SUMMARY
The mRNA translational control protein, Musashi, plays a critical role in cell fate determination through sequence-specific interactions with select target mRNAs. In proliferating stem cells, Musashi exerts repression of target mRNAs to promote cell cycle progression. During stem cell differentiation, Musashi target mRNAs are de-repressed and translated. Recently, we have reported an obligatory requirement for Musashi to direct translational activation of target mRNAs during Xenopus oocyte meiotic cell cycle progression. Despite the importance of Musashi in cell cycle regulation, only a few target mRNAs have been fully characterized. In this study, we report the identification and characterization of a new Musashi target mRNA in Xenopus oocytes. We demonstrate that progesterone-stimulated translational activation of the Xenopus Musashi1 mRNA is regulated through a functional Musashi binding element (MBE) in the Musashi1 mRNA 3′ untranslated region (3′ UTR). Mutational disruption of the MBE prevented translational activation of Musashi1 mRNA and its interaction with Musashi protein. Further, elimination of Musashi function through microinjection of inhibitory antisense oligonucleotides prevented progesterone-induced polyadenylation and translation of the endogenous Musashi1 mRNA. Thus, Xenopus Musashi proteins regulate translation of the Musashi1 mRNA during oocyte maturation. Our results indicate that the hierarchy of sequential and dependent mRNA translational control programs involved in directing progression through meiosis are reinforced by an intricate series of nested, positive feedback loops, including Musashi mRNA translational autoregulation. These autoregulatory positive feedback loops serve to amplify a weak initiating signal into a robust commitment for the oocyte to progress through the cell cycle and become competent for fertilization.
INTRODUCTION
The mRNA translational control protein Musashi plays a critical role in promoting physiological stem cell self-renewal, and has been implicated in the development and progression of neural, colon, breast, and hematopoietic cancers (Kanemura et al., 2001; Toda et al., 2001; Hemmati et al., 2003; Okano et al., 2005; Sureban et al., 2008; Ito et al., 2010; Kharas et al., 2010; Wang et al., 2010). In mammalian stem and progenitor cells, Musashi represses translation of the mRNA encoding Numb, an inhibitor of Notch, Hedgehog and p53 signalling pathways, and of the mRNA encoding p21WAF, an inhibitor of cyclin-dependent kinases (Imai et al., 2001; Jafar-Nejad et al., 2002; Le Borgne et al., 2005; Battelli et al., 2006; Di Marcotullio et al., 2006; Colaluca et al., 2008; Ito et al., 2010; Kharas et al., 2010). By repressing the translation of these inhibitory proteins, Musashi functions to maintain cells in an undifferentiated state capable of self-renewal. The de-repression and translation of Numb and p21WAF mRNAs induces cell differentiation (Yan and Ziff, 1995; Erhardt and Pittman, 1998; Hughes et al., 2000; Battelli et al., 2006; Ito et al., 2010). In fully differentiated cells, Musashi protein levels are very low, suggesting that target mRNAs are de-repressed simply through the loss of Musashi protein (Sakakibara and Okano, 1997). Analysis of the kinetics of target mRNA de-repression, however, indicates that de-repression precedes loss of Musashi protein (MacNicol et al., 2011). The molecular basis for this functional switch in mammalian Musashi activity has not yet been elucidated.
In immature, stage-VI Xenopus laevis oocytes, a number of mRNAs encoding proteins necessary for maturation are repressed and are subsequently translationally activated in response to progesterone-induced meiotic cell cycle resumption (Radford et al., 2008). We recently demonstrated that, in addition to a role in stem cell self-renewal, Musashi is critical for the meiotic maturation of Xenopus oocytes, where it functions to promote rather than repress target mRNA translation (Charlesworth et al., 2006; Arumugam et al., 2010). Hormone-stimulated polyadenylation and translational activation of Xenopus maternal mRNAs occurs in a distinct, temporal pattern and are classed as “early” (prior to germinal vesicle (nuclear) breakdown (GVBD), e.g. Mos) or “late” (coincident with, or after,GVBD e.g. cyclin B1; Ballantyne et al., 1997). The strict temporal order of early- and late-class mRNA translation is mediated through mRNA-binding proteins that are sequentially activated by progesterone-stimulated signal transduction pathways, and are directed to short sequence elements that are generally located within the target mRNA 3′ untranslated region (3′ UTR; MacNicol and MacNicol, 2010). Musashi function is essential to mediate progesterone-dependent, early-class mRNA translational activation whereas the cytoplasmic polyadenylation element binding protein (CPEB) directs late-class mRNA translation (Charlesworth et al., 2006; Arumugam et al., 2010). Interestingly, Musashi function is required for progesterone-dependent CPEB activation (Charlesworth et al., 2006; Arumugam et al., 2010), consistent with prior observations that late-class mRNAs were dependent upon prior translational activation of early-class mRNAs (Ballantyne et al., 1997; de Moor and Richter, 1997).
Musashi has been shown to regulate the translation of target mRNAs through a Musashi binding element (MBE) within the 3′ UTR of transcripts. The general consensus sequence of the MBE is (G/A)U1–3AGU (Imai et al., 2001), although tolerance for substitution of either flanking nucleotide has been reported (Ohyama et al., 2012). Musashi plays an evolutionarily conserved role in mRNA translational regulation from Drosophila to mammalian stem cells (Imai et al., 2001; Okabe et al., 2001; Okano et al., 2002, 2005; Battelli et al., 2006; Charlesworth et al., 2006). To date, four mammalian (Numb, p21, dcx and APC) and two Xenopus (Mos and cyclin B5) mRNAs have been shown to be direct targets of Musashi regulation (Imai et al., 2001; Battelli et al., 2006; Charlesworth et al., 2006; Horisawa et al., 2009; Arumugam et al., 2010; Spears and Neufeld, 2011). A number of mRNAs have been also implicated as targets based on co-immunoprecipitation with Musashi1 or on their behaviour after attenuation of Musashi function (Charlesworth et al., 2006; de Sousa Abreu et al., 2009). An important issue is to determine whether these are direct or indirect targets, as well as the relative importance of Musashi-dependent regulation of the mRNA to the control of cellular physiology.
In addition to the Mos and cyclin B5 mRNAs, translation of endogenous Musashi mRNA is also important for Xenopus meiotic cell cycle progression (Arumugam et al., 2010). Injection of antisense oligonucleotides that prevent translation of the Musashi1 (Nrp1) or Musashi2 (Xrp1) mRNAs delayed progesterone-stimulated maturation, while combinatorial attenuation of both Musashi1 and Musashi2 mRNAs blocked maturation. In this study, we investigated the regulation of Musashi mRNA translation in response to progesterone treatment. We report that the Musashi1 mRNA 3′ UTR contains a functional MBE, and that translational activation of the endogenous Musashi1 mRNA is dependent on functional Musashi activity. Our results indicate that translation of the Xenopus Musashi1 mRNA is subject to autoregulation in response to meiotic cell cycle progression.
RESULTS
The Translation of the Xenopus mRNA Encoding Musashi1 Is Activated During Oocyte Maturation
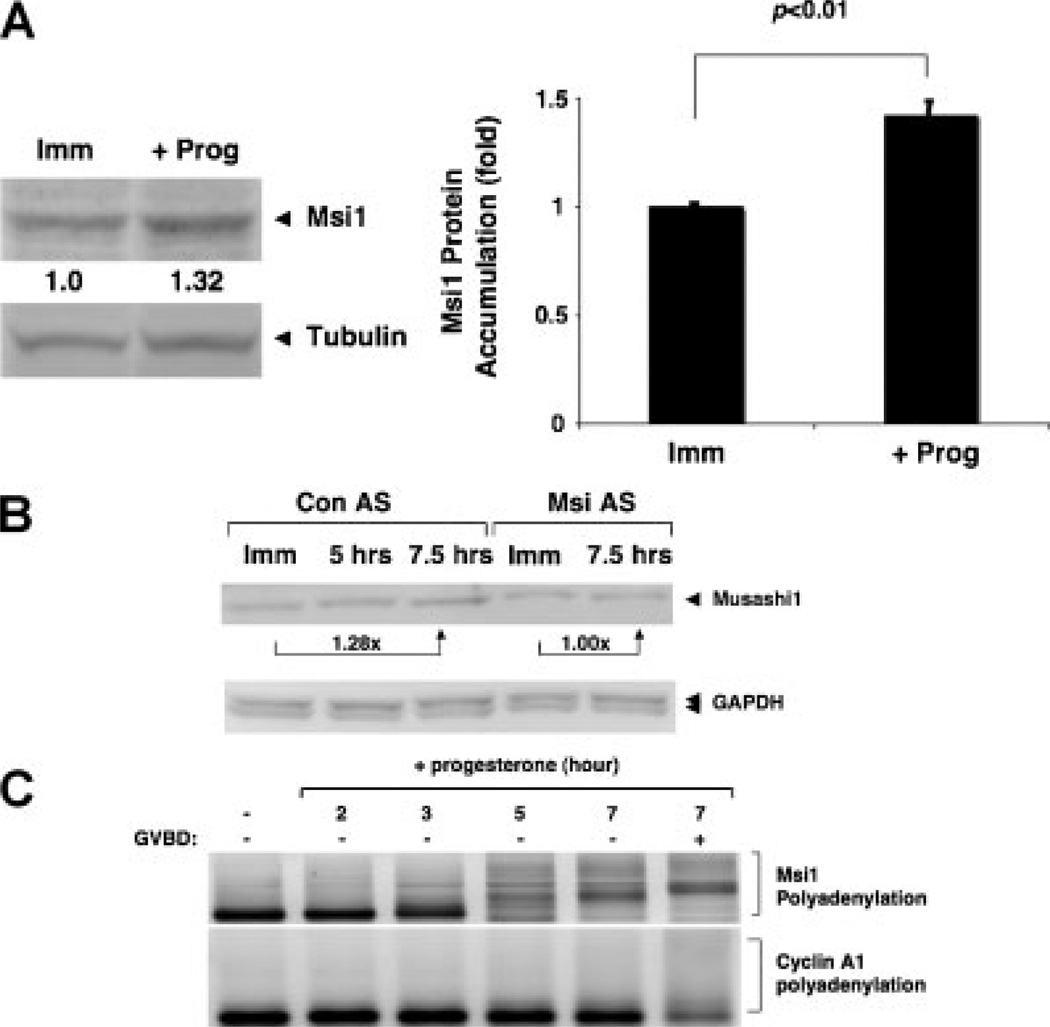
Downregulation of Musashi1 function through antisense oligonucleotide treatment delays progesterone-stimulated oocyte maturation (Arumugam et al., 2010). This treatment was shown to reduce Musashi1 protein accumulation in progesterone-stimulated oocytes, suggesting that translation of Musashi1 mRNA may be required to mediate normal progression through oocyte maturation. To examine this possibility directly, we performed Western blot analyses to assess Musashi1 protein levels during progesterone-stimulated oocyte maturation. We reproducibly observed an approximate 1.4-fold increase of Musashi1 protein in progesterone-stimulated oocytes when normalized to tubulin protein levels in the same lysates (Fig. 1A). Consistent with our earlier work (Arumugam et al., 2010), Musashi antisense oligonucleotides attenuated Musashi1 protein accumulation in response to progesterone stimulation (Fig. 1B; 21 ± 2% SEM n = 3, inhibition of maximum levels seen in control progesterone-stimulated oocytes). Since the antisense oligonucleotides block translation of the Musashi1 mRNA, we conclude that synthesis of Musashi1 protein, rather than enhanced stability, account for the progesterone-dependent accumulation. Examination of the Xenopus Musashi1 mRNA 3′ UTR revealed the presence of a consensus MBE and a cytoplasmic polyadenylation element (CPE) in addition to a canonical polyadenylation hexanucleotide element (Fig. 2A). The CPE does not overlap the polyadenylation hexanucleotide, suggesting that the upstream MBE would act dominantly to the late-acting CPE to direct early, pre-GVBD polyadenylation in response to progesterone stimulation (MacNicol and MacNicol, 2010). Consistent with this interpretation, the endogenous Xenopus Musashi1 mRNA undergoes progesterone-stimulated polyadenylation prior to oocyte GVBD (Fig. 1C; 3, 5 and 7 hr [−GVBD] time points). By contrast, the late-class, CPE-controlled endogenous cyclin A1 mRNA (Charlesworth et al., 2004) undergoes polyadenylation after GVBD in the same RNA samples (Fig. 1C, 7 hr [+GVBD] time point). We conclude that Musashi1 mRNA polyadenylation and translation is activated early in response to progesterone stimulation.
Figure 1.
Xenopus Musashi1 mRNA is translationally activated in response to progesterone stimulation. A: Immature, stage-VI oocytes were left untreated (Imm) or stimulated with progesterone (+prog), and were analysed for endogenous Musashi1 (Msi1) protein accumulation by Western blot. When 50% of the progesterone-treated oocyte population reached GVBD (GVBD50), the oocytes were segregated and those that had not yet completed GVBD were analysed as representative of early pre-GVBD events (Charlesworth et al., 2002). Quantitations of fold-changes in Musashi1 levels (as indicated below Western blot) were normalized to tubulin from the same sample, and levels in time-matched, immature oocytes (Imm) were arbitrarily set to 1.0. The bar graph represents data from three independent experiments, with SEM indicated. Student’s t-test confirmed the significance of the differences between the sample sets (P < 0.01). B: Immature, stage-VI oocytes were injected with control antisense oligonucleotides (Con AS) or antisense oligonucleotides targeting both endogenous Musashi1 and Musashi2 mRNAs (Msi AS), and cultured overnight. Oocytes were then either left untreated (Imm) or stimulated with progesterone. In this experiment, oocytes reached GVBD50 at 7.5 hr and were segregated based on whether or not they completed GVBD. Those oocytes that had not completed GVBD were analysed, along with oocytes harvested 2.5 hr earlier. Msi AS-injected oocytes did not mature in response to progesterone, and time-matched samples were prepared at the Con AS 7.5 hr time point. Levels of endogenous Musashi1 (upper panel) and GAPDH (lower panel) protein were analysed by Western blot. Fold-change in Musashi1 protein between immature and 7.5 hr of progesterone is indicated. Similar results were seen in two additional experiments. C: Oocytes treated with or without progesterone for the indicated times were analysed for endogenous Musashi1 mRNA polyadenylation by RNA ligation-coupled PCR. In this experiment, oocytes reached GVBD50 at 7 hr and were segregated into those that had not (−) or had (+) completed GVBD. An increase in size of the PCR products is indicative of polyadenylation (Charlesworth et al., 2002; Charlesworth et al., 2004). Polyadenylation of the endogenous late class cyclin A1 mRNA in the same samples occurred after completion of GVBD, as expected.
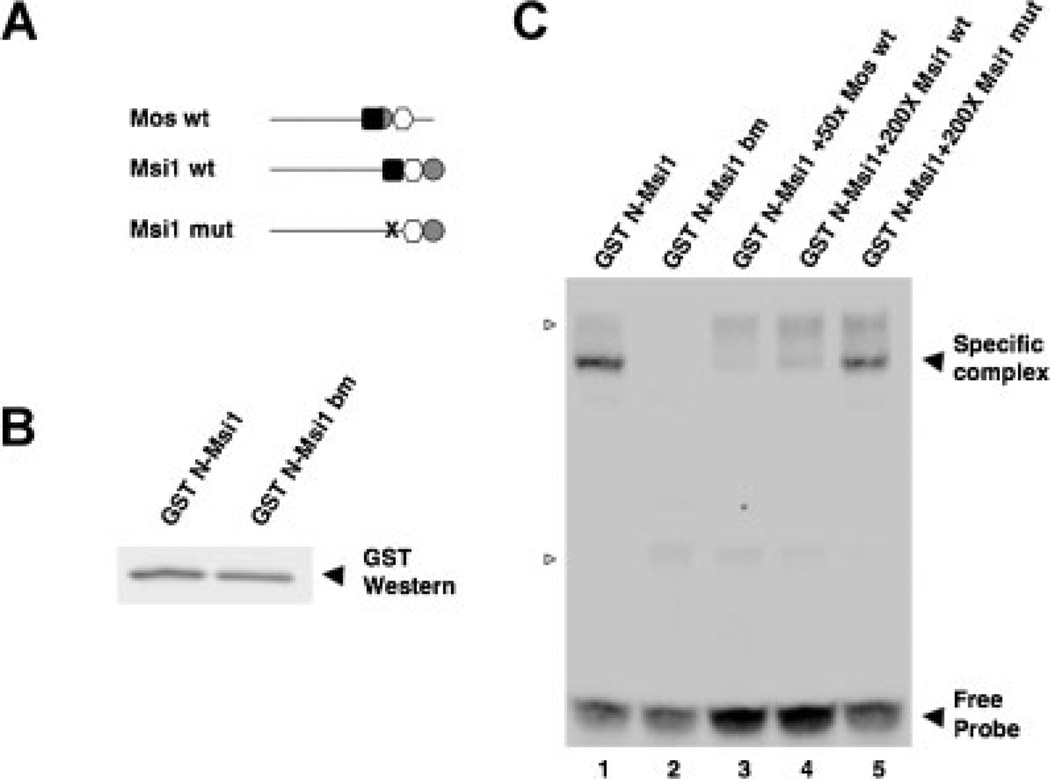
Figure 2.
The Musashi1 protein binds specifically to the MBE in the Xenopus Musashi1 mRNA 3′ UTR. A: Schematic representation of the Mos and Musashi1 3′ UTR constructs employed. Within the wildtype (wt) 3′ UTRs, elements are indicated by a black square (consensus Musashi binding site); white circle (consensus CPE); and grey hexagon (consensus polyadenylation hexanucleotide). The disrupted mutant MBE (AUAGU → AUccU) in the Musashi binding mutant (mut) UTR is shown as an “X”. B: The N-terminal mRNA binding domain of Musashi1 (N-Msi) or an RNA binding mutant variant (N-Msi bm) were expressed as GST fusion proteins in rabbit reticulocyte lysates for use in the EMSA reactions. A GST Western blot of the programmed lysates confirmed that each protein was expressed to comparable levels. C: RNA electrophoretic mobility shift assays using the indicated, unlabelled RNA probes to compete Mos 3′ UTR interaction with Musashi1. The Mos 3′ UTR MBE has been shown previously to be bound specifically by the N-terminal domain of Musashi1 (N-Msi) but not to an RNA-binding mutant of this protein (Charlesworth et al., 2006) as reproduced here (lanes 1 and 2, respectively). The wildtype Musashi1 3′ UTR (lane 4), like the wildtype Mos 3′ UTR (lane 3), efficiently competed with the biotinylated Mos 3′ UTR probe to prevent formation of a specific complex with N-Msi. By contrast, the MBE mutant Musashi 3′ UTR (lane 5) was not able to efficiently compete for the biotinylated Mos 3′ UTR probe binding to N-Msi. Several non-specific complexes, detected with unprogrammed reticulocyte lysate are indicated by open arrowheads. A representative experiment is shown.
The Xenopus Musashi1 Protein Interacts Specifically With the MBE in the Musashi1 mRNA 3′ UTR
In order to determine if the MBE was capable of interacting with Musashi1 protein, the last 100 nucleotides of the Musashi1 3′ UTR were cloned with either a wildtype MBE (Msi1 WT) or with a mutationally disrupted MBE [Msi1 mut, where the sequence AUAGU was changed to AUccU (Charlesworth et al., 2006)], and assayed by RNA electrophoretic mobility shift assay (EMSA; Fig. 2). The Mos 3′ UTR MBE has been shown previously to bind specifically to the N-terminus of Musashi1 (N-Msi) but not to an RNA-binding mutant of this protein (N-Msi bm; Charlesworth et al., 2006). We reproduce these findings here as positive and negative controls, respectively, for our analysis of the Musashi1 mRNA 3′ UTR MBE (Fig. 2C, lanes 1 and 2, respectively). The Musashi1 3′ UTR mRNA constructs were used as unlabelled competitors against a biotin-labelled Mos 3′ UTR in a reticulocyte lysate containing either N-Msi1 or N-Msi bm proteins (Fig. 2B). The unlabelled, wildtype Musashi1 3′ UTR, like the wild-type Mos 3′ UTR, efficiently competed with the labelled Mos 3′ UTR to prevent formation of a specific complex with N-Msi1 (Fig. 2C; lanes 4 and 3, respectively). By contrast, the MBE-disrupted Musashi1 3′ UTR failed to compete for complex formation (Fig. 2C, lane 5). We conclude that the Musashi1 protein binds specifically to the MBE in the Musashi1 mRNA 3′ UTR.
The MBE Is Necessary for the Translational Activation Exerted by the Musashi1 3′ UTR
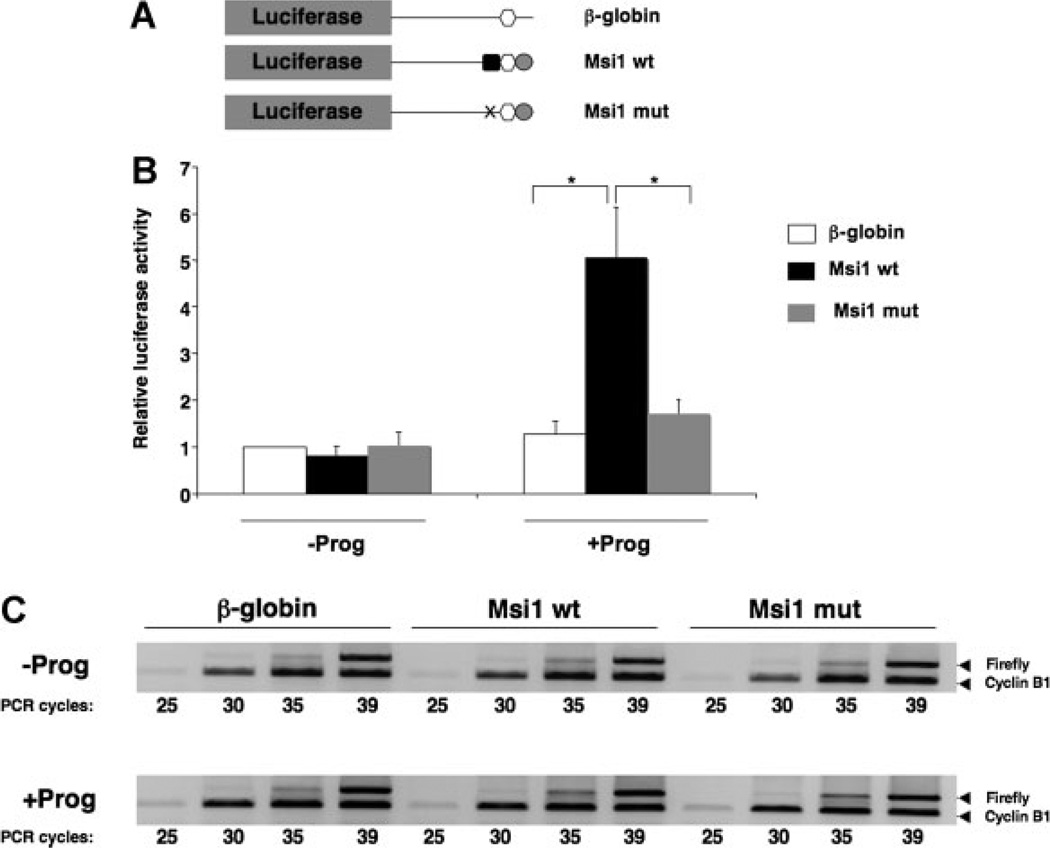
To directly address if the MBE was responsible for early translational activation of the Musashi1 mRNA, the wildtype or MBE mutant Musashi1 3′ UTR constructs were fused to a luciferase reporter and injected into immature oocytes (Fig. 3A). Progesterone stimulation induced a significant increase in luciferase activity associated with wildtype Musashi1 UTR, normalized to a co-injected Renilla luciferase control mRNA (Fig. 3B). By contrast, the Musashi-binding mutant 3′ UTR (Msi1 mut) did not significantly increase luciferase activity, and was similar to the activity of the unregulated, control β-globin UTR in either untreated or progesterone-treated oocytes (Fig. 3B). We see no evidence of MBE-exerted translational repression in immature oocytes (Fig. 3B). The stability of the MBE mutant 3′ UTR was similar to the wildtype 3′ UTR (Fig. 3C), indicating that the difference in luciferase activity is a consequence of reporter mRNA translation. Our data indicate that the early translational activation of the Musashi1 mRNA requires the MBE and is independent of the CPE in the 3′ UTR.
Figure 3.
The MBE is necessary for the Musashi1 3′ UTR to direct progesterone-dependent translational activation. A: Schematic representation of 3′ UTR constructs fused to a Firefly luciferase reporter mRNA. Symbols represent elements as described in the legend to Figure 2A. B: Oocytes were injected with mRNA encoding Renilla luciferase, and the indicated Musashi1 (Msi1) 3′ UTR Firefly luciferase reporter constructs and incubated for 16 hr. Subsequently, time matched immature (−Prog) and progesterone treated (+Prog) oocytes were lysed when progesterone-treated samples had reached GVBD and analysed for Renilla and Firefly luciferase activity. The plot shows an average ratio of Firefly luciferase activity derived from the Musashi1 3′ UTR reporter mRNAs relative to the co-injected Renilla luciferase mRNA, from three independent experiments. All ratios were normalized to the Firefly reporter mRNA fused to the unregulated β-globin UTR without progesterone (arbitrarily set to 1.0). Error bars indicate the SEM and differences were significant, as assessed by a Bonferroni test (*P < 0.01). C: The levels of each Firefly reporter mRNA were determined using semi-quantitative PCR. PCR-amplification of Firefly luciferase and cyclin B1 mRNA in the same samples was performed for different cycle numbers, as indicated. The PCR products were visualized after separation through a 2% agarose gel. No significant differences in stability of the different constructs were detected with or without progesterone treatment.
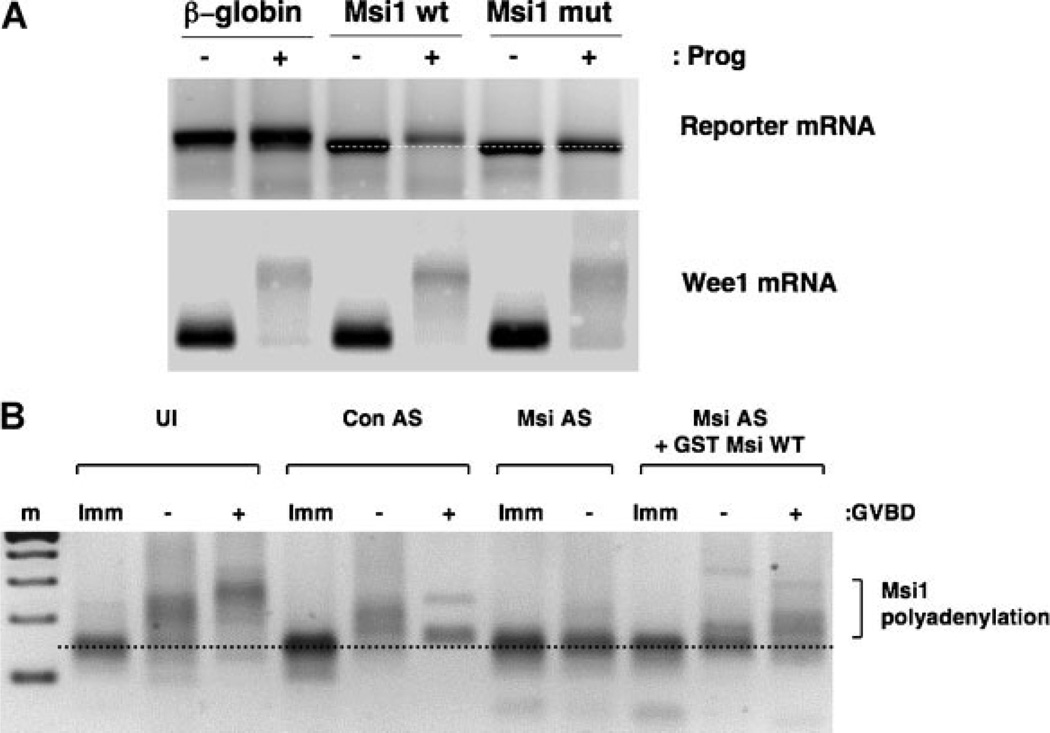
Consistent with the requirement for the MBE for translational activation of the Xenopus Musashi1 mRNA, we found that progesterone-stimulated polyadenylation of the Musashi1 3′ UTR was dependent upon the MBE (Fig. 4A). In addition to a functional cis MBE element in the Musashi1 mRNA 3′ UTR, Musashi protein function is required for the early progesterone-dependent polyadenylation of the endogenous Musashi1 mRNA. In contrast to control oligonucleotide-injected oocytes, those injected with antisense oligonucleotides targeting both Musashi1 and Musashi2 mRNAs failed to mature and failed to mediate early-class polyadenylation of the endogenous Musashi1 mRNA (Fig. 4B, Msi AS-). It should be noted that the antisense oligonucleotides target nucleotides 20–44 in the 5′ end of Musashi1 mRNA, and result in cleavage but not degradation of the larger mRNA fragment (see below). We have previously demonstrated the specificity of this inhibition by demonstrating that ectopic expression of wildtype Xenopus Musashi1 protein could reconstitute progesterone- dependent mRNA translational activation and overcome the block to cell cycle progression (Arumugam et al., 2010). We recapitulate these findings, and extend them to demonstrate that the inhibitory effect of oligonucleotide treatment on endogenous Musashi1 mRNA polyadenylation was reversible through ectopic expression of GST-tagged, wildtype Musashi1 (Fig. 4B). Thus, while effectively blocked for translation, the larger cleaved Musashi1 mRNA fragment retains 3′ UTR MBE responsiveness to cues triggering progesterone-dependent polyadenylation. Indeed, early-class Musashi1 mRNA polyadenylation was observed prior to GVBD in the rescue experiment (Fig. 4B, Msi AS + GST Msi WT). The timing of and dependence upon Musashi function for translational control of the endogenous Musashi1 mRNA is similar to what we observed with translational activation of the early-class Mos and cyclin B5 mRNAs (Charlesworth et al., 2006; Arumugam et al., 2010) and indicate that translational activation of the Musashi mRNA is mediated through activation of pre-existing Musashi protein.
Figure 4.
The MBE is necessary for progesterone-dependent polyadenylation of the Musashi1 3′ UTR. A: The indicated β-globin or Musashi1 3′ UTRs (see Fig. 3A) were fused downstream of the GST open reading frame and in vitro transcribed mRNA prepared. The GST reporter constructs were injected into immature oocytes and incubated overnight. Half of the injected oocytes were stimulated with progesterone (+) for 6 hr. Progesterone-treated oocytes were lysed at GVBD, total RNA prepared, analysed for polyadenylation of reporter mRNA constructs using RNA-ligation coupled RT-PCR, and compared to time-matched, untreated samples (−). The forward primer targeted the GST coding region to specifically amplify the reporter constructs. An increase in PCR product size above that seen in immature oocytes (dotted reference line) is indicative of polyadenylation. Polyadenylation of the endogenous, late class Wee1 mRNA(Charlesworth et al., 2000; Charlesworth et al., 2004) was used as a control. B: Immature, stage-VI oocytes were injected with control antisense oligonucleotides (Con AS) or antisense oligonucleotides targeting both endogenous Musashi1 and Musashi2 mRNAs (Msi AS), and cultured overnight. The next morning, a portion of the Msi AS-injected oocytes were re-injected with RNA encoding a GST tagged form of the wildtype Musashi1 (Msi AS + GST Msi WT). Oocytes were then either left unstimulated (Imm) or treated with progesterone. When 50% of the progesterone-treated population matured, oocytes were segregated into those that had not (−) or had (+) completed GVBD, and total RNA was isolated. Time-matched samples were also prepared from progesterone-stimulated Msi AS oocytes (no rescue), which did not mature, and from immature oocytes. Samples were analysed for endogenous Musashi1 mRNA polyadenylation by RNA ligation-coupled PCR. Uninjected (UI) control oocytes were also analysed as indicated. An increase in PCR product size above that seen in immature oocytes (dotted reference line) is indicative of polyadenylation.
DISCUSSION
We report that the polyadenylation and translational activation of the Xenopus Musashi1 mRNA is regulated by a MBE within the Musashi1 3′ UTR. Further, we show that translational activation of endogenous Musashi1 mRNA is dependent on functional Musashi activity. Taken together, our results indicate that the translation of the Xenopus Musashi1 mRNA is subject to autoregulation in response to meiotic cell cycle progression. The MBE sequence and position is conserved within the Xenopus tropicalis Musashi1 mRNA 3′ UTR (Accession NM_001011470), suggesting that autoregulation may be a shared control mechanism between the two amphibian species. The possible contribution of autoregulation to the control of mammalian Musashi mRNA translation will be an interesting avenue for future study.
Feedback mechanisms play a crucial role during meiotic maturation, especially in maintaining the kinetics necessary for the temporal hierarchy of progesterone-induced signalling events. Active maturation promoting factor (MPF) can attenuate the function of the cyclin-dependent kinase inhibitor Myt1 and target other kinases that can activate MPF, thus generating an auto-amplification loop that leads to the robust activation of MPF (Nebreda and Ferby, 2000; Karaiskou et al., 2001). Such activation is necessary to generate a bi-stable, on/off switch wherein all of MPF is active upon progesterone stimulation, compared to no active MPF in immature oocytes (Ferrell, 2008). Computational analyses have shown that in the presence of a noisy stimulus, a “slow” feedback loop (translational stimulation) in association with a “fast” feedback loop (phosphorylation/ dephosphorylation) provides a robust on/off switch for MPF activation (Brandman et al., 2005). In this regard, mitogen-activated protein (MAP) kinase signalling attenuates Myt1 function (Palmer et al., 1998). While Mos is the primary activator of MAP kinase signalling in oocytes, the polyadenylation and translational activation of the Mos mRNA was attenuated upon inhibition of MAP kinase (Howard et al., 1999; Gross et al., 2000). Thus, MAP kinase provides a positive feedback loop for a robust Mos mRNA translation that contributes to subsequent MPF activation. Another example of a Xenopus mRNA translation feedback loop is exemplified by GLD2, an evolutionarily conserved, non-canonical poly(A) polymerase (Wang et al., 2002; Barnard et al., 2004; Rouhana et al., 2005; Nakanishi et al., 2006; Benoit et al., 2008). GLD2 acquires substrate specificity by interacting with sequence-specific RNA binding proteins, and so is only recruited to a subset of mRNAs (Wang et al., 2002; Barnard et al., 2004; Rouhana et al., 2005; Suh et al., 2006). Recently, GLD-2 protein was shown to autoregulate the activation of GLD-2 mRNA during Xenopus oocyte maturation, and this mechanism was also implied to play a role in the rapid accumulation of GLD-2 protein in the mammalian brain during synaptic stimulation (Rouhana and Wickens, 2007). In addition, mRNA translational autoregulation has also been described for the Drosophila CPEB family member, Orb2, which stimulates translation of the Orb2 mRNA in the developing oocyte (Tan et al., 2001).
Musashi proteins have been predominantly thought to act to promote self-renewal and to oppose differentiation by repressing select mRNA target translation in mammalian stem and progenitor cells (reviewed in Okano et al., 2005; MacNicol et al., 2008; Nishimoto and Okano, 2010). In the immature, stage-VI Xenopus oocyte, however, we find no evidence for Musashi-dependent repression of the Musashi1mRNA(Fig. 3B), nor the previously characterized Musashi-target mRNA encoding Mos (Charlesworth et al., 2006). The molecular basis for the differential ability of Musashi to mediate repression remains to be elucidated. Translational activation, on the other hand, may be a common feature of Musashi function in both stem cells and oocytes. Indeed, we have recently found that Musashi converts from a repressor to an activator of target mRNA translation as mammalian stem/progenitor cells initiate neuronal differentiation (MacNicol et al., 2011). The progesterone-stimulated activation of Musashi-dependent mRNA translation in Xenopus oocytes would appear to parallel the differentiation-induced switch in mammalian Musashi function, and similar regulatory mechanisms may underlie these translation-promoting responses to extracellular cues (Arumugam et al., 2012).
Our findings indicate that Musashi1 autoregulates translation of its own mRNA, resulting in a progesterone-stimulated increase in Musashi1 protein that is required for timely progression through oocyte maturation. Presumably, the pre-existing pool of Musashi protein in the immature oocyte is targeted by progesterone signalling to activate translation of Musashi target mRNAs (Fig. 5). Injected Musashi antisense DNA oligonucleotides do not appear to eliminate the pre-existing Musashi1 protein (Arumugam et al., 2010; and Fig. 1B), suggesting that the inhibitory effect on cell cycle progression may be a consequence of reducing functional Musashi below a critical threshold for cell cycle progression and disrupting the feedback amplification pathways.
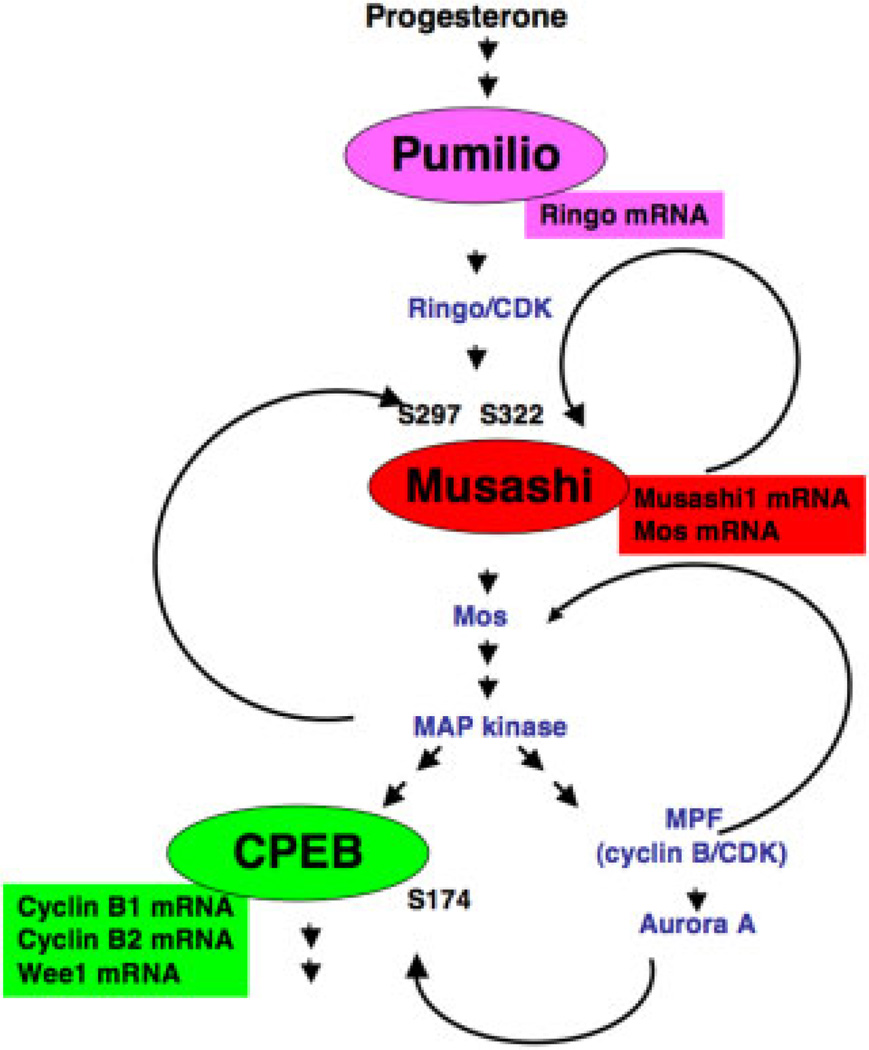
Figure 5.
Multiple feedback loops contribute to commitment and progression through meiotic cell-cycle progression. A schematic representation of the hierarchy of translational regulatory pathways and feedback loops that function sequentially to control commitment and progression through Xenopus oocyte maturation. Major translational control proteins (Pumilio, Musashi and CPEB) are represented by ovals while their specific target mRNAs are represented by rectangles. These mRNAs represent a selection of known target mRNAs, and so should not be considered an inclusive list for each factor. Key signalling components and their relative position with regard to activation timing are shown within the network. MPF activation usually coincides with oocyte GVBD. Within this network, translation of Ringo and activation of Ringo/CDK trigger phosphorylation of Musashi1 on serine-297 and -322 (S297 and S322, respectively) (Arumugam et al., 2012). Musashi then activates translation of the endogenous Musashi1 mRNA to establish a positive feedback loop (this study), as indicated. Musashi-dependent translation of the Mos mRNA leads to activation of MAP kinase signalling, which occupies a multi-nodal hub in the pathway. MAP kinase signalling can trigger phosphorylation of additional Musashi1 protein in a positive feedback loop (Arumugam et al., 2012), can phosphorylate CPEB to prime it for activation by a serine-174 (S174) kinase (Keady et al., 2007), and can phosphorylate and inhibit Myt1, leading toMPF activation (Palmer et al., 1998). MPF activation leads to activation of Aurora A (Frank-Vaillant et al., 2000; Maton et al., 2003), which can further phosphorylate CPEB S174 (Mendez et al., 2000). MPF has also been reported to phosphorylate and stabilize Mos protein (Castro et al., 2001). For the sake of clarity, two additional pathways where Ringo/CDK phosphorylates and inhibits Myt1 (Ruiz et al., 2008) and MPF phosphorylates and targets degradation of CPEB (Mendez et al., 2002) have been omitted. See text for further details.
We have recently demonstrated that Musashi1 function is regulated in response to progesterone stimulation through phosphorylation of two conserved serine residues in the C-terminal domain, serine-297 and serine-322 (Arumugam et al., 2012). Specifically, progesterone triggers phosphorylation and activation of Musashi-dependent, early-class mRNA translation via Ringo/cyclin-dependent kinase (CDK) signalling. Initial Ringo/CDK activation is mediated by translational control independent of Musashior CPEB-dependent mechanisms. Ringo mRNA is translationally repressed by the Pumilio proteins, Pumilio1 and/or Pumilio2, in immature oocytes (Padmanabhan and Richter, 2006; Cao et al., 2010; Ota et al., 2011). An immediate-early response to progesterone stimulation (within 15–30 min) is the inactivation of Pumilio as a repressor protein and translation of the Pumilio target mRNA encoding Ringo/Speedy, an atypical activator of cyclin-dependent kinases 1 and 2 (Cdk1 and Cdk2) (Lenormand et al., 1999; Ferby et al., 1999; Karaiskou et al., 2001; Padmanabhan and Richter, 2006). Phosphorylation and activation of Musashi1 then allows the polyadenylation and translation of direct Musashi target mRNAs, including Mos, cyclin B5 and Musashi1 (which typically occur 2–3 hr after stimulation). Musashi1 phosphorylation and activation is subsequently augmented by MAP kinase signalling, a downstream effector of Musashi-dependent Mos mRNA translation (Arumugam et al., 2012). Thus, Ringo/CDK phosphorylation of Musashi1 initiates two nested feedback loops to positively reinforce Musashi activation: (i) autoregulation of endogenous Musahsi1 mRNA translation (as reported in this study); and (ii) Mos-dependent activation of MAP kinase signalling leading to further activation of Musashi1 protein via serine-297 and serine-322 phosphorylation.
In addition to direct early-class mRNA targets, Musashi also indirectly regulates subsequent late-class, CPE-dependent mRNAs (Charlesworth et al., 2006; Arumugam et al., 2010; and Fig. 5). This indirect dependence may be mediated in part by Mos-dependent MAP kinase activation. Indeed, activation of CPEB requires permissive MAP kinase phosphorylation followed by phosphorylation of serine-174 by a MAP kinase-independent mechanism (Keady et al., 2007). The characterized CPEB serine-174 kinase, Aurora A, is activated after GVBD in an MPF-dependent manner (Frank-Vaillant et al., 2000; Castro et al., 2003; Maton et al., 2003; Keady et al., 2007).
Collectively, these studies illuminate a complex regulatory hierarchy involving the sequential activation of distinct mRNA translational control programs (Pumilio, Musashi and CPEB, respectively) during progesterone-stimulated, Xenopus oocyte maturation (Fig. 5). Within this schema, Musashi1 is a critical component of both the initiation and the subsequent signal amplification step. While the role of Musashi has not been addressed, recent work has revealed a sequential hierarchy involving CPEB and deleted in azoospermia-like (DAZL) translational control programs during murine oocyte maturation (Chen et al., 2011). The emerging logic is that the translational control programs operate sequentially in response to distinct signalling pathways as a means to select and enforce the correct temporal order of mRNA translation necessary for meiotic cell cycle progression (MacNicol and MacNicol, 2010). As evidenced by the work presented in this study, the sequential translational control programs employ a series of nested, positive feedback loops that serve to amplify the initial weak trigger and thereby generate a robust output that commits the oocyte to progress through meiosis and to become competent for fertilization.
MATERIALS AND METHODS
Plasmid Constructions and RNA Synthesis
The Firefly luciferase vector pGEM_luc2 was constructed by cloning the luciferase 2 gene from pGL4.20[luc2/Puro] (Promega, Madison, WI) into the pGEM-4Z vector (Promega), as previously described (Arumugam et al., 2010). The pGEMFluc Xenopus β-globin 3′ UTR reporter has been previously described (Arumugam et al., 2010).
pGEMFluc Xenopus Musashi1 wt 3′ UTR reporter
PCR primers were designed to amplify the last 100 nucleotides of NRP-1B (Xenopus Musashi1) mRNA 3′ UTR, with a 5′ SacI site (5′-CGGAGCTCCAATACTGCAATGTACAATGTACTGC) and a 3′ BamHI site (5′-GCGGGATCCTGAATAAAATTCAATTTATTTTG). cDNA was prepared using RNA extracted from immature Xenopus oocytes using the reverse PCR primer and Superscript III (Invitrogen, Carlsbad, CA). The 100-nucleotide portion of NRP-1B 3′ UTR was amplified using Platinum Taq (Invitrogen), the PCR product digested with SacI and BamHI, and ligated into SacI/BamHI-digested pGEM_luc2.
pGEMFluc Xenopus Msi1 mbm 3′ UTR reporter
The Musashi-binding mutant of NRP-1B 3′ UTR (AUAGU → AUccU) was made by site-directed mutagenesis of the pGEMFluc Musashi1 wt 3′ UTR reporter.
pGEMGST xMsi1 wt 3′ UTR reporter
The Musashi1 wt UTR was cloned by cutting the pGEMFluc Musashi1 3′ UTR construct at the 5′ EcoRI site and a 3′ BamHI site, isolating the Musahsi1 3′ UTR, blunting with Klenow and ligation into a XbaI digested, Klenow-treated pGEM GST vector.
pGEMGST Musashi1 mbm 3′ UTR reporter
The Musashi-binding mutant Musashi1 3′ UTR was cloned by cutting it from the pGEMFluc xMsi1 mbm 3′ UTR construct, and cloned into the pGEM GST vector as described for the Musashi1 wt construct above.
RNA Electrophoretic Mobility Shift Assay Competition Constructs
For Electrophoretic Mobility Shift Assay competition assays, the pGEMFluc Musashi1 wt UTR (Msi1 wt) and Musashi-binding mutant UTR (Msi1 mbm) constructs were digested with EcoRI to excise the luciferase gene, re-ligated, and used to generate unlabelled competitor RNA. The Mos UTR probe has been described previously (Charlesworth et al., 2006).
All constructs were transcribed with SP6 RNA Polymerase (Promega), as previously described (Melton et al., 1984).
Oocyte Culture and Microinjections
Immature, stage-VI Xenopus oocytes were isolated and cultured as described previously (Machaca and Haun, 2002). Oocytes were injected with 23 ng RNA/oocyte, unless otherwise noted. Oocytes were induced to mature with 2 µg/ml progesterone. Animal protocols were approved by the UAMS Institutional Animal Care and Use committee, in accordance with Federal regulations.
Luciferase Reporter Assays
Stage-VI oocytes were injected with 0.1 ng of Firefly luciferase mRNA and 0.35 pg of Renilla luciferase control mRNA (Minshall et al., 2001), and incubated at 18°C for 18 hr. Injected oocytes were split into two pools, where one was left untreated while the other was stimulated with progesterone. Three pools of five oocytes were lysed when progesterone samples reached GVBD50 along with three pools of five oocytes of the time-matched, untreated samples. For each experimental point, the oocytes were lysed in 50 µl of Passive lysis buffer (Promega). A 10-µl portion of lysate was analysed for Renilla and Firefly luciferase activity using the Dual-Luciferase Assay System (Promega) on a TD-20/20 Turner Designs luminometer. Mean values and standard deviation were determined for each experimental point, with the ratio of Firefly to Renilla luciferase for each construct normalized to the values obtained with the Firefly β-globin 3′ UTR reporter construct (set to 1.0).
RNA Electrophoretic Mobility Shift Assays
GST fusion proteins were in vitro transcribed/translated using TNT SP6-coupled Reticulocyte Lysate System (Promega). A 5′ biotin-labelled, Mos UTR RNA oligonucleotide probe was synthesized by Integrated DNA Technologies, as previously described (Charlesworth et al., 2006). Unlabelled competitor mRNAs were transcribed in vitro. An 80 fmol portion of labelled probe was incubated with 1 µl of reticulocyte lysate and 200 molar excess of unlabelled mRNA in binding buffer [50mM Tris pH 7.5, 20mM KCl, 150mM NaCl, 2mM EGTA, 0.05% NP-40, 6mM DTT, 8U RNaseOUT (Okabe et al., 2001)] in a final volume of 20 µl. The binding reaction was incubated at room temperature for 20 min, and then 1 µl of 20 mg/ml heparin was added and incubated for a further 20 min. A 5-µl volume of the binding reaction was run on a 6% DNA retardation gel (Invitrogen) and transferred to Biodyne B membranes (Pierce, Rockford, IL) according to the manufacturer’s instructions. Biotinylated RNA was detected using Chemiluminescent Nucleic Acid Detection Module (Pierce) according to the manufacturer’s directions, with the modification that incubation with the streptavidin–HRP conjugate was for 40 min. Image collection was performed using an AlphaInnotech ChemiImager (San Leandro, CA).
Antisense Oligodeoxynucleotide Injections
Antisense oligodeoxynucleotides 5′-GCGCTTCTGTCTCCATTCGGTCTCT and 5′-CCCATCTGCCTCCATAGCCTTCTC were designed to target endogenous Xenopus Musashi1 and Musashi2 mRNAs, respectively (Arumugam et al., 2010). For knockdown experiments, oocytes were injected with 50 ng of Musashi1 and 50 ng of Musashi2 antisense oligonucleotides, and the control oocytes were injected with 100 ng of control antisense oligonucleotide 5′-TAGAGAAGATAATCGTCATCTTA (Ferby et al., 1999). Oocytes were incubated at 18°C for 16 hr, followed by injection of mRNA encoding GST-Musashi1 wildtype rescue protein (Arumugam et al., 2010), as indicated, and/or progesterone treatment.
Western Blotting
Oocytes were lysed in NP40 lysis buffer containing sodium vanadate and a protease inhibitor cocktail (Sigma, St. Louis, MO). The lysate was then spun, clarified and transferred immediately to 1 × LDS sample buffer (Nupage). The lysates were run on a 10% Nupage gel and transferred to a 0.2-µm-pore-size nitrocellulose filter (Protran; Midwest Scientific, Valley Park, MO). The membrane was blocked with5%non-fat, dry milk in Tris-buffered saline with 0.1% Tween20 (TBST) for 60 min at room temperature. Abcam antibodies to Msi1 (Ab33251) were used at 1:1,000; Sigma antibodies to Tubulin and GAPDH were used at 1:20,000; and Santa Cruz Biotechnology (Santa Cruz, CA) antibodies to GST were used at 1:5,000. The filters were visualized with horseradish peroxidase- conjugated, anti-rabbit antibody using enhanced chemiluminescence in a Fluorchem 8000 Advanced Imager (Alpha Innotech Corp.).
Polyadenylation Assays
cDNAs for polyadenylation assays were synthesized using RNA ligation-coupled RT-PCR as described previously (Charlesworth et al., 2004). For polyadenylation of reporter constructs, oocytes were injected with reporter mRNAs at a concentration of 1 ng/oocyte. The primers used to analyse the GST reporters were targeted to the GST region, as described previously (Charlesworth et al., 2004). The primer used to analyse endogenous Xenopus Musashi1 mRNA polyadenylation was 5′-CAATACTGCAATGTACAATGTACTGC. The primers for Wee1 and Cyclin A1 have been described previously (Charlesworth et al., 2004).
Semi-Quantitative PCR
cDNAs for semi-quantitative PCR analysis were synthesized using RNA ligation-coupled RT-PCR, as described previously (Charlesworth et al., 2004). For the PCR reaction, 2 µl worth cDNA was used with 3mM MgCl2, 1mM Firefly luciferase primers (forward, 5′-GAGTACTTCGAGATGAGC; reverse, 5′-CACGAAGTCGTACTCGTT) and 0.25mM cyclin B1 primers (forward, 5′-GGCTTGAGACCTCGTACAGC; reverse, 5′-CAGGGAGGCAACCAGATG).
ACKNOWLEDGMENTS
We thank Chad E. Cragle for helpful discussions. This work was supported by NIH grant RO1 HD35688 (to A.M.M.) and NIH grant RR020146 (to M.C.M.). DNA sequencing was provided by the UAMS Translational Research Institute supported by the National Institutes of Health National Center for Research Resources grant UL1 RR029884.
Grant sponsor: NIH; grant number: RO1 HD35688; Grant sponsor: NIH; Grant number: RR020146; Grant sponsor: National Institutes of Health National Center for Research Resources; Grant number: UL1 RR029884.
Abbreviations
- 3′ UTR
3′ untranslated region
- CPE
cytoplasmic polyadenylation element
- CPEB
CPE-binding protein
- GVBD
germinal vesicle breakdown
- MAP kinase
mitogen activated protein kinase
- MBE
Musashi binding element
- MPF
maturation promoting factor.
REFERENCES
- Arumugam K, MacNicol MC, Wang Y, Cragle CE, Tackett AJ, Hardy LL, MacNicol AM. Ringo/CDK and MAP kinase regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J Biol Chem. 2012;287:10639–10649. doi: 10.1074/jbc.M111.300681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell cycle progression. EMBO J. 2010;29:387–397. doi: 10.1038/emboj.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne S, Daniel DLJ, Wickens M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol Biol Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21(WAF-1) Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP-and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A, Mandart E, Lorca T, Galas S. Involvement of Aurora A kinase during meiosis I–II transition in Xenopus oocytes. J Biol Chem. 2003;278:2236–2241. doi: 10.1074/jbc.M207894200. [DOI] [PubMed] [Google Scholar]
- Castro A, Peter M, Magnaghi-Jaulin L, Vigneron S, Galas S, Lorca T, Labbe JC. Cyclin B/cdc2 induces c-Mos stability by direct phosphorylation in Xenopus oocytes. Mol Biol Cell. 2001;12:2660–2671. doi: 10.1091/mbc.12.9.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in xenopus oocytes. J Biol Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Ridge JA, King LA, MacNicol MC, MacNicol AM. A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 2002;21:2798–2806. doi: 10.1093/emboj/21.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Welk J, MacNicol A. The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′ untranslated region. Dev Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Richter JD. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le SY, Shapiro BA, Penalva LO. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- Erhardt JA, Pittman RN. Ectopic p21WAF1 expression induces differentiation-specific cell cycle changes in PC12 cells characteristic of nerve growth factor treatment. J Biol Chem. 1998;273:23517–23523. doi: 10.1074/jbc.273.36.23517. [DOI] [PubMed] [Google Scholar]
- Ferby I, Blazquez M, Palmer A, Eritja R, Nebreda AR. A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 1999;13:2177–2189. doi: 10.1101/gad.13.16.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE., Jr Feedback regulation of opposing enzymes generates robust, all-or-none bistable responses. Curr Biol. 2008;18:R244–R245. doi: 10.1016/j.cub.2008.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant M, Haccard O, Thibier C, Ozon R, Arlot-Bonnemains Y, Prigent C, Jessus C. Progesterone regulates the accumulation and the activation of Eg2 kinase in Xenopus oocytes. J Cell Sci. 2000;113:1127–1138. doi: 10.1242/jcs.113.7.1127. [DOI] [PubMed] [Google Scholar]
- Gross SD, Schwab MS, Taieb FE, Lewellyn AL, Qian YW, Maller JL. The critical role of the MAP kinase pathway in meiosis II in Xenopus oocytes is mediated by p90(Rsk) Curr Biol. 2000;10:430–438. doi: 10.1016/s0960-9822(00)00425-5. [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisawa K, Imai T, Okano H, Yanagawa H. 3′-Untranslated region of doublecortin mRNA is a binding target of the Musashi1 RNA-binding protein. FEBS Lett. 2009;583:2429–2434. doi: 10.1016/j.febslet.2009.06.045. [DOI] [PubMed] [Google Scholar]
- Howard EL, Charlesworth A, Welk J, MacNicol AM. The mitogen-activated protein kinase signaling pathway stimulates mos mRNA cytoplasmic polyadenylation during Xenopus oocyte maturation. Mol Cell Biol. 1999;19:1990–1999. doi: 10.1128/mcb.19.3.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Gollapudi L, Sladek TL, Neet KE. Mediation of nerve growth factor-driven cell cycle arrest in PC12 cells by p53. Simultaneous differentiation and proliferation subsequent to p53 functional inactivation. J Biol Chem. 2000;275:37829–37837. doi: 10.1074/jbc.M003146200. [DOI] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, Goldman J, Goh H, Kim SH, Kim DW, Chuah C, Oehler VG, Radich JP, Jordan CT, Reya T. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafar-Nejad H, Norga K, Bellen H. Numb: “Adapting” notch for endocytosis. Dev Cell. 2002;3:155–156. doi: 10.1016/s1534-5807(02)00228-9. [DOI] [PubMed] [Google Scholar]
- Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, Fushiki S, Nakamura Y, Yamasaki M, Okano H, Arita N. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68:141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- Karaiskou A, Perez LH, Ferby I, Ozon R, Jessus C, Nebreda AR. Differential regulation of Cdc2 and Cdk2 by RINGO and cyclins. J Biol Chem. 2001;276:36028–36034. doi: 10.1074/jbc.M104722200. [DOI] [PubMed] [Google Scholar]
- Keady BT, Kuo P, Martinez SE, Yuan L, Hake LE. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci. 2007;120:1093–1103. doi: 10.1242/jcs.03416. [DOI] [PubMed] [Google Scholar]
- Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, Gozo M, Einhorn W, Lane SW, Scholl C, Frohling S, Fleming M, Ebert BL, Gilliland DG, Jaenisch R, Daley GQ. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Lenormand JL, Dellinger RW, Knudsen KE, Subramani S, Donoghue DJ. Speedy: A novel cell cycle regulator of the G2/M transition. EMBO J. 1999;18:1869–1877. doi: 10.1093/emboj/18.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machaca K, Haun S. Induction of maturation-promoting factor during Xenopus oocyte maturation uncouples Ca(2+) store depletion from store-operated Ca(2+) entry. J Cell Biol. 2002;156:75–85. doi: 10.1083/jcb.200110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNicol AM, Wilczynska A, MacNicol MC. Function and regulation of the mammalian Musashi mRNA translational regulator. Biochem Soc Trans. 2008;36:528–530. doi: 10.1042/BST0360528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. doi: 10.4161/cc.10.1.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNicol MC, MacNicol AM. Developmental timing of mRNA translation—Integration of distinct regulatory elements. Mol Reprod Dev. 2010;77:662–669. doi: 10.1002/mrd.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maton G, Thibier C, Castro A, Lorca T, Prigent C, Jessus C. Cdc2-Cyclin B triggers H3 kinase activation of aurora-A in xenopus oocytes. J Biol Chem. 2003;278:21439–21449. doi: 10.1074/jbc.M300811200. [DOI] [PubMed] [Google Scholar]
- Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Nebreda AR, Ferby I. Regulation of the meiotic cell cycle in oocytes. Curr Opin Cell Biol. 2000;12:666–675. doi: 10.1016/s0955-0674(00)00150-2. [DOI] [PubMed] [Google Scholar]
- Nishimoto Y, Okano H. New insight into cancer therapeutics: Induction of differentiation by regulating the Musashi/Numb/ Notch pathway. Cell Res. 2010;20:1083–1085. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Nagata T, Tsuda K, Kobayashi N, Imai T, Okano H, Yamazaki T, Katahira M. Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 2012;40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- Okano H, Imai T, Okabe M. Musashi: A translational regulator of cell fate. J Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Ota R, Kotani T, Yamashita M. Biochemical characterization of Pumilio1 and Pumilio2 in Xenopus oocytes. J Biol Chem. 2011;286:2853–2863. doi: 10.1074/jbc.M110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A, Gavin A-C, Nebreda AR. A link between MAP kinase and p34cdc2/cyclin B during oocyte maturation: p90rsk phosphorylates and inactivates the p34cdc2 inhibitory kinase Myt1. EMBO J. 1998;17:5037–5047. doi: 10.1093/emboj/17.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wickens M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA. 2007;13:188–199. doi: 10.1261/rna.333507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz EJ, Hunt T, Nebreda AR. Meiotic inactivation of Xenopus Myt1 by CDK/XRINGO, but not CDK/cyclin, via site-specific phosphorylation. Mol Cell. 2008;32:210–220. doi: 10.1016/j.molcel.2008.08.029. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J Biol Chem. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci USA. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- Tan L, Chang JS, Costa A, Schedl P. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development. 2001;128:1159–1169. doi: 10.1242/dev.128.7.1159. [DOI] [PubMed] [Google Scholar]
- Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Ziff EB. NGF regulates the PC12 cell cycle machinery through specific inhibition of the Cdk kinases and induction of cyclin D1. J Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]