Abstract
Both Glis, the downstream effectors of hedgehog signaling, and Zic transcription factors are required for Myf5 expression in the epaxial somite. Here we demonstrate a novel synergistic interaction between members of both families and Pax3, a paired-domain transcription factor that is essential for both myogenesis and neural crest development. We show that Pax3 synergizes with both Gli2 and Zic1 in transactivating the Myf5 epaxial somite (ES) enhancer in concert with the Myf5 promoter. This synergy is dependent on conserved functional domains of the proteins, as well as on a novel homeodomain motif in the Myf5 promoter and the essential Gli motif in the ES enhancer. Importantly, overexpression of Zic1 and Pax3 in the 10T1/2 mesodermal cell model results in enrichment of these factors at the endogenous Myf5 locus and induction of Myf5 expression. In our previous work, we showed that by enhancing nuclear translocation of Gli factors, Zics provide spatiotemporal patterning for Gli family members in the epaxial induction of Myf5 expression. Our current study indicates a complementary mechanism in which association with DNA-bound Pax3 strengthens the ability of both Zic1 and Gli2 to transactivate Myf5 in the epaxial somite.
Keywords: Myf5, myogenesis, Pax, Zic, Gli, skeletal muscle
Introduction
Development of skeletal muscle is controlled by the myogenic regulatory factors (MRFs): Myf5, MyoD, Myogenin, and MRF4. These transcription factors are essential for the determination and differentiation of skeletal muscle during embryogenesis (18, 45), and display the unique ability to convert non-muscle cell types to skeletal muscle (58). Myf5 is the first MRF to be expressed, at E8 in the mouse (42). Transcripts are first detectable in the dermomyotome of the earliest somites, then in the newly formed myotome, followed by expression in the ventral dermomyotome and branchial arches (42). Following migration of muscle precursor cells from the somites to the limb buds, Myf5 is also transiently activated in the developing limbs (42, 54). In the adult, Myf5 expression is downregulated and only maintained in muscle satellite cells and spindles (61). Targeted disruption of Myf5, MyoD, and MRF4 in the mouse results in a complete absence of myoblasts, underscoring the importance of these factors for myogenic commitment (9, 29, 47).
Myf5 and Mrf4 are linked, and the shared locus is subject to complex transcriptional regulation (18). Manipulation of the locus in transgenic reporter mice has uncovered a number of discrete enhancers that direct expression of each gene in specific progenitor cell populations in the embryo (12). One of the best characterized enhancers in the locus is the Myf5 epaxial somite (ES) enhancer. This region lies ~6.6 kb upstream of the Myf5 transcription start site and controls the expression of Myf5 in the epaxial muscle progenitors of the dorsal somite (53, 55). Interestingly, while the Myf5 ES enhancer activates its own promoter, it cannot engage productively with the closer Mrf4 promoter or with several cryptic promoters in the locus (12, 56), suggesting that expression of Myf5 in the epaxial somite requires specific interactions with its homologous promoter.
Several signaling pathways and their downstream effectors have been implicated in activity of the Myf5 ES enhancer. Correct expression of Myf5 in the epaxial somite requires Sonic hedgehog (Shh) signaling via a conserved Gli motif, Wnt signaling through multiple TCF/LEF motifs, and Dmrt2 motifs in the ES enhancer (5, 21, 51, 56). Much less is known regarding control of the Myf5 promoter, but FoxD3 binding to a conserved motif in the Myf5 promoter of zebrafish is required for maintenance of Myf5 expression in the somites (32).
Glis and Zics are closely related zinc-finger transcription factors, shown to have antagonistic effects in neural patterning (10), and cooperative effects in skeletal patterning and myogenesis (2, 43). Mutations in both families result in a range of developmental abnormalities (25, 28), and members of both families are important for Myf5 expression in the epaxial somite (6, 35, 43). Expression of Pax3, a paired-domain transcription factor that is essential for both myogenesis and neural crest development, also overlaps with that of Myf5 in myogenic progenitors in the dermomyotome and limb buds. Pax3 has been shown to activate several Myf5 enhancers, both directly (3, 14) and indirectly (51), and mutations in Pax3 lead to Waardenburg syndrome types I and III, diseases characterized by defects in muscle and neural crest derivatives (24).
Given the overlapping roles of Gli, Zic, and Pax transcription factors in somite myogenesis, we asked whether these factors are capable of synergizing in activating the Myf5 ES enhancer and homologous promoter. Here we demonstrate novel synergistic interactions between Gli2 and Pax3, and Zic1 and Pax3. This synergy is dependent on conserved functional domains of the proteins, as well as on a novel homeodomain motif in the Myf5 promoter and the essential Gli motif in the ES enhancer. Importantly, overexpression of Zic1 and Pax3 in the 10T1/2 mesodermal cell model results in the enrichment of these factors at the endogenous Myf5 locus and induction of Myf5 expression. Unlike Gli2 and Pax3, Zic1 is expressed exclusively in epaxial muscle progenitors within the dermomyotome. In our previous work, we showed that Zics provide spatiotemporal patterning for Gli family members in the induction of Myf5 expression (43). Here we show that in addition to enhancing the nuclear translocation of Gli factors (30, 43), Zic1 also associates with Pax3 on the Myf5 promoter to drive Myf5 expression. Likewise, the ability of Gli2 to transactivate Myf5 is strengthened by a synergistic association with Pax3. Collectively, our data indicate novel interactions that link several well-established myogenic pathways.
Materials and Methods
Plasmids and antibodies
Mammalian expression plasmids Gli1 and Gli2 in pcDNA3.1-His, and Gli2 lacking the C-terminal domain (Gli2[ΔC2] and Gli2[ΔC4]) have been described previously (50). Zic1 and Zic2 in pCS2FLAG have also been described (43). Pax3-HA in pcDNA3 and Pax7-FLAG in pBRIT were purchased from Addgene (#27319 and #17521, respectively). Zic1 lacking the ZOC domain (Zic1[ΔZOC]) or zinc fingers (Zic1[ΔZF]), and Pax3 lacking the homeodomain (Pax3[ΔHD]) or transactivation domain (Pax3[ΔTD]) were made using standard site-directed mutagenesis on the plasmids described above. The Myf5 ES enhancer, EpExt in (5), and Myf5 promoter (56) were amplified from mouse genomic DNA and cloned into the firefly luciferase reporter vector pGL3-Basic (Promega) to generate E-P-luc. Mutations in the ES enhancer Gli motif (Gli-mt) (21) and Myf5 promoter homeodomain motif (HD-mt) were made using standard site-directed mutagenesis of E-P-luc. (Gli)8-TK-luc, containing eight wild-type Gli binding sites from the Myf5 ES enhancer, has been described (21). Antibodies used in this study were: anti-HA monoclonal antibody (sc-7392, Santa Cruz Biotechnology, Inc.), anti-FLAG M2 monoclonal antibody (#F3165, SIGMA), anti-Gli2 polyclonal antibody (ab7195, abcam), anti-V5 monoclonal antibody (#R960-25, Invitrogen), and normal mouse IgG (sc-2025, Santa Cruz Biotechnology, Inc.).
Cell culture
NIH 3T3 mouse embryonic fibroblasts and C3H/10T1/2 mouse embryonic mesenchymal stem cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech, Inc.) + 10% fetal bovine serum (FBS) (Denville) and antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin). 10T1/2 cells were also supplemented with 10 mM HEPES.
Transient transfections and reporter assays
Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol, and harvested at 48 hours post6-transfection. For co-immunoprecipitation assays, gel-shift assays, RT-PCR, and ChIP assays, cells were transfected with expression constructs in 10 cm plates. For reporter assays, cells were reverse transfected with expression and reporter constructs in 96-well plates according to the manufacturer’s protocol. To aid cell lysis, plates were frozen at −70° for 1 h, followed by 10 min at 25°, 15 min at 37°, and 10 min at 25°. Firefly and renilla luciferase activities were measured using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol.
Co-immunoprecipitation and Western analysis
10T1/2 cells transfected with Pax3-HA and FLAG-Zic1 expression plasmids were harvested 48 h after transfection in PBS + protease inhibitors (#P8340, SIGMA). Cells were pelleted, washed with PBS + protease inhibitors, then lysed in lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 1 mM Na orthovanadate, 1 mM DTT, and protease inhibitors) for 30 min on ice. Lysates were mixed with equal parts adjustment buffer (50 mM Tris, 150 mM NaCl), then cell debris was pelleted and lysates transfered to new tubes. Lysates were incubated with or without antibodies for 2 h at 4°C with rotation, then incubated with Protein A Sepharose beads (#17-5280-01, Amersham) for 2 h at 4°C with rotation. Immunoprecipitates were washed 3X with wash buffer (1:1 lysis buffer + adjustment buffer), then resuspended in 2X SDS loading dye and boiled for 5 min before storing at −20°C. Proteins were separated by SDS-polyacrylamide gel electrophoresis, transfered to PVDF membranes, blocked 1 hr at RT with 5% milk diluted in PBS, and incubated at 4°C overnight with anti-HA antibody diluted 1/100 in 5% milk-PBS. Membranes were washed in PBS and incubated with a horseradish peroxidase-conjugated anti-mouse antibody (GE Healthcare). SuperSignal West Pico Chemiluminescent Substrate (Pierce) was used for detection.
Preparation of nuclear extracts
Crude nuclear extracts from 10T1/2 cells overexpressing Pax3-HA were prepared as previously described (15). Total protein in the extracts was quantitated by the Bradford method (8).
Gel mobility shift assays
Gel-shift assays were carried out as previously described (22) using nuclear extracts prepared as above. Incubations with antibodies or unlabeled oligonucleotide competitors were carried out at room temperature for 20 min prior to the addition of probe. Forward sequences of oligonucleotides used as probe/competitors are: consensus Pax3 motif from the −58/−56 kb distal Myf5 enhancer: 5’-GCATGACTAATTGCATGGTAACTGGAGAAA-3’ (11); wt Pax3 HD motif from the Myf5 promoter: 5’-CTGGGCGTTATTAGCATATCCCACC-3’; mt Pax3 HD motif from the Myf5 promoter: 5’- CTGGGCGTTATGAGGATCTACCACC-3’. Mutated bases are underlined.
RT-PCR
RNA was extracted using the Qiagen RNeasy kit, according to the manufacturer’s instructions. RNA was DNase-treated and reverse-transcribed as previously described (22). PCR was performed using 20–50 ng cDNA and Pfu DNA polymerase with the following cycling conditions: 95°C for 5 min, followed by 38 cycles of 95°C for 1 min, 51°C for 1 min, 72°C for 45 sec, and a final extension at 72°C for 10 min. Primer sequences for amplifying Myf5 and GAPDH are as described (43).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed with 10T1/2 cells overexpressing FLAG-Zic1 and Pax3-HA using the Fast ChIP method (40) with some modifications. Cells were fixed in 1% formaldehyde in DMEM for 10 min and dounced 10X prior to sonication. Cells were sonicated for 8 rounds of 15-sec pulses at 90% power output on a Branson Sonifier 450 (VWR Scientific) to shear the DNA to a ladder of ~200–800 bp, and efficiency of shearing was verified by agarose gel electrophoresis. Chromatin was immunoprecipitated using 2 µg of specific antibodies or normal rabbit IgG. Quantitative PCR was performed using forward and reverse primers (300 nM) and the QuantitTect SYBR Green PCR Kit (Qiagen). Reaction conditions were 40 cycles of: 94°C for 15 sec, 55°C for 30 sec, and 72°C for 30 sec. Sequences of primers are as follows: Myf5 promoter: F: 5’- GTCAAAGGGACCAGTAAAC-3’; R: 5’-GGGGCTCTTTATATATTCCTG-3’; ES enhancer: F: 5’-CAAAGCCCCAGAGAGAGCCGGA-3’; R: 5’-CCTGGCGTGCTTTGCTCTGC-3’. PCR products were analyzed on a 1.5% agarose gel to verify correct size of product and specificity of primer annealing.
Results and Discussion
Pax3 synergizes with Gli2 and Zic1 in transactivating the Myf5 ES enhancer and promoter
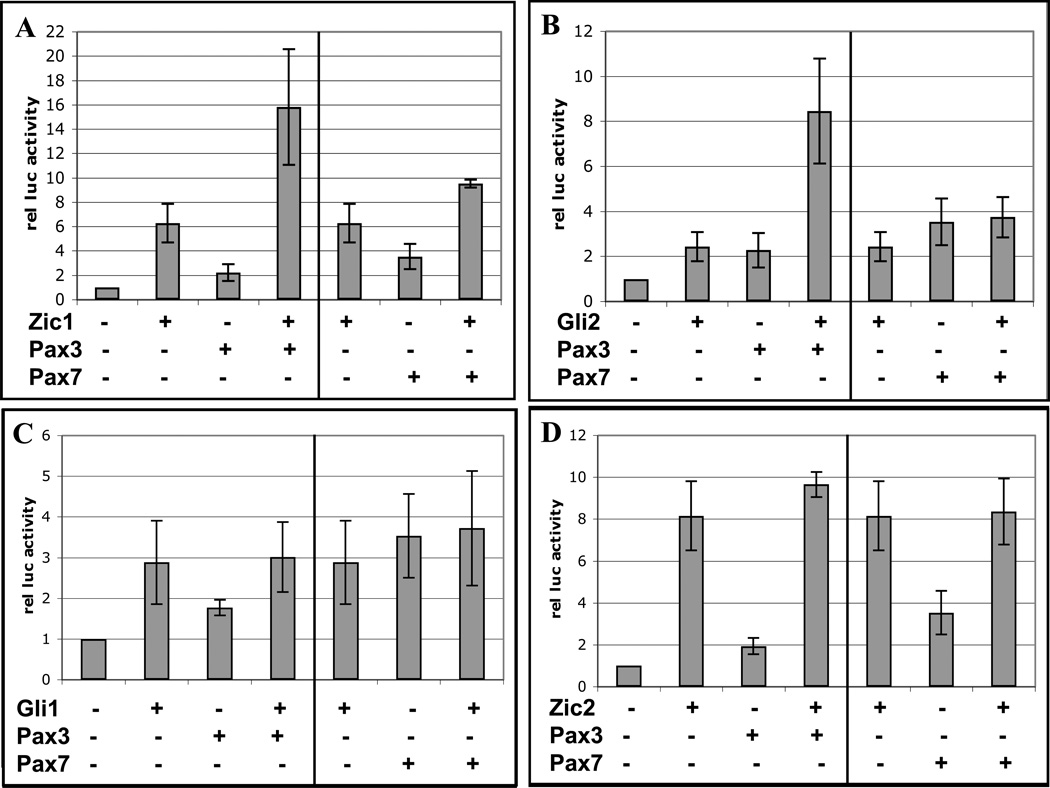
It has been demonstrated that promoter context is critical in determining the behavior of enhancers, and the Myf5 ES enhancer is no exception (21, 56). To rule out potentially spurious results following the use of a non-homologous promoter, we cloned the mouse ES enhancer and Myf5 promoter (56) upstream of the firefly luciferase reporter for use in these studies. To test whether Gli and Zic transcription factors cooperate with members of the Pax family in activating the Myf5 ES enhancer, we co-transfected expression constructs for these factors with the reporter construct into 3T3 cells, and assayed luciferase activity (Fig 1). 3T3 fibroblasts represent a convenient system in which to test these potential interactions, since these cells express low levels of Gli2 and no Pax3 or Zic factors. While overexpression of Zic1 or Pax3 alone increased activity of the reporter construct, the combination of factors displayed a modest, but statistically significant synergy (Fig. 1A). Likewise, Gli2 and Pax3 synergized in activating the ES enhancer-promoter (Fig. 1B). Interestingly, other family members (Gli1, Zic2, and Pax7) showed no synergistic effects (Fig. 1C & D), suggesting that factor-specific contacts are required for recruitment/stabilization of coactivators.
Fig. 1. Pax3 synergizes with Gli2 and Zic1 in transactivating the Myf5 ES enhancer and promoter.
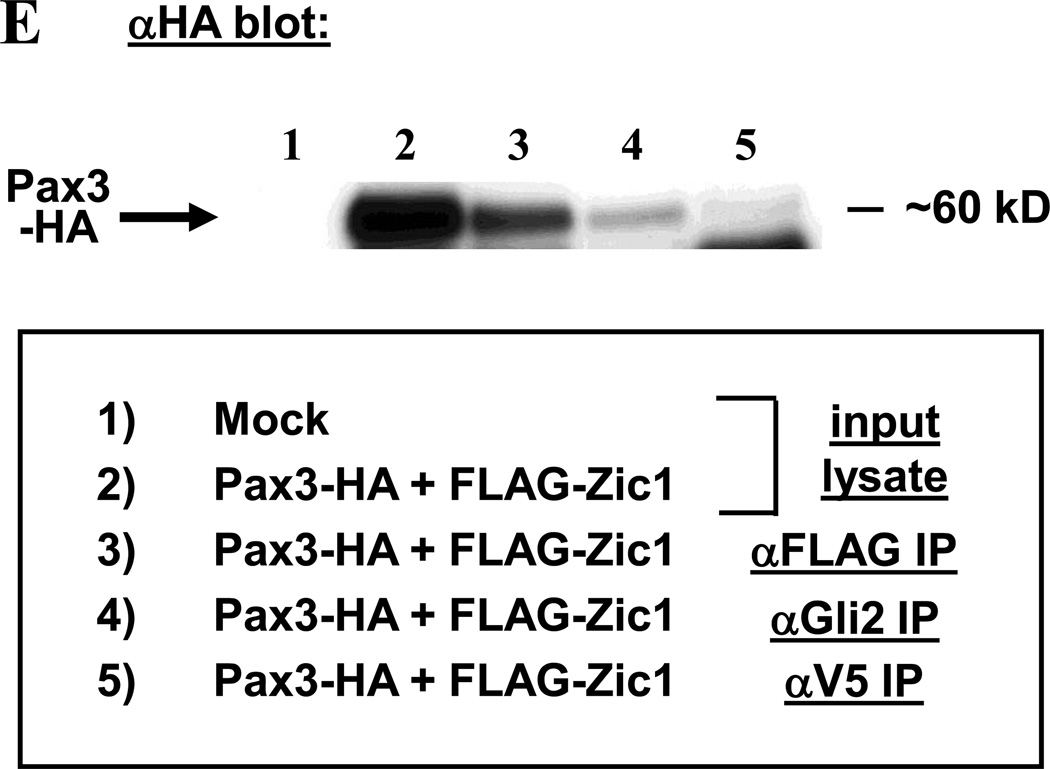
A–D. A luciferase reporter plasmid containing the Myf5 ES enhancer and minimal promoter was transiently transfected into 3T3 cells with or without expression plasmids for Zic1, Zic2, Gli1, Gli2, Pax3, or Pax7. Cells were harvested 48 h post-transfection and assayed for luciferase activity. Data are plotted as the mean value and standard deviation of relative luciferase activity, with activity of the reporter construct alone set at 1. Pax3 synergizes with Zic1 (A, asterisk = p < 0.01) and Gli2 (B, asterisk = p < 0.05) in transactivating the reporter (one-tailed, one-step t-test comparing activity of factors in combination to the sum of individual factors), whereas other combinations of factors show no synergistic effects (C-D). E. 10T1/2 cells were transfected with Pax3-HA and FLAG-Zic1 expression plasmids, and cell extracts were immunoprecipitated with antibodies to FLAG, Gli2, or V5 (as a negative control). Immunoprecipitates were subjected to Western analysis using HA antibodies.
To determine whether Gli2 and Zic1 can physically associate with Pax3 in the absence of DNA, we made cytoplasmic extracts from 10T1/2 cells transfected with FLAG-Zic1 and Pax3-HA. 10T1/2 cells were used because they produced high levels of the overexpressed proteins for co-immunoprecipitaton assays. Co-immunoprecipitations were performed using antibodies to FLAG or endogenous Gli2, and immunoprecipitated proteins were probed with HA antibodies. Pax3-HA, which runs at ~60 kD, was not detected in mock-transfected lysates, only in transfected cells (Fig. 1E). Pax3 was precipitated with antibodies to FLAG or Gli2; by contrast, only a very faint band was detected using antibodies to an unrelated V5 epitope (Fig. 1E). These results indicate a specific physical interaction between Zic1-Pax3 and Gli2-Pax3.
A novel homeodomain motif in the Myf5 promoter is required for Pax3 synergy with Gli2 and Zic1
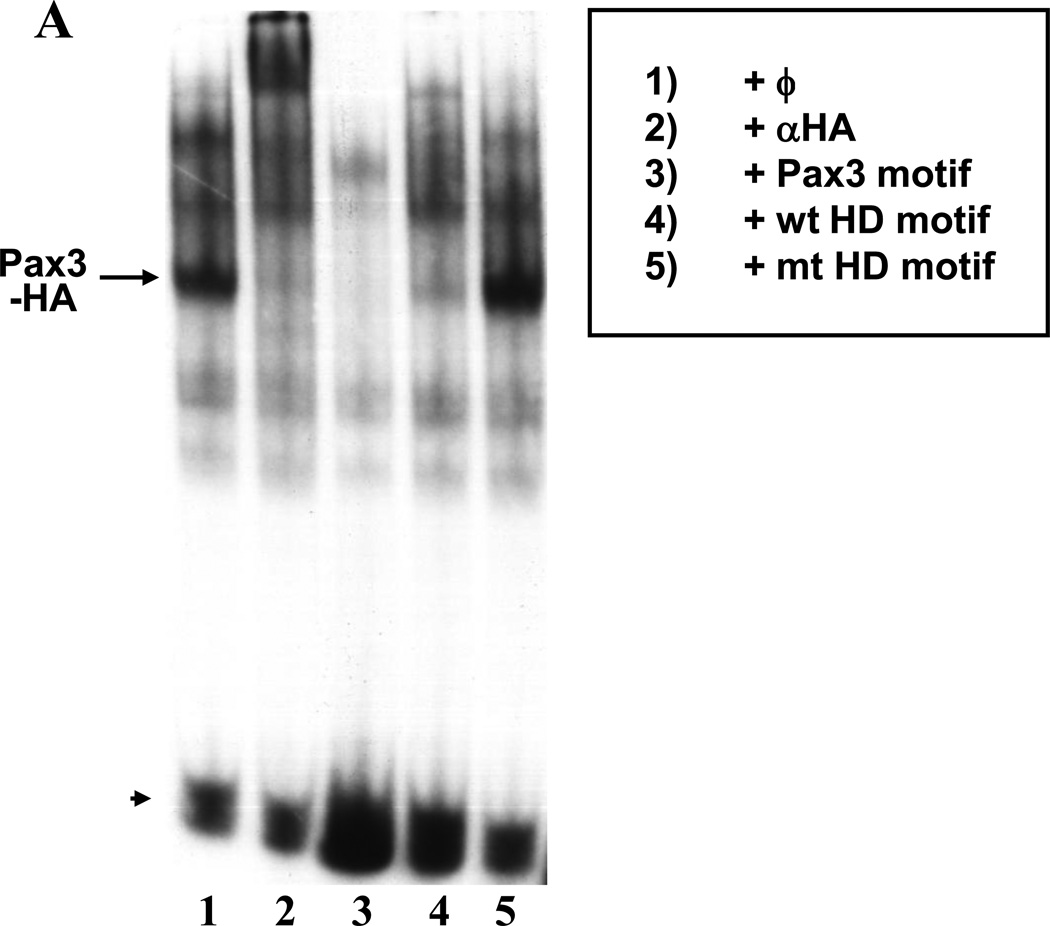
The Pax family is structurally defined by the presence of a DNA-binding motif called a paired domain (PD). A subset of family members, including Pax3, contain an additional DNA8-binding domain known as a paired-type homeodomain (HD) (33, 41). The PD and HD are functionally interdependent and capable of modifying Pax binding to DNA (13). Although the Myf5 ES enhancer and promoter do not contain a consensus PD motif, multi-species sequence alignments revealed a highly conserved single HD motif (TAAT) at −79 relative to the transcription start site in mouse. To determine whether Pax3 can bind this sequence, we performed gel-shift assays using nuclear extracts from 10T1/2 cells transfected with Pax3-HA. Pax3-HA bound to the labeled probe containing a consensus Pax3 motif from the −58/−56 kb distal Myf5 enhancer (11), and this complex was supershifted with antibodies to HA (Fig. 2A, lanes 1–2). An excess of cold competitor oligonucleotides containing the consensus Pax3 motif competed away this complex (Fig. 2A, lane 3). Importantly, the wild-type, but not the mutant Myf5 promoter HD motif also competed for Pax3 binding (Fig. 2A, lanes 3–5), although not as well as the consensus Pax3 sequence, which contains both a paired motif and a HD motif. This indicates that Pax3 can recognize the HD motif from the Myf5 promoter, although binding is not as strong in the absence of a paired motif.
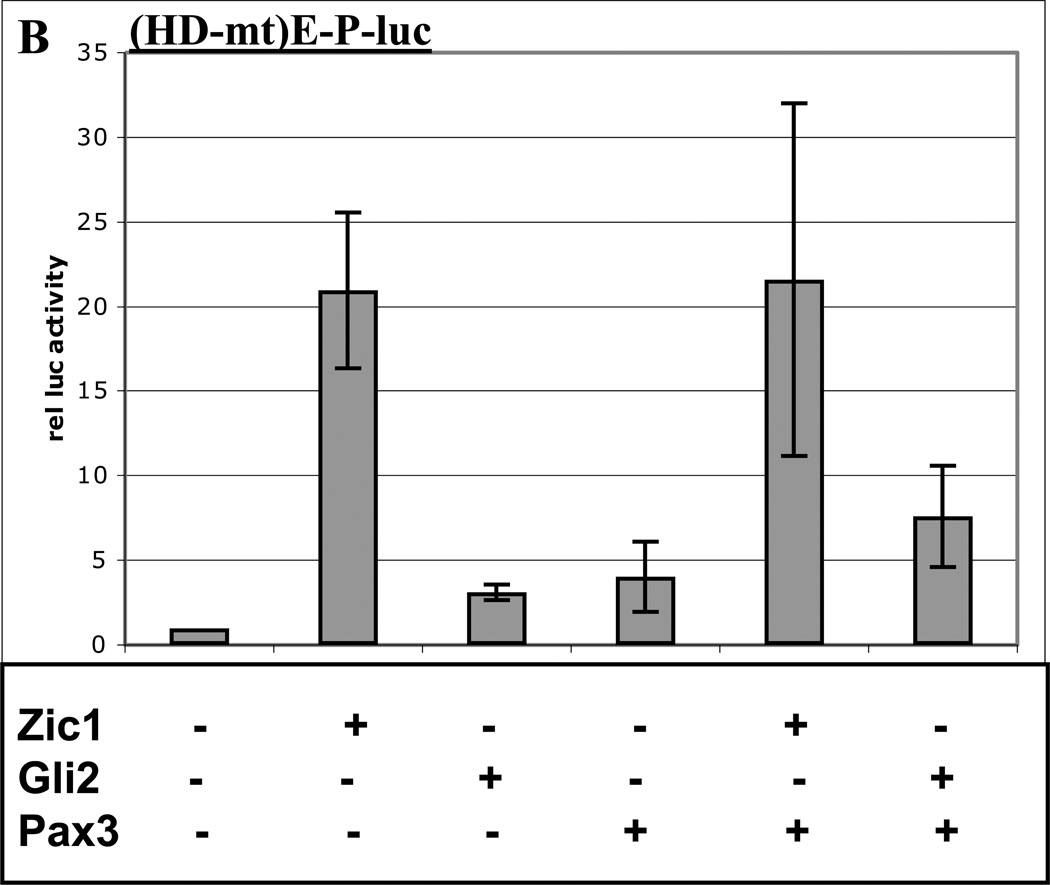
Fig. 2. Sequences in the Myf5 ES enhancer and promoter are required for Pax3 synergy with Gli2 and Zic1.
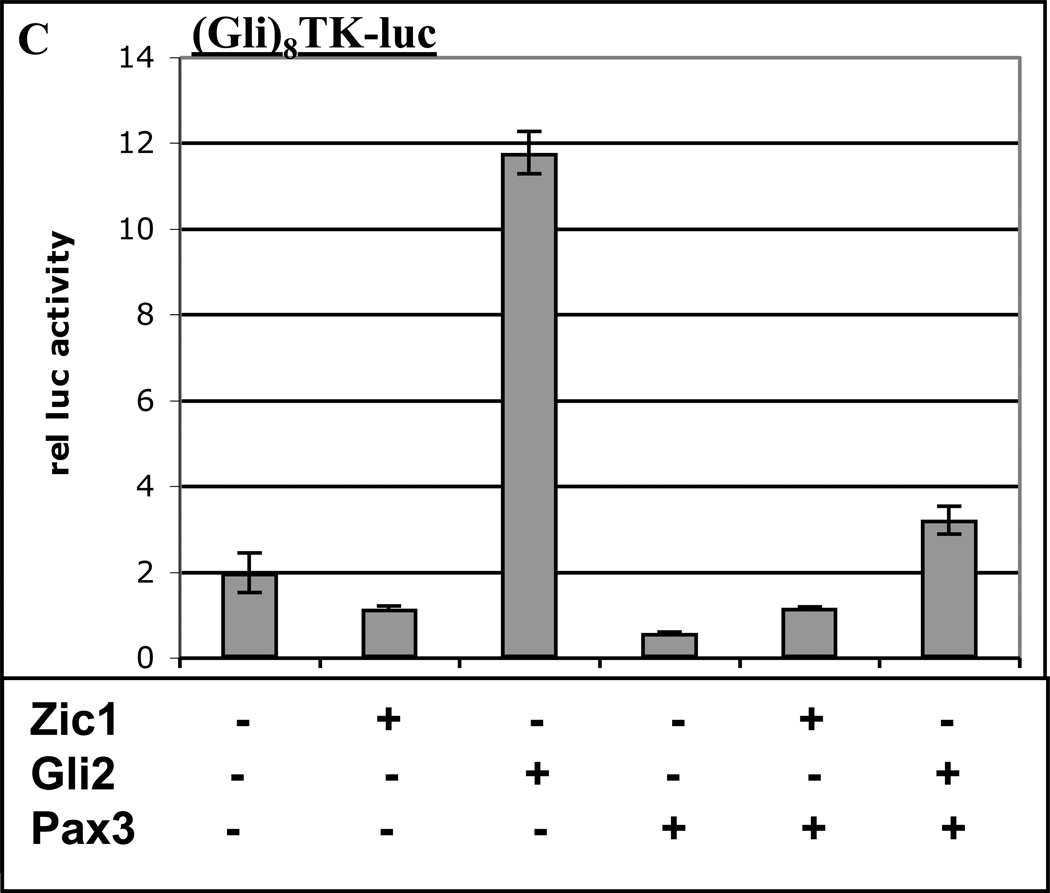
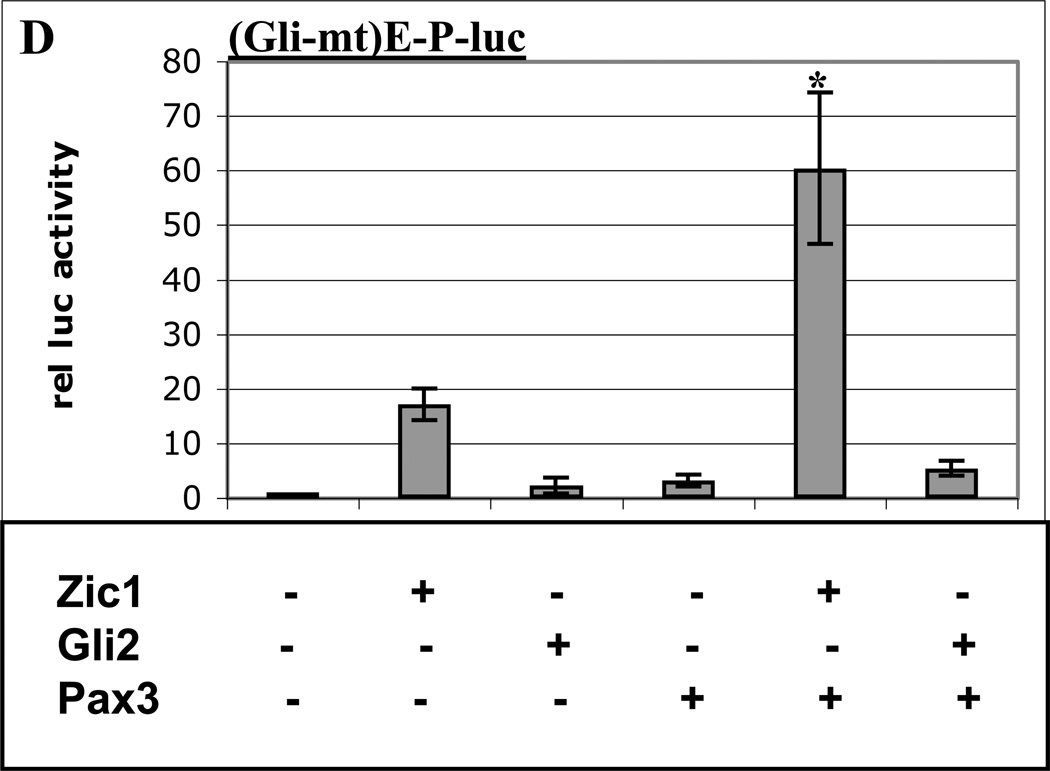
A. Labeled probe containing a consensus Pax3 binding site from the −58/−56 kb distal Myf5 enhancer (11) was mixed with nuclear extracts from 10T1/2 cells overexpressing Pax3-HA, and analyzed via gel-shift assays. Antibodies (αHA, lane 2) or competitor oligos (Pax3 consensus motif, lane 3; wt HD motif in Myf5 promoter, lane 4, and mt HD motif in Myf5 promoter, lane 5) are indicated. The complex containing Pax3-HA bound to the probe (supershifted with αHA in lane 2) is labeled. Arrowhead indicates free probe. B. A luciferase reporter plasmid containing the Myf5 ES enhancer and promoter with a mutation in the promoter HD motif (HD-mt) was transiently transfected into 3T3 cells with or without expression plasmids for Zic1, Gli2, or Pax3. C. A luciferase reporter plasmid containing 8 consensus Gli motifs upstream of the thymidine kinase (TK) promoter was transiently transfected into 3T3 cells with or without expression plasmids for Zic1, Gli2, or Pax3. D. A luciferase reporter plasmid containing the Myf5 ES enhancer and promoter with a mutation in the ES enhancer Gli motif (Gli-mt) was transiently transfected into 3T3 cells with or without expression plasmids for Zic1, Gli2, or Pax3. For B–D, cells were harvested and assayed, and data was analyzed as in Fig. 1. Asterisk in D indicates synergy between Zic1 and Pax3 (p < 0.05, one-tailed, one-step t-test comparing activity of factors in combination to the sum of individual factors).
To determine whether the HD motif is required for Pax3 synergy with Zic1 and Gli2, we performed cotransfection experiments with the Myf5 reporter construct containing a mutated HD motif, as described above. Surprisingly, Zic1 appears to be a stronger activator of the ES enhancer-promoter construct when the HD motif is mutated, and Pax3 is still able to activate this construct, indicating an indirect effect of Pax3 on the ES enhancer or Myf5 promoter (Fig. 2B). Importantly, despite the higher individual activity of these factors, Zic1-Pax3 synergy is completely abrogated in the absence of a functional HD motif (Fig. 2B). Likewise, Gli2-Pax3 synergy is lost on the HD-mutated construct (Fig. 2B). These results indicate that the novel HD motif in the Myf5 promoter is required for Pax3 to synergize with both Zic1 and Gli2, in spite of the fact that these factors can interact in the absence of DNA. It is possible that association between Zic1/Gli2 and Pax3 helps to stabilize Pax3 binding to the HD motif in the absence of a paired motif in the Myf5 promoter.
To confirm that Pax3 does not cooperate with Gli2 in the absence of a Pax binding site, we tested Pax3-Gli2 interactions on a reporter construct containing 8 Gli binding sites upstream of the Thymidine kinase (TK) promoter. As expected, Gli2 strongly activates this construct, whereas Pax3 does not (Fig. 2C). The combination of Gli2 and Pax3 is less potent than Gli2 alone, demonstrating that Pax3 does not behave as a cofactor for Gli2 (Fig. 2C).
The essential Gli motif in the Myf5 ES enhancer is required for Pax3-Gli2 synergy
The conserved variant Gli motif in the ES enhancer is required for maintenance of Myf5 expression in the epaxial somite via Shh signaling (21, 56), and Glis have been demonstrated to bind this essential site (21). To verify that this sequence is required for Gli2-Pax3 synergy, we performed cotransfection experiments with a Myf5 reporter construct containing a mutation in the ES enhancer Gli motif. As expected, synergy between Gli2 and Pax3 is abolished in the absence of a functional Gli binding site (Fig. 2D), indicating that Gli2 binding to the ES enhancer is required for synergy with Pax3.
Both Zic and Gli family members bind to DNA via five C2H2-type zinc fingers, and Zics have been shown to recognize Gli binding sites, albeit with much lower affinity than Gli factors (37). To test whether Zic1 synergizes with Pax3 via binding to the Gli motif in the ES enhancer, we tested Zic1-Pax3 interactions on the Gli-mt reporter construct. Zic1 and Pax3 are still capable of synergizing in the absence of a functional Gli binding site, indicating that Zic1 does not require the Gli motif in the ES enhancer to synergize with Pax3 (Fig. 2D). Furthermore, Zic1-Pax3 synergy does not take place on a construct driven by multiple Gli motifs (Fig. 2C), providing further evidence that Zic1 does not synergize with Pax3 via binding to Gli motifs.
In addition to recognizing Gli motifs, Zic family members have been shown to bind a wide range of GC-rich sequences in their target genes (17, 37, 48, 60). Two conserved candidate sequences in the Myf5 promoter were able to compete for Zic1 binding in gel-shift assays; however, mutation of either sequence had no effect on Zic1-Pax3 synergy (data not shown). This suggests that Zic1 synergizes with Pax3 by binding to functionally redundant motifs in the Myf5 promoter; however, we cannot rule out that Zic1 acts as a transcriptional cofactor for Pax3. This is particularly difficult to test in light of the fact that Zic1 activates a wide variety of promoters through binding to degenerate GC-rich motifs (36, 37).
Conserved regions of Pax3, Gli2, and Zic1 are required for synergy
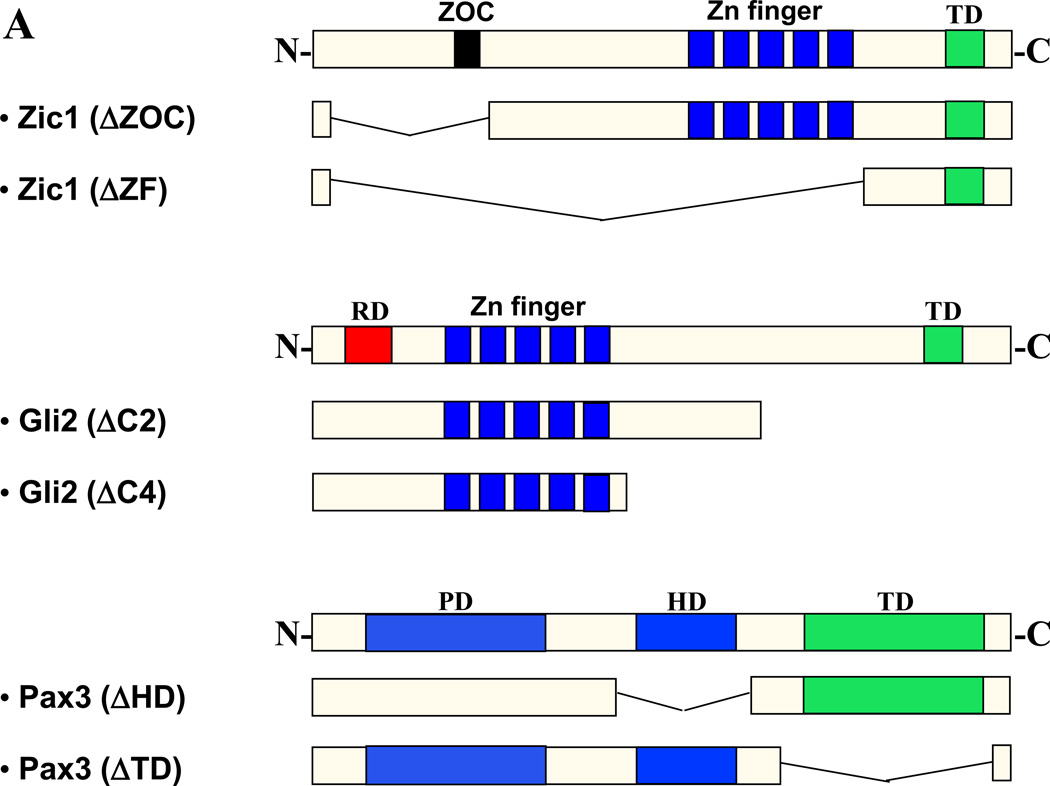
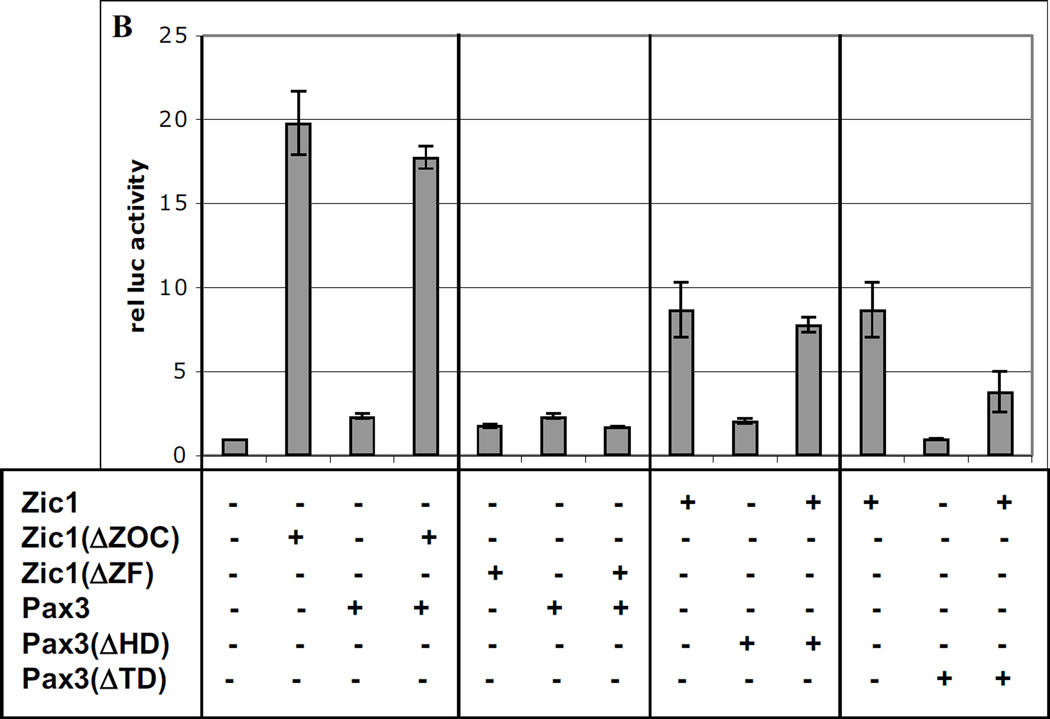
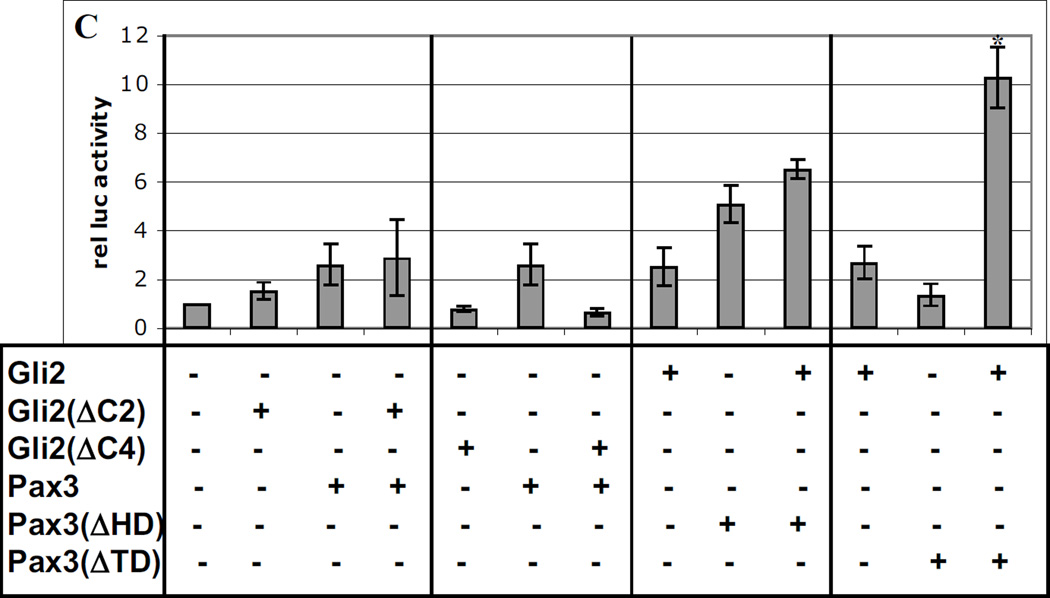
To determine regions of the proteins required for synergy, we tested various truncated forms of the three factors (Fig. 3A) in co-transfection experiments with the Myf5 ES enhancer-promoter reporter. Interestingly, Zic1 lacking the Zic-Opa conserved motif (Zic1ΔZOC) was a more potent transactivator than full-length Zic1, suggesting that the ZOC motif may serve a repressive function (Fig. 3B). However, despite its higher activity, Zic1ΔZOC was unable to synergize with Pax3, indicating that in addition to repressing Zic1 activity, the ZOC motif is also required for cooperative interactions with other factors (Fig. 3B). This is consistent with previous studies indicating that this protein domain behaves as a context-dependent activator or repressor of transcription (38). Zic factors associate with DNA via their zinc fingers; as expected, when the zinc fingers of Zic1 were removed (Zic1ΔZF), the protein had little effect on reporter activity and was incapable of synergizing with Pax3 (Fig. 3B). Truncation of the Pax3 HD also resulted in a loss of synergy with Zic1, further confirming that DNA-binding of Pax3 is required for synergy (Fig. 3B). Likewise, when the transactivation domain (TD) of Pax3 was deleted, the remaining protein was incapable of synergizing with Zic1, suggesting that the ability of Pax3 to recruit coactivators is critical for synergy (Fig. 3B).
Fig. 3. Conserved regions of Pax3, Gli2, and Zic1 are required for synergy.
A. Diagram of truncations in expression constructs encoding Zic1, Gli2, and Pax3. ZOC = ZOC motif; ZF = zinc finger domain; HD = homeodomain; TD = transactivation domain; C2 & C4 = C terminal domains. B–C. Truncated constructs in A were tested for synergy in transactivating the Myf5 ES enhancer and promoter as in Fig 1–2. Asterisk in C indicates synergy (p = 0.01) between Gli2 and Pax3(ΔTD) (one-tailed, one-step t-test comparing activity of factors in combination to the sum of individual factors).
Interestingly, truncation of the C-terminal TD of Gli2 (ΔC2 and ΔC4) also abrogated synergy with Pax3, but deletion of the Pax3 TD had no effect on synergy with Gli2 (Fig. 3C), suggesting that in the context of Gli2-Pax3 interactions, it is the Gli2 TD that is competent to recruit transcriptional coactivators. This is consistent with studies indicating that Gli2 acts primarily as a transcriptional activator (16, 34) and is the major transducer of Shh signaling in the mouse (35, 44). As with Pax3-Zic1, Pax3-Gli2 synergy is dependent on the conserved HD of Pax3 (Fig. 3C).
Pax3 and Zic1 are enriched at the ES enhancer during induction of Myf5 transcription
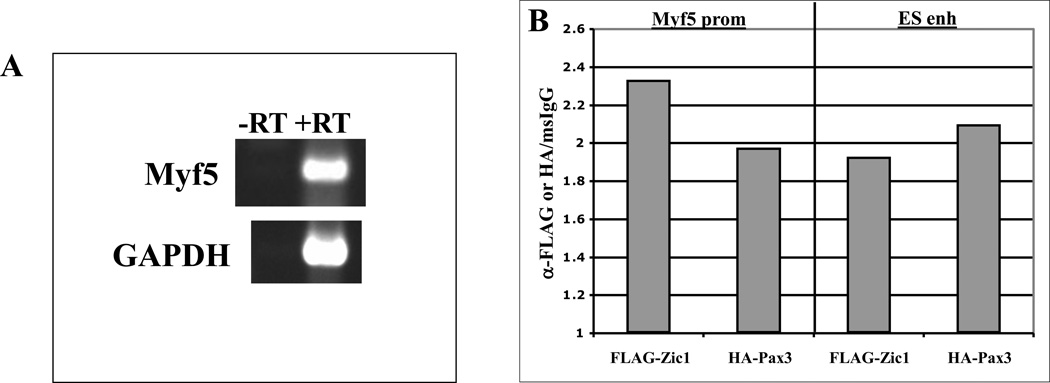
Zic1 has been shown to initiate expression of Myf5 in 10T1/2 cells, which can be induced to form skeletal muscle in response to myogenic cues (43). To confirm that Zic1 and Pax3 occupy the endogenous Myf5 promoter during activation of Myf5 expression, we performed chromatin immunoprecipitation (ChIP) assays on chromatin from 10T1/2 cells overexpressing FLAG-Zic1 and Pax3-HA. Myf5 induction in the transfected cells was confirmed by RT-PCR (Fig. 4A). Immunoprecipitation with either FLAG- or HA-specific antibodies yielded ~2-fold enrichment of the Myf5 promoter over that obtained with non-immune IgG (Fig. 4B), demonstrating that these factors occupy the endogenous Myf5 promoter during induction of Myf5 expression. Interestingly, we also observed enrichment of FLAG-Zic1 and Pax3-HA at the ES enhancer, likely due to enhancer-promoter looping interactions (Fig. 4B) (49).
Fig. 4. Pax3 and Zic1 are enriched at the ES enhancer during activation of Myf5 transcription.
10T1/2 cells were transiently transfected with FLAG-Zic1 and Pax3-HA expression plasmids. RNA was isolated for RT-PCR or cells were fixed for chromatin immunoprecipitation (ChIP) analysis. A. RT-PCR was performed using primers specific for Myf5 and GAPDH. B. ChIP assays were performed using antibodies specific for FLAG or HA or normal mouse IgG. Immunoprecipitated chromatin was analyzed by qPCR using primers specific for the Myf5 promoter or ES enhancer. Data are represented as fold enrichment of the Myf5 promoter or ES enhancer by αFLAG or αHA relative to normal mouse IgG.
Cooperation among several distinct pathways promotes myogenesis in the epaxial somite
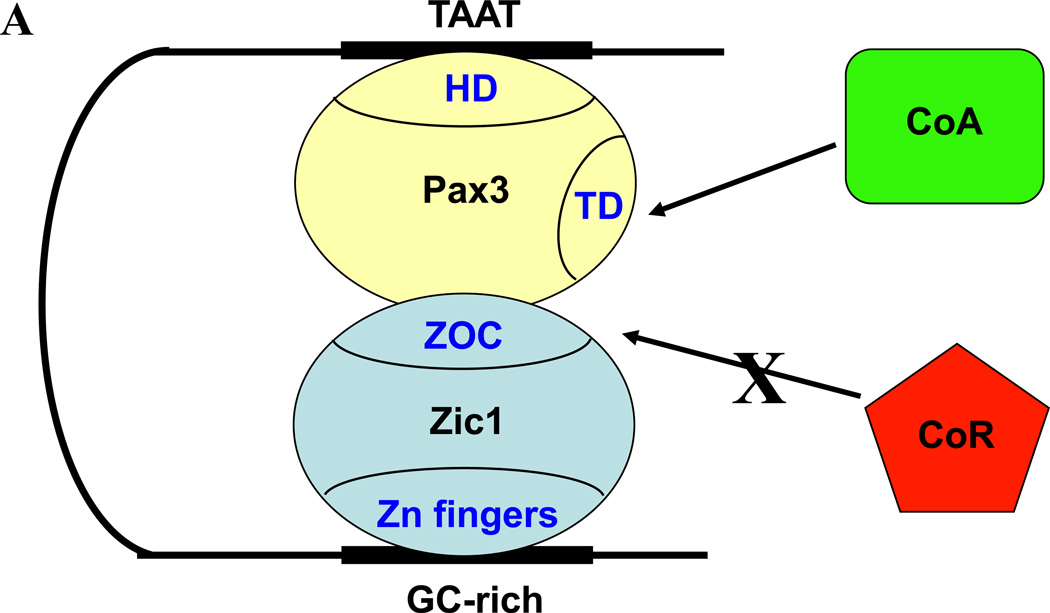
Taken together, our data suggest an intriguing new model for cooperative interactions between Pax3 and Zic1, in which Pax3 binds its recognition motif in the Myf5 promoter via the HD and Zic1 binds GC-rich sequences via its zinc finger domain (Fig. 5A). Contacts between Pax3 and the ZOC motif of Zic1 (which is normally repressive in the absence of Pax3) prevent Zic1 from recruiting transcriptional co-repressors, while the transactivation domain (TD) of Pax3 serves to recruit transcriptional co-activators for gene expression (Fig. 5A).
Fig. 5. Cooperation among several distinct pathways promotes myogenesis in the epaxial somite.
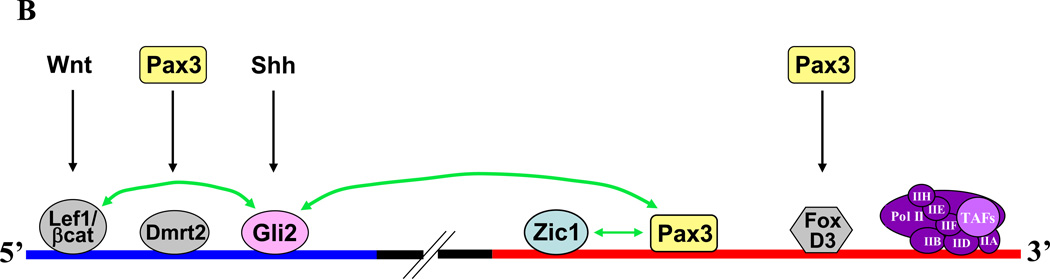
A. Model of Pax3-Zic1 synergy. Pax3 binds the homeodomain motif in the Myf5 promoter (TAAT) via its homeodomain (HD), and Zic1 binds GC-rich sequences via its zinc finger domain. Interaction between Pax3 and the ZOC domain of Zic1 prevents the latter from recruiting transcriptional co-repressors (CoR). The transactivation domain (TD) of Pax3 recruits transcriptional co-activators (CoA). B. Transcription factors and upstream signaling pathways regulating Myf5 expression in the epaxial somite. The blue bar indicates the Myf5 ES enhancer, and the red bar indicates the Myf5 promoter. Green arrows indicate synergistic interactions between transcription factors. Shh signaling is required for Gli activation of the ES enhancer via a single essential motif (21, 56). Wnt and Shh signaling culminate in synergy between LEF1/β-catenin bound to three motifs (shown as a single motif for simplicity) in the ES enhancer and Gli bound downstream (5). Pax3 activates Myf5 in the epaxial somite indirectly, by driving expression of the Dmrt2 transcription factor, which activates the ES enhancer (51), and in zebrafish, by driving expression of FoxD3, which activates the Myf5 promoter (32). In the current work, Pax3 also activates epaxial Myf5 expression directly, by binding a homeodomain motif in the Myf5 promoter and synergizing with both Gli2, bound to the ES enhancer, and Zic1, which recognizes GC-rich motifs in the promoter. Refer to the text for more details.
Our work places Zic and Pax genes in the broader context of several well-established signaling pathways that regulate myogenesis (Fig. 5B). As shown in our earlier work, by enhancing nuclear translocation of Gli factors, Zics provide spatial patterning for the Gli family, which is expressed throughout the somite, to activate Myf5 expression in epaxial muscle progenitors (43). In contrast to this, our current study indicates a different mechanism of cooperativity between Zic1 and Pax3, through the establishment of interactions that require DNA-binding and likely help to recruit or stabilize coactivator proteins. Interestingly, the combination of Pax3/7 and Zic genes is sufficient to induce neural crest formation in Xenopus (52), and in the mouse, Pax3 and Zic1 are both expressed in the dorsal neural tube (20, 43). This indicates that in addition to driving commitment of cells to other lineages, the presence of these factors is not sufficient to induce Myf5 expression in non-muscle tissues.
Pax3 serves to regulate Myf5 in the epaxial somite at multiple levels – as we have shown, through direct binding of a homeodomain motif within the promoter, as well as indirectly, via upregulation of FoxD3 and Dmrt2 (32, 51) (Fig. 5B). Although the BMP antagonist Noggin is required for Zic2 expression in the epaxial somite (43), the positive signals mediating expression of Zics in this compartment are still unknown, although Wnts secreted from the dorsal neural tube and surface ectoderm are likely candidates. Signaling by both canonical and non-canonical Wnts plays an important role in myogenesis (57), and direct binding by LEF1/β-catenin is required for full activity of the ES enhancer via synergy with Gli (5) (Fig. 5B).
It will be interesting to determine if the novel synergistic interactions described here extend to Myf5 activation/maintenance in other muscle lineages. During embryogenesis, Pax3 is expressed in the dorsal neural tube and PSM, followed by expression throughout the somites which is subsequently restricted to the dermomyotome (19, 20). Following this, Pax3 expression is decreased in the epaxial somite and maintained in hypaxial precursors (4, 19, 59). At E12.5, Pax3 continues to be expressed in MyoD-positive regions in the trunk and the limbs before expression is lost at later stages (23). Pax3 directly regulates Myf5 expression in the hypaxial somite and some hindlimb muscle precursors via binding to a −57.5 kb upstream enhancer (3). Recently, Pax3 was also shown to be a direct regulator of the −111 kb enhancer, which regulates expression of Myf5 in the ventral somite and a subset of limb muscle precursors (14). Since Shh was recently shown to be required for Myf5 expression in limb muscle progenitor cells (1, 26), it will be important to determine whether Gli2 and Pax3 synergize in driving Myf5 expression in this muscle lineage. Although Shh is not required for Myf5 activation in hypaxial progenitors (7, 31), Gli2 expression overlaps that of Myf5 in this domain (35). This raises the possibility that Gli2 (activated independent of Shh signaling; possibly via FGF and PKCδ/MEK1 (27, 46)) and Pax3 synergize in activating hypaxial Myf5 expression. Likewise, while the strong epaxial expression of Zics closely mimics that of Pax3 at E9.5 (43), Zic2/3 are also expressed in the limb buds and in the developing limbs at later stages (39). Thus, it will also be important to determine whether Zics synergize with Pax3 in activating the hypaxial Myf5 enhancers. Understanding the spatiotemporal dynamics of these interactions and the mechanisms by which these factors cooperate to drive myogenesis remains a significant challenge for future studies.
Highlights.
Pax3 synergizes with Gli2 and Zic1 in transactivating Myf5.
Pax3 synergy with Gli2/Zic1 requires a novel Myf5 promoter homeodomain motif.
Pax3 synergy with Gli2 requires the essential Gli motif in the Myf5 ES enhancer.
Zic1 and Pax3 are enriched at the ES enhancer during Myf5 induction.
Acknowledgements
We thank H. Sasaki for providing all full-length and truncated Gli expression constructs, K. Hanger and P. Zhu for technical assistance, and J. Chen for critical reading of the manuscript. This study was supported by NIH RO1HD007796.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson C, Williams VC, Moyon B, Daubas P, Tajbakhsh S, Buckingham ME, Shiroishi T, Hughes SM, Borycki AG. Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 2012;26:2103–2117. doi: 10.1101/gad.187807.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aruga J, Mizugishi K, Koseki H, Imai K, Balling R, Noda T, Mikoshiba K. Zic1 regulates the patterning of vertebral arches in cooperation with Gli3. Mech Dev. 1999;89:141–150. doi: 10.1016/s0925-4773(99)00220-8. [DOI] [PubMed] [Google Scholar]
- 3.Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham ME. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20:2450–2464. doi: 10.1101/gad.382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 5.Borello U, Berarducci B, Murphy P, Bajard L, Buffa V, Piccolo S, Buckingham M, Cossu G. The Wnt/beta-catenin pathway regulates Glimediated Myf5 expression during somitogenesis. Development. 2006;133:3723–3732. doi: 10.1242/dev.02517. [DOI] [PubMed] [Google Scholar]
- 6.Borycki A, Brown AM, Emerson CP., Jr Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development. 2000;127:2075–2087. doi: 10.1242/dev.127.10.2075. [DOI] [PubMed] [Google Scholar]
- 7.Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP., Jr Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126:4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- 8.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 10.Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- 11.Buchberger A, Freitag D, Arnold HH. A homeo-paired domain-binding motif directs Myf5 expression in progenitor cells of limb muscle. Development. 2007;134:1171–1180. doi: 10.1242/dev.02798. [DOI] [PubMed] [Google Scholar]
- 12.Carvajal JJ, Keith A, Rigby PW. Global transcriptional regulation of the locus encoding the skeletal muscle determination genes Mrf4 and Myf5. Genes Dev. 2008;22:265–276. doi: 10.1101/gad.442408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corry GN, Raghuram N, Missiaen KK, Hu N, Hendzel MJ, Underhill DA. The PAX3 paired domain and homeodomain function as a single binding module in vivo to regulate subnuclear localization and mobility by a mechanism that requires base-specific recognition. J Mol Biol. 2010;402:178–193. doi: 10.1016/j.jmb.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Daubas P, Buckingham ME. Direct molecular regulation of the myogenic determination gene Myf5 by Pax3, with modulation by Six1/4 factors, is exemplified by the-111kb-Myf5 enhancer. Dev Biol. 2013;376:236–244. doi: 10.1016/j.ydbio.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 17.Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, Liu Y, Hunsaker TL, Johnson JE. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130:1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- 18.Francetic T, Li Q. Skeletal myogenesis and Myf5 activation. Transcription. 2011;2:109–114. doi: 10.4161/trns.2.3.15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 1994;120:957–971. doi: 10.1242/dev.120.4.957. [DOI] [PubMed] [Google Scholar]
- 20.Goulding MD, Chalepakis G, Deutsch U, Erselius JR, Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. Embo J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP., Jr Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev. 2002;16:114–126. doi: 10.1101/gad.940702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himeda CL, Ranish JA, Hauschka SD. Quantitative proteomic identification of MAZ as a transcriptional regulator of muscle-specific genes in skeletal and cardiac myocytes. Mol Cell Biol. 2008;28:6521–6535. doi: 10.1128/MCB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horst D, Ustanina S, Sergi C, Mikuz G, Juergens H, Braun T, Vorobyov E. Comparative expression analysis of Pax3 and Pax7 during mouse myogenesis. Int J Dev Biol. 2006;50:47–54. doi: 10.1387/ijdb.052111dh. [DOI] [PubMed] [Google Scholar]
- 24.Hoth CF, Milunsky A, Lipsky N, Sheffer R, Clarren SK, Baldwin CT. Mutations in the paired domain of the human PAX3 gene cause Klein- Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I) Am J Hum Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- 25.Houtmeyers R, Souopgui J, Tejpar S, Arkell R. The ZIC gene family encodes multi-functional proteins essential for patterning and morphogenesis. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu JK, McGlinn E, Harfe BD, Kardon G, Tabin CJ. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 2012;26:2088–2102. doi: 10.1101/gad.187385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang R, Stolte D, Kurz H, Ehehalt F, Cann GM, Stockdale FE, Patel K, Christ B. Ventral axial organs regulate expression of myotomal Fgf-8 that influences rib development. Dev Biol. 2003;255:30–47. doi: 10.1016/s0012-1606(02)00051-9. [DOI] [PubMed] [Google Scholar]
- 28.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 29.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 30.Koyabu Y, Nakata K, Mizugishi K, Aruga J, Mikoshiba K. Physical and functional interactions between Zic and Gli proteins. J Biol Chem. 2001;276:6889–6892. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- 31.Kruger M, Mennerich D, Fees S, Schafer R, Mundlos S, Braun T. Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development. 2001;128:743–752. doi: 10.1242/dev.128.5.743. [DOI] [PubMed] [Google Scholar]
- 32.Lee HC, Huang HY, Lin CY, Chen YH, Tsai HJ. Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev Biol. 2006;290:359–372. doi: 10.1016/j.ydbio.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 33.Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 34.Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 35.McDermott A, Gustafsson M, Elsam T, Hui CC, Emerson CP, Jr, Borycki AG. Gli2 and Gli3 have redundant and context-dependent function in skeletal muscle formation. Development. 2005;132:345–357. doi: 10.1242/dev.01537. [DOI] [PubMed] [Google Scholar]
- 36.Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- 37.Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276:2180–2188. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- 38.Mizugishi K, Hatayama M, Tohmonda T, Ogawa M, Inoue T, Mikoshiba K, Aruga J. Myogenic repressor I-mfa interferes with the function of Zic family proteins. Biochem Biophys Res Commun. 2004;320:233–240. doi: 10.1016/j.bbrc.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 39.Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- 40.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 41.Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 42.Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 43.Pan H, Gustafsson MK, Aruga J, Tiedken JJ, JC JC, Emerson CP., Jr A role for Zic1 and Zic2 in Myf5 regulation and somite myogenesis. Dev Biol. 2011;351:120–127. doi: 10.1016/j.ydbio.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 45.Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 46.Riobo NA, Haines GM, Emerson CP., Jr Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 47.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 48.Salero E, Perez-Sen R, Aruga J, Gimenez C, Zafra F. Transcription factors Zic1 and Zic2 bind and transactivate the apolipoprotein E gene promoter. J Biol Chem. 2001;276:1881–1888. doi: 10.1074/jbc.M007008200. [DOI] [PubMed] [Google Scholar]
- 49.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 51.Sato T, Rocancourt D, Marques L, Thorsteinsdottir S, Buckingham M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010;6:e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- 53.Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S, Rigby PW. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000;127:3745–3757. doi: 10.1242/dev.127.17.3745. [DOI] [PubMed] [Google Scholar]
- 54.Tajbakhsh S, Buckingham ME. Mouse limb muscle is determined in the absence of the earliest myogenic factor myf-5. Proc Natl Acad Sci U S A. 1994;91:747–751. doi: 10.1073/pnas.91.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teboul L, Hadchouel J, Daubas P, Summerbell D, Buckingham M, Rigby PW. The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somitic myogenesis. Development. 2002;129:4571–4580. doi: 10.1242/dev.129.19.4571. [DOI] [PubMed] [Google Scholar]
- 56.Teboul L, Summerbell D, Rigby PW. The initial somitic phase of Myf5 expression requires neither Shh signaling nor Gli regulation. Genes Dev. 2003;17:2870–2874. doi: 10.1101/gad.1117603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012;22:602–609. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell TK, Turner D, Rupp R, Hollenberg S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 59.Williams BA, Ordahl CP. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development. 1994;120:785–796. doi: 10.1242/dev.120.4.785. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y, Hwang CK, Junn E, Lee G, Mouradian MM. ZIC2 and Sp3 repress Sp1-induced activation of the human D1A dopamine receptor gene. J Biol Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- 61.Zammit PS, Carvajal JJ, Golding JP, Morgan JE, Summerbell D, Zolnerciks J, Partridge TA, Rigby PW, Beauchamp JR. Myf5 expression in satellite cells and spindles in adult muscle is controlled by separate genetic elements. Dev Biol. 2004;273:454–465. doi: 10.1016/j.ydbio.2004.05.038. [DOI] [PubMed] [Google Scholar]