Abstract
To enhance web-based exploration of Medicare data, we present a unique query interface merging ontology navigation with query construction, for cohort discovery based on demographics, disease classification codes, medication and other types of clinical data. Our interface seamlessly blends query construction with functions for hierarchical browsing and rendering of terms and associated codes from vocabulary systems and ontologies, such as International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM). By unifying ontology navigation activities with query widget generation, a user can perform fine-tuned full boolean queries based on the substructure of the ontology, with flexibility to enable or disable subsumption-based queries. Query performance were evaluated on top disease subtypes of Centers for Medicare and Medicaid Services data, consisting of 5% of 2009 Limited Data Set files (inpatient and outpatient). Such interfaces will help moving the data access paradigm from a hypothesis-driven style to a data-driven one, while improving efficiency as a collective “secondary-use user community.”
Introduction
In 2008, the US healthcare spending accounted for over 16% of the nation’s GDP ( http://www.kaiseredu.org/ ). Government programs, such as Medicare and Medicaid, account for a significant portion of this expenditure. With the healthcare cost growing nearly 5% annually and almost quadrupling every decade since 1980, this nation’s economic stability and collective wellbeing hinge upon the success of healthcare reform and particularly the national implementation of electronic medical records (EMR). The secondary use of clinical data is recognized as one of the most promising ways to improve health outcomes and costs [ 1 ].
The Centers for Medicare and Medicaid Services (CMS) collects and compiles beneficiary data from all health care providers who provide services each year. While Medicaid provides funding for medical and health-related services for low-income people in the United States, Medicare is a federal health insurance program for citizens ages 65 and older and younger people with disabilities, as well as dialysis patients. CMS data consists of three distinct categories according to the level of privacy review and its own process for requesting data: (i) Identifiable Data Files , which contain actual beneficiary-specific and physician-specific information; (ii) Limited Data Set Files , which contain beneficiary level health information but exclude specified direct identifiers as outlined in the HIPAA Privacy Rule; and (iii) Non-Identifiable Data Files , which contain non-identifiable aggregated information and are within the public domain. Due to the size and complex nature of such data, their secondary use is a daunting task for both uninitiated as well as experienced users. The Research Data Assistance Center (ResDAC), a CMS contractor based at the University of Minnesota, provides free assistance to academic, government and non-profit researchers interested in using Medicare and Medicaid data for their research. Many state governments also have health departments that offer process for policy-compliant access to such data.
However, the existing process to access CMS data is inefficient, since the main steps of Institutional Review Board (IRB), Data Use Agreement (DUA), Statistical Analysis System (SAS) data extraction, reconstruction, re-extraction, and statistical and domain-specific analyses are time consuming and unnecessarily manually intensive (top of Fig. 1 ). It presents two areas for enhancement (regulatory matters are beyond the scope of the current paper):
Transforming from a hypothesis-driven paradigm to a discovery-driven paradigm. Existing access to Medicare and Medicaid data requires a pre-determined “hypothesis” before approval by a local IRB to begin the data request process. The data obtained, after a typically lengthy process (in months), may or may not be sufficient for the initial hypothesis, making revisions necessary. A discovery-driven paradigm would preempt the need for some of the avoidable revisions by allowing feasibility-oriented, limited data exploration (e.g. getting counts) to occur before hand. A better sense about study feasibility should be obtained before setting the IRB-DUA-SAS-extraction process into full motion.
Pushing common, shared post-processing tasks upstream, to the data source side. By appropriately reconstructing source data structure, it should be possible to reduce or eliminate the need for SAS-based data-extraction at the downstream investigator’s end, allowing the use of generic XML-based data format for immediate porting to multitudes of statistical software packages (such as R and S-plus, in addition to SAS). Eliminating redundant pre-processing tasks this way will help improve the overall data access efficiency as a community.
Figure 1:
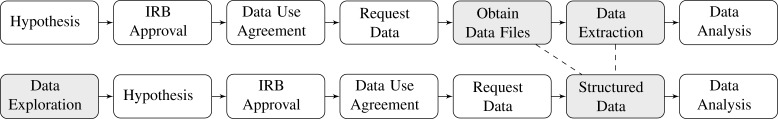
Existing data access steps (top) vs. the suggested new data access paradigm (bottom), where data exploration to assess feasibility is the initial step, and the data extraction step is eliminated.
This paper focuses on interfaces that are needed to provide better support for the discovery-driven paradigm (bottom of Fig. 1 ). In designing interfaces for querying curated healthcare data, we found challenges in managing disease classification codes, medication history and nomenclature, amplified by the longitudinal nature of clinical care. In order to address such challenges, we developed a query interface combining ontology browsing with query construction, for cohort discovery based on demographics, disease classification codes, medication and other types of clinical data.
Our work is a continuation of the Physio-MIMI project – Multi-modality, Multi-resource Environment for Physiological and Clinical Research [ 2 ]. Physio-MIMI is a collaborative project among four CTSA institutions to create an integrated informatics infrastructure for accessing distributed data. Physio-MIMI provides an ontology-driven federated data integration solution allowing sleep researchers to seamlessly query and access clinical and physiological data across different institutions (with approved data use agreements and ethics approvals). The reusability of the PhysioMIMI framework is embodied in its architecture using domain ontologies to directly drive the user interface called Visual Aggregator and Explorer (VISAGE [ 3 ]) and the data federation mapping interface called PhysioMap [ 4 ].
The main contribution of this work is an ontology-driven interface reusing large, existing standardized terminological systems as plug-and-play components to facilitate discovery-driven data exploration and help transform the existing data access paradigm. The multi-functional interface unifies the browsing and navigation of ontological terms with the steps for incremental construction of visualized query widgets to make the interface easier and effective to use by both novices and experts alike. The ability to allow user enable or disable subsumption-based queries (see Section 3.1) provides additional flexibility for query construction. These interface features and implemented functionalities are tested and validated on a data source consisting of 5% of all 2009 CMS limited data, with queries for top primary diagnoses and their associated costs for impatient and outpatient services.
1. Materials
1.1. Medicare Data
The CMS collects and compiles beneficiary data on a yearly basis. Limited Data Set (LDS) files from year 1999 up to 2011 without direct identifiers are available for research purposes. Among all LDS files, seven Standard Analytical Files (SAFs) for each institutional (inpatient, outpatient, skilled nursing facility, hospice, or home health agency) and non-institutional (physician and durable medical equipment providers) claim types are commonly used for statistical analysis. The SAF data is organized at the claim level and includes basic beneficiary demographic information, date of service, diagnosis and procedure code, provider number, and reimbursement amount. Each dataset has an associated Data Dictionary specifying the files’ data elements. After completing Data Use Agreement and study protocol, we obtained 5% of Denominator File, Inpatient Standard Analytic File and Outpatient Standard Analytic File for beneficiaries in fiscal year 2009 across 50 states ( http://www.cms.gov ). The Denominator File contains beneficiary demographic and enrollment. The Inpatient file contains final action fee-for-service claims data submitted by inpatient care providers for reimbursement of facility costs. This file includes diagnosis (ICD-9 diagnosis), procedure (ICD-9 procedure code), and other information. The SAF file contains claims data submitted by outpatient providers.
1.2. International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM)
The ICD is a standardized coding system classifying diseases and other health problems. It is considered the global health information standard for mortality and morbidity statistics. It is commonly used in healthcare for reimbursement and resource allocation decision-making. The ICD has been revised periodically and published in a series of editions by the World Health Organization (WHO). Its ninth revision, ICD-9, was extended to ICD-9-CM [ 5 ] by the U.S. National Center for Health Statistics (NCHS) with “CM” standing for “Clinical Modification.” The ICD-9-CM is based on the ICD-9 but including procedure codes and additional morbidity details. It is the official system of assigning diagnostic and procedure codes associated with inpatient, outpatient, and physician office utilization in the United States. The ICD-9-CM codes have been required for Medicare and Medicaid claims in the U.S. since 1979.
1.3. VISAGE: Visual Aggregator and Explorer
VISAGE [ 3 ] is a domain ontology-driven interface for a federated approach to data integration. Interface design features for VISAGE include auto-generated slider bar, selection boxes, and built-in charting. Administrative functionalities include role-based access control, auditing, and query lifecycle management functionalities include Query Builder, Query Manager, and Query Explorer.
Work reported in this paper is an extension of VISAGE, with new features developed for enabling the reuse of standardized terminological systems. The interface allows hierarchical browsing and lookup of terms and associated codes by novice as well as expert users from vocabulary systems (ontologies) such as ICD-9-CM. A key distinction is that such ontology exploration activities are seamlessly embedded, not merely co-existent, within query construction in VISAGE, whereby a user can perform fine-tuned full boolean queries based on the substructure of the ontology, with flexibility to enable or disable subsumption-based queries (Section 3.1).
2. Methods
Our approach consists of the steps of importing ICD-9-CM, constructing a CMS database, mapping CMS data to ICD-9-CM, and query generation.
Importing ICD-9-CM
The generic ICD-9-CM classification scheme [ 5 ] does not readily lend itself for use as an ontology. We used a form of the ICD-9-CM classification scheme with a better structure, the “ICD-9 for Health Ontology Mapper,” an OWL ontology written for the i2b2 platform ( http://bioportal.bioontology.org/ontologies/2059 ). The Apache Jena Ontology API [ 6 ] was used for parsing the ICD-9-CM OWL file to extract all the classes while preserving the hierarchy. The extracted classes and their hierarchical information were saved in a CSV file and imported to the VISAGE extension using a Ruby script. This resulted in 22,202 classes and 21,998 distinct codes.
Constructing a CMS Database
The CMS data files consist of 5% de-identified claim data in the U.S. in 2009. The data files were provided as a set of three CSV files totaling 17GB, with more than 10M records and about 3,000 data fields. A selection of 123 relevant fields was determined by reviewing the data dictionaries shipped with the CMS data files. Three tables were created in a MySQL database to store this selection of data. An AWK script pre-processed the original CSV files in a form more suitable for database import. This script performed cleanup tasks such as converting quarter numbers to pseudo-month numbers and reformatting dates. A separate script imported the pre-processed CSV files into a MySQL database.
Mapping CMS Data to ICD-9-CM
Physio-MIMI is a data integration framework capable of linking multiple independent data sources in a single instance using a domain ontology as a plug-and-play component in VISAGE. Therefore, mapping table columns in the CMS database to the ICD-9-CM concepts in the VISAGE extension is a part of the plug-and-play process to properly generate query statements corresponding to query widgets. This is a one-time effort in setup, and does not involve the end user. For example, in the CMS database, the table about the inpatient claims has a column named “claim_principal_diagnosis_code,” which can take any ICD-9-CM diagnosis code as a column value. One way to handle this is to map the column “claim_principal_diagnosis_code” to the ICD-9-CM concept “ICD-9-CM” and then exhaustively mapping each code appearing in the column to its ICD-9-CM concept counterpart. Instead of exhaustively mapping each code, the VISAGE extension uses a manual, seed map between from “claim_principal_diagnosis_code” and the concept “ICD-9-CM.” The rest of mapping is performed dynamically during the Query Generation phase.
Query Generation
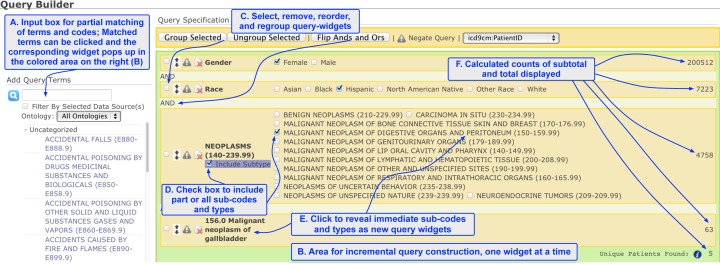
The query widgets are translated into SQL fragments which are executed over the relational database storing the Medicare data. For each query widget, the VISAGE extension records the query terms as well as the associated options. It relies on the database to ontology mapping to generate the appropriate query. The VISAGE extension does not require each ontology concept to be mapped to a database component. Instead, it leverages the ontology class structure to search for the closest mapped ancestor of the concept. For example, the term “Neoplasms” (see Fig. 2 ) is not mapped to a database column. The mapping interface automatically finds its closest ancestor “ICD-9-CM” which is mapped to the data source column called “claim_principal_diagnosis_code,” and an appropriate SQL fragment is generated.
Figure 2:
A screenshot of the general layout of Query Builder, with ontological browsing and query generation support. The left, unshaded area supports the searching of terms and browsing of the ontology (A). The light-green shaded area in the middle and right for query construction (B), with automatically generated widgets for slider bars and selection boxes. Query widgets can be reordered, re-grouped with different combination of boolean operations (C). The immediate sub-codes or subcategories of a term are displayed together with the leading term, allowing fine-tuned selection of a subgroup of the sub-codes as well as checking the box to include all subtypes, transitively (D). Clicking a subcategory generates a new query widget with that subcategory as the leading term, again with all of its sub-codes displayed in a new area (E). A live count of 5 unique beneficiaries of Hispanic female with gallbladder cancer is displayed, among a total of 200,512 female and 7,223 Hispanic patients (F).
3. Results
3.1. Functional Features of the Interface
As shown in the upper part of the (smaller) left column in Fig. 2 , a user can input either an ICD-9 code such as “156.0” or a search term “neoplasm of gallbladder” to locate the terms in the ontology display area. Once the term is located, a user can “click” the term, just as what couple be done with other demographic items such as “Gender” and “Race” to create the corresponding query widget in the shaded, query construction area (middle right column).
If the clicked term is a primitive ICD-9 code, then it is automatically selected as part of the query (for “156.0,” 63 results are displayed in Fig. 2 - bottom right ). If, however, a composite ICD-9 term is clicked, such as “Neoplasms,” then all of its immediate subtypes are displayed as selectable boxes in corresponding query widget.
A user can then perform one of several actions ( Fig. 2 ): (1) select one or more subtypes to query, by checking the corresponding boxes, with the logical relationship automatically set to “or” among the selected subtypes; (2) click one of the subtype terms to reveal all of its immediate children, if the current term is no already primitive; (3) check the box marked “Include Subtype” to query all subtypes as an aggregated result, to perform a subsumption-based query ; (4) delete a query widget or re-order and regroup them, or apply the appropriate logical operations on the subgroups (see options in the upper bar of shaded query construction area in Fig. 2 ). The combination of such possibilities queries provides substantial flexibility and improved resolution for query construction.
3.2. Sample Queries and Their Performance
To test the performance of the interface, we created queries on the most common claimed diseases and highest costs, respectively (5% of 2009). The most common diseases include Pneumonia organism unspecified (22,219 claims; 20,299 patients), Care involving other specified rehabilitation procedure (18,074 claims; 16,282 patients), and Obstructive chronic bronchitis with acute exacerbation (15,199 claims; 11,883 patients). The aggregated most expensive inpatient claims are Upper respiratory inflammation due to fumes and vapors ($339,860), Closed fracture of fifth cervical vertebra ($228,120), and Aftercare for healing traumatic fracture of upper leg ($222,230). The aggregated most expensive outpatient claims are Mechanical complication of automatic implantable cardiac defibrillator ($20,780), Fitting and adjustment of automatic implantable cardiac defibrillator ($19,190), and Urinary frequency ($17,500).
Performance is assessed on sample simple and complex queries. Ten simple queries for the top 10 principal diagnosis codes took average times in less than a half second. More complex queries, such as the one shown in Fig. 2 , took 1.90 seconds if the checkbox “include subtype” is selected for “150–159.99,” and 0.16 seconds otherwise.
3.3. Evaluation
We consider evaluation of the interface in two ways: query construction effort, and feature comparison. Query construction effort was evaluated using a side-by-side comparison of i2b2 and VISAGE with sample queries. Results for such an evaluation have been reported in [ 3 ] and [ 7 ].
A general comparison of features was given in [ 8 ]. Among the systems compared (ePCRN, i2b2/SHRINE, FARSITE, VISAGE, and TRANSFoRm Workbench), our merging of terminology-neighborhood navigation with query widget generation and construction provides efficient, in-place and fine-tuned capabilities for performing full boolean queries based on the inclusion and exclusion of sub-codes and categories. This is a unique feature not seen in other systems.
4. Discussion
Our study is limited in several ways. First, we only deal with a small percentage of CMS data. Second, the interface has not been simultaneously accessed by a large number of users so the performance data is primitive. Third, our user evaluation is incomplete; its unique features have not been evaluated by the intended user of clinical investigators. Fourth, the ICD-9 sub-categories and sub-codes may not faithfully reflect such at the valid conceptual level, in that the sub-categories may not be inclusive, because of the imposed tree structure. This ICD-9 limitation propagates to our interface as well. Finally, we have not addressed data security, regulatory and policy-compliant aspects, which are beyond the scope of the current paper.
Our approach is general in the sense that it is applicable to other ontological systems such as the Epilepsy and Seizure Ontology (EpSO) for the Prevention and Risk Identification of SUDEP Mortality (PRISM) project, funded by the National Institute of Neurological Disorders and Stroke (NINDS) [ 9 , 10 ].
In sum, we have presented the initial steps of developing a user interface that unifies ontology browsing with query construction. Such an interface would be needed to transform the existing Medicare data access paradigm, to facilitate hypothesis-generation and cohort discovery, and reduce the data-extraction burden currently placed on the end user side. The most time- and effort-intensive task in the currently data access paradigm consists of developing an analytic data set that contains one record per study subject, a data set that carries all of the variables of interest, ready for statistical analysis. The placement of this process upstream would therefore improve data access efficiency and lead to increased productivity in their secondary use.
Acknowledgments
This publication was made possible by the following NIH awards: Clinical and Translational Science Collaborative (CTSC) of Cleveland, UL1TR000439, UL1RR024989-05, and UL1 RR024989-05S.
References
- 1. Safran C , Bloomrosen M , Hammond W , Labkoff S , Markel-Fox S , Tang PC , et al. Toward a National Framework for the Secondary Use of Health Data: An American Medical Informatics Association . J Am Med Inform Assoc . 2007 ; 14 : 1 – 9 . doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Three New Informatics Pilot Projects to Aid Clinical and Translational Scientists Nationwide . http://www.nih.gov/news/health/jan2009/ncrr-26.htm .
- 3. Zhang GQ , Siegler T , Saxman P , Sandberg N , Mueller R , Johnson N , et al. VISAGE: A Query Interface for Clinical Research . Proc AMIA CRI ; 2010 ; pp. 76 – 80 . [PMC free article] [PubMed] [Google Scholar]
- 4. Mueller R , Sahoo S , Dong X , Redline S , Arabandi S , Luo L , Zhang GQ . Mapping Multi-institution Data Sources to Domain Ontology for Data Federation: the Physio-MIMI Approach . Proc. 2011 AMIA CRI ; p. 38 . [Google Scholar]
- 5. ICD-9-CM. http://www.cdc.gov/nchs/icd/ICD-9cm.htm
- 6. Apache Jena Ontology API . http://jena.apache.org/documentation/ontology/index.html .
- 7. Mueller M . Case Western Reserve University ; 2011 . Ontology-driven Data Integration for Clinical Sleep Research . PhD Thesis, [Google Scholar]
- 8. Zhao L , Keung LC , Rossiter J , Arvanitis TN . Query Formulation Workbench, EU TRANSFoRm Project Report D5.3 . 2012 .
- 9. Zhang GQ , Sahoo SS , Lhatoo SD . From Classification to Epilepsy Ontology and Informatics . Epilepsia . 2012 ; 53 : 28 – 32 . doi: 10.1111/j.1528-1167.2012.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui L , Bozorgi A , Lhatoo SD , Zhang GQ , Sahoo SS . EpiDEA: Extracting Structured Epilepsy and Seizure In-formation from Patient Discharge Summaries for Cohort Identification . AMIA Annual Symp Proc ; 2012 ; pp. 1191 – 1200 . [PMC free article] [PubMed] [Google Scholar]