severity, cause, exacerbation and/or other aspects of asthma and CF were studied. With a questionnaire developed by our collaborators, more than 100 medical features for these saliva sample donors were also collected in the clinic, which include the patients’ disease history, disease control self-evaluations, clinical measurements performed by the attending physicians, the diagnosis and medical treatment by the clinicians, etc. Our current and future work includes more sophisticated analysis of the assay results and correlating them to different medical features along with their clinical significance.
Conclusion
In this paper, we described a fiber-optic based multiplexed protein microarray platform for profiling biomarkers in human saliva samples. Thorough optimizations have been performed for all aspects of the assay including microsphere encoding, antibody coupling, saliva sample analysis, image processing, and data analysis. The performance of the platform has been characterized using recombinant human proteins as well as with 291 human saliva samples collected from patients with respiratory diseases as well as healthy control individuals. The concentrations of protein biomarkers in these three groups showed significant differences, which demonstrated the possibility of using this platform as a tool for respiratory disease research. This platform is flexible and should be applicable to other protein biomarkers. In addition, the basic array design should make it possible to be transferred to other platforms such as microfluidic chips.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (grant 08UDE017788-05). Dr. Elena Benito-Peña also acknowledges support from the Spanish Foundation for Science and Technology (FECYT). We thank Dr. Kathryn Mayer for insightful discussions during the preparation of this manuscript.
Footnotes
Associated content
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- (1).Mandel ID. J. Oral Pathol. Med. 1990;19:119–125. doi: 10.1111/j.1600-0714.1990.tb00809.x. [DOI] [PubMed] [Google Scholar]
- (2).Streckfus CF, Bigler LR. Oral Dis. 2002;8:69–76. doi: 10.1034/j.1601-0825.2002.1o834.x. [DOI] [PubMed] [Google Scholar]
- (3).Kaufman E, Lamster IB. Crit. Rev. Oral Biol. Med. 2002;13:197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- (4).Chiappin S, Antonelli G, Gatti R, De Palo EF. Clin. Chim. Acta. 2007;383:30–40. doi: 10.1016/j.cca.2007.04.011. [DOI] [PubMed] [Google Scholar]
- (5).Fahy JV, Liu J, Wong H, Boushey HA. Am. Rev. Respir. Dis. 1993;147:1126–1131. doi: 10.1164/ajrccm/147.5.1126. [DOI] [PubMed] [Google Scholar]
- (6).Fahy JV, Wong H, Liu J, Boushey HA. Am. J. Respir. Crit. Care Med. 1995;152:53–58. doi: 10.1164/ajrccm.152.1.7599862. [DOI] [PubMed] [Google Scholar]
- (7).Keatings VM, Collins PD, Scott DM, Barnes PJ. Am. J. Respir. Crit. Care Med. 1996;153:530–534. doi: 10.1164/ajrccm.153.2.8564092. [DOI] [PubMed] [Google Scholar]
- (8).Saha S, Doe C, Mistry V, Siddiqui S, Parker D, Sleeman M, Cohen ES, Brightling CE. Thorax. 2009;64:671–676. doi: 10.1136/thx.2008.108290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Robinson DS, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley AM, Corrigan C, Durham SR, Kay AB. N. Engl. J. Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- (10).Walker C, Bode E, Boer L, Hansel TT, Blaser K, Virchow JC. Am. Rev. Respir. Dis. 1992;146:109–115. doi: 10.1164/ajrccm/146.1.109. [DOI] [PubMed] [Google Scholar]
- (11).Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DWH. Am. J. Respir. Crit. Care Med. 1995;151:1075–1082. doi: 10.1164/ajrccm/151.4.1075. [DOI] [PubMed] [Google Scholar]
- (12).Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. J. Pediatr. 2001;138:699–704. doi: 10.1067/mpd.2001.112897. [DOI] [PubMed] [Google Scholar]
- (13).Paggiaro PL, Chanez P, Holz O, Ind PW, Djukanovic R, Maestrelli P, Sterk PJ. Eur. Respir. J. 2002;37:3s–8s. doi: 10.1183/09031936.02.00000302. [DOI] [PubMed] [Google Scholar]
- (14).Pizzichini E, Pizzichini MMM, Leigh R, Djukanovic R, Sterk PJ. Eur. Respir. J. 2002;37:9s–18s. doi: 10.1183/09031936.02.00000902. [DOI] [PubMed] [Google Scholar]
- (15).Mangos JA, McSherry NR, Benke PJ. Pediatr. Res. 1967;1:436. doi: 10.1203/00006450-196711000-00002. [DOI] [PubMed] [Google Scholar]
- (16).Ryberg M, Moller C, Ericson T. J. Dent. Res. 1987;66:1404–1406. doi: 10.1177/00220345870660082401. [DOI] [PubMed] [Google Scholar]
- (17).Schmekel B, Ahlner J, Malmstrom M, Venge P. Respir. Med. 2001;95:670–675. doi: 10.1053/rmed.2001.1123. [DOI] [PubMed] [Google Scholar]
- (18).Gaber F, Daham K, Higashi A, Higashi N, Gulich A, Delin I, James A, Skedinger M, Gyllfors P, Nord M, Dahlen SE, Kumlin M, Dahlen B. Thorax. 2008;63:1076–1082. doi: 10.1136/thx.2008.101196. [DOI] [PubMed] [Google Scholar]
- (19).Koka S, Beebe TJ, Merry SP, DeJesus RS, Berlanga LD, Weaver AL, Montori VM, Wong DT. J. Am. Dent. Assoc. 2008;139:735–740. doi: 10.14219/jada.archive.2008.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Mullen JBM, Wright JL, Wiggs BR, Pare PD, Hogg JC. Br. Med. J. 1985;291:1235–1239. doi: 10.1136/bmj.291.6504.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wardlaw AJ, Dunnette S, Gleich GJ, Collins JV, Kay AB. Am. Rev. Respir. Dis. 1988;137:62–69. doi: 10.1164/ajrccm/137.1.62. [DOI] [PubMed] [Google Scholar]
- (22).Djukanovic R, Roche WR, Wilson JW, Beasley CRW, Twentyman OP, Howarth PH, Holgate ST. Am. Rev. Respir. Dis. 1990;142:434–457. doi: 10.1164/ajrccm/142.2.434. [DOI] [PubMed] [Google Scholar]
- (23).Beasley R, Keil U, von Mutius E, Pearce N, Ait-Khaled N, Anabwani G, Anderson HR, Asher MI, Bjorkstein B, Burr ML, Clayton TO, Crane J, Ellwood P, Lai CKW, Mallol J, Martinez FD, Mitchell EA, Montefort S, Robertson CF, Shah JR, Sibbald B, Stewart AW, Strachan DP, Weiland SK, Williams HC. Int Study Asthma Allergies, C. Lancet. 1998;351:1225–1232. [Google Scholar]
- (24).Rackemann FM. Am. J. Med. 1947;3:601–606. doi: 10.1016/0002-9343(47)90204-0. [DOI] [PubMed] [Google Scholar]
- (25).Cartier A, Thomson NC, Frith PA, Roberts R, Hargreave FE. J. Allergy Clin. Immunol. 1982;70:170–177. doi: 10.1016/0091-6749(82)90038-0. [DOI] [PubMed] [Google Scholar]
- (26).WHO [accessed Feb 3, 2013];Global serverillance, prevention and control of chronic respritatory disease: A comprehensive approach. http://www.who.int/gard/publications/GARD_Manual/en/index.html.
- (27).Andersen DH. Am. J. Dis. Child. 1938;56:344–399. [Google Scholar]
- (28).Collins FS. Science. 1992;256:774–779. doi: 10.1126/science.1375392. [DOI] [PubMed] [Google Scholar]
- (29).Ratjen F, Doring G. Lancet. 2003;361:681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- (30).Cystic fibrosis foundation [accessed May 9, 2013]; www.cff.org.
- (31).Wadsworth S, Sin D, Dorscheid D. J Asthma Allergy. 2011;4:77–86. doi: 10.2147/JAA.S15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Yamada N, Yuji R, Suzuki E. J Health Sci. 2009;55:682–688. [Google Scholar]
- (33).MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- (34).MacBeath G. Nat. Genet. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- (35).Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Trends Biotechnol. 2002;20:160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- (36).Luminex Corporation [accessed May 4, 2013]; www.luminexcorp.com.
- (37).Illumina [accessed May 3, 2013]; www.illumina.com.
- (38).Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Clin. Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- (39).Carson RT, Vignali DAA. J. Immunol. Methods. 1999;227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- (40).Yamamoto K, Ito S, Yasukawa F, Konami Y, Matsumoto N. Anal. Biochem. 2005;336:28–38. doi: 10.1016/j.ab.2004.09.030. [DOI] [PubMed] [Google Scholar]
- (41).Leng SX, McElhaney JE, Walston JD, Xie D, Fedarko NS, Kuchel GA. J Gerontol a-Biol. 2008;63:879–884. doi: 10.1093/gerona/63.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- (43).Weigl B, Domingo G, LaBarre P, Gerlach J. Lab Chip. 2008;8:1999–2014. doi: 10.1039/b811314a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Anal. Chem. 2012;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- (45).Blicharz TM, Siqueira WL, Helmerhorst EJ, Oppenheim FG, Wexler PJ, Little FF, Walt DR. Anal. Chem. 2009;81:2106–2114. doi: 10.1021/ac802181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Masseyeff R, Albert W, Staines NA. Methods of Immunological Analysis. 1st ed. Wiley; Blackwell: 1992. pp. 655–671. [Google Scholar]
- (47).Esser D, Alvarez-Llamas G, de Vries MP, Weening D, Vonk RJ, Roelofsen H. Biomark Insights. 2008;3:25–27. doi: 10.4137/bmi.s607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Blicharz TM. Ph.D. Dissertation. Tufts University; Medford, MA: 2009. Fiber-Optic Microsphere-Based antibody Arrays for use in Salivary Diagnostics. [Google Scholar]
- (49).Thomadaki K, Helmerhorst EJ, Tian N, Sun X, Siqueira WL, Walt DR, Oppenheim FG. J. Dent. Res. 2011;90:1325–1330. doi: 10.1177/0022034511420721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Holgate ST, Polosao R. Nat Rev Immunol. 2008;8:218–230. doi: 10.1038/nri2262. [DOI] [PubMed] [Google Scholar]
- (51).Barnes PJ. Respir Res. 2001;2:64–65. doi: 10.1186/rr39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Barnes PJ. J. Clin. Invest. 2008;118:3546–3556. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Lee KS, Min KH, Kim SR, Park SJ, Park HS, Jin GY, Lee YC. Am. J. Respir. Crit. Care Med. 2006;174:161–170. doi: 10.1164/rccm.200510-1558OC. [DOI] [PubMed] [Google Scholar]
- (54).March CJ, Mosley B, Larsen A, Cerretti DP, Braedt G, Price V, Gillis S, Henney CS, Kronheim SR, Grabstein K, Conlon PJ, Hopp TP, Cosman D. Nature. 1985;315:641–647. doi: 10.1038/315641a0. [DOI] [PubMed] [Google Scholar]
- (55).Bangslabs [accessed April 13, 2013]; http://www.bangslabs.com/sites/default/files/bangs/docs/pdf/205.pdf.
- (56).R&D sytems [accessed Apr 1, 2013]; http://www.rndsystems.com/Products/MAB002.
- (57).Andor Technology [accessed May 11, 2013]; http://www.andor.com/pdfs/specifications/Andor_iKon-L_936_Specifications.pdf.
- (58).Weisberg HF. Central Tendency and Variability. SAGE Publications; 1991. [Google Scholar]
- (59).Ellington AA, Kullo IJ, Bailey KR, Klee GG. Clin. Chem. 2010;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Wong J, Sibani S, Lokko NN, LaBaer J, Anderson KS. J. Immunol. Methods. 2009;350:171–182. doi: 10.1016/j.jim.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wild DG. The Immunoassay Handbook: Theory and applications of ligand binding, ELISA and related techniques. 4th ed. Elsevier Science; 2013. [Google Scholar]
- (62).Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP. Cytom Part B-Clin Cy. 2004;61B:35–39. doi: 10.1002/cyto.b.20021. [DOI] [PubMed] [Google Scholar]
- (63).Elshal MF, McCoy JP. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Trune DR, Larrain BE, Hausman FA, Kempton JB, MacArthur CJ. Hear. Res. 2011;275:1–7. doi: 10.1016/j.heares.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).duPont NC, Wang KH, Wadhwa PD, Culhane JF, Nelson EL. J. Reprod. Immunol. 2005;66:175–191. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Dossus L, Becker S, Achaintre D, Kaaks R, Rinaldi S. J. Immunol. Methods. 2009;350:125–132. doi: 10.1016/j.jim.2009.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.