Abstract
In biomedical research, network analysis provides a conceptual framework for interpreting data from high-throughput experiments. For example, protein-protein interaction networks have been successfully used to identify candidate disease genes. Recently, advances in clinical text processing and the increasing availability of clinical data have enabled analogous analyses on data from electronic medical records. We constructed networks of diseases, drugs, medical devices and procedures using concepts recognized in clinical notes from the Stanford clinical data warehouse. We demonstrate the use of the resulting networks for clinical research informatics in two ways—cohort construction and outcomes analysis—by examining the safety of cilostazol in peripheral artery disease patients as a use case. We show that the network-based approaches can be used for constructing patient cohorts as well as for analyzing differences in outcomes by comparing with standard methods, and discuss the advantages offered by network-based approaches.
Introduction
Data mining of electronic health records (EHRs) is on the rise due to increasing availability of the data as well as to advances in clinical text processing 1 – 3 . EHR mining has been used for detecting adverse drug event associations 4 – 6 and for profiling off-label use of drugs 7 , among other uses 8 , 9 . Simultaneously, in bioinformatics research, network analysis has provided the conceptual framework to interpret protein-protein interaction, gene-disease association and drug-target associations. For example, network analysis of protein-protein associations led to the identification of candidate genes for inherited Ataxias 10 , and revealed clusters of highly interacting proteins of functional relevance 11 . Similarly, EHRs can be used to construct a large-scale network representation of associations among clinical entities. For example, a network representation of disease associations learned from clinical problem summary lists has been used to uncover unexpected associations 12 and a network representation of patients based on their associated ICD9 codes has been used for patient stratification 13 . Both types of networks have even been combined, for instance the integration of cellular networks with comorbidity networks constructed from Medicare data could generate hypotheses about disease mechanisms 14 .
However, most approaches are restricted to associations among drugs and diseases, and are based on the structured clinical data. In contrast with existing uses of network analysis in EHR mining, we construct networks from the unstructured clinical text and we include medical devices and procedures in addition to drugs and diseases. We hypothesize that we can use the resulting networks for accurate control group construction and outcomes analysis. In particular, we apply our network-based approaches to examine the safety of cilostazol in peripheral artery disease (PAD) patients, and we compare our results with standard methods used in comparative effectiveness research. We believe that network analysis offers ways to visualize and analyze associations mined from EHRs, which can provide novel insights into the data by taking into account the connectivity of the clinical events and entities.
Methods
Data Collection and Processing:
We used data from the Stanford Translational Research Integrated Database Environment (STRIDE), which spans 17-years of data from 1.7 million patients with over 10.5 million unstructured clinical notes. We processed the clinical notes as described previously 6 . In brief, we used an optimized version of the NCBO Annotator 3 , 6 with a set of 19 clinically relevant ontologies, removed ambiguous terms 15 – 18 , and flagged negated terms and terms attributed to family history 19 , 20 . We normalized all drugs to their ingredients using RxNorm, such that the terms “pletal” and “cilostazol” are both normalized to the ingredient “cilostazol.” In addition, we made use of the hierarchies of the ontologies to aggregate concepts. More details on the data processing can be found in 21 .
Study and control group construction:
Using the timestamp of the clinical note in which the treatment concept (cilostazol) was first mentioned, we defined an index time point of treatment for all patients and grouped the annotations into two groups: concepts associated with clinical events that happened before treatment (which can be used for matching control groups) and concepts associated with events that happened after treatment (which can be interpreted as outcomes). We defined a cohort of 11,547 PAD patients using the expert-selected concepts listed in Table 1 . To reduce bias introduced by difference in data coverage, we excluded patients having less than one year’s worth of data after their first PAD mention, reducing the number of PAD patients to 5,746. For the cilostazol study group, we selected 223 PAD patients who had a cilostazol mention after or at the same time as their first PAD mention.
Table 1:
Peripheral artery disease definition
| Concept | UMLS CUI |
|---|---|
| Peripheral arterial diseases | C1704436 |
| Peripheral vascular diseases | C0085096 |
| Peripheral arterial occlusive disease | C1306889 |
| Intermittent claudication | C0021775 |
| Claudication (finding) | C1456822 |
Propensity score matching (PSM):
We used PSM to address biases introduced by confounding variables. We manually defined 15 potential confounders including demographic variables such as gender, race, age at the first PAD mention, and co-morbidities as well as co-prescriptions. All covariates except for the demographic variables were defined by the first mention of the respective concept before the index time point. We used the Matching package for R 22 to perform PSM and to check balance in the variables between the cilostazol and control groups.
Patient-patient similarity:
We used all concepts before the index time point and constructed a patient-feature matrix, where each row represents a patient and each column a concept. The cells indicate the presence or absence of the concept in any clinical note before the index time point. To reduce dimensionality, we used the Human Disease Ontology to aggregate disease concepts and SNOMED CT to aggregate procedures. For drugs, we used the ATC hierarchy to group drugs into classes at the third level (therapeutic subgroup). We used the patient-feature matrix to compute pairwise patient similarity ( s p ) using Jaccard index and visualized the resulting patient-patient similarity matrix as a network using Cytoscape 23 . We then applied 1:1 nearest neighbor matching to construct a control group.
Outcome analysis:
For analyzing the outcomes of patients, we used only the concepts after the index time point.
Logistic regression analysis:
We defined a set of 9 expert-selected outcome variables. These outcome variables include major adverse cardiovascular events (MACE), major limb events (MALE) and cardiac arrhythmias. We computed odds ratios and 95% confidence intervals using logistic regression.
| Equation 1 |
Concept-concept association network:
We used co-occurrence statistics ( Equation 1 ) to compute the association ( s c ) between different concepts ( cid1 , cid2 ) from the clinical notes. We filtered scores based on the strength (s c >= 2) and significance (p <= 0.05, one-sided Fisher’s exact test) of the association. The nodes in the networks represent the clinical concepts (drugs, diseases, devices and procedures) and the edges their associations. We used associations between concepts for the cilostazol and control groups separately to construct a network for each group. To compare the outcomes in the cilostazol group with those in the control group, we merged the two concept-concept networks and only kept those nodes and edges that are over or under-represented in the cilostazol group compared to the control (p <= 0.05, one-sided Fisher’s exact test). We then visualized the resulting network using Cytoscape.
Results
Our results show that network-based methods are at par with standard methods in comparative effectiveness research in terms of control group construction and analysis of clinical outcomes for the case of cilostazol use for PAD. PAD is a major health problem affecting millions of patients worldwide. It is defined by obstruction of infrarenal aorta and lower-extremity arteries leading to claudication and leads to a significant impairment of quality of life. Cilostazol is the most effective treatment for PAD and has a black-box warning for cardiovascular events, especially in patients with congestive heart failure due to prior experience with a similar drug 24 .
Control group construction:
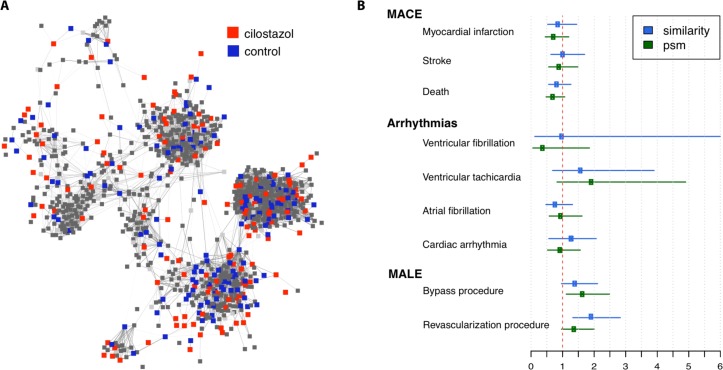
Figure 1A shows a network representation of the 223 cilostazol patients (red nodes) and their most similar PAD patients ( s p >= 0.36). The thickness of the edges corresponds to the similarity between patients. Patients who share multiple disease, drug, device and procedure annotations are similar and thus closer in the network. We constructed a control group of 223 patients by choosing the nearest neighbor (1:1 match) or most similar patient (blue nodes) for each cilostazol patient. We also applied PSM on 15 expert-selected variables (potential confounders) such as age, gender, several co-morbidities and co-prescriptions.
Figure 1:
(A) Patient-patient similarity network for constructing a control group and (B) outcome analysis based on network-based control group (similarity) and propensity-score matched control group (psm).
Comparing the cilostazol group with all other PAD patients (before matching), we observe a significant difference or imbalance in several variables including gender (there are more males in the cilostazol group), co-prescriptions (cilostazol patients are taking more drugs), vascular surgical and bypass procedures (cilostazol patients undergo more procedures) ( Table 2 ). These variables are likely to be correlated with the outcome, so in a properly matched cohort the imbalance would be averted. As expected, using PSM removes the imbalance in all selected variables. In comparison, the network-based matching removes most of the imbalance, except for ACE inhibitors and Cardiac arrhythmias (p < 0.05). Interestingly, the network-based matching introduces imbalance in the ‘history of cardiac arrhythmias’ variable. However, a slightly stricter level of significance (p < 0.01) would result in perfect balance.
Table 2 :
Balance in expert-selected variables before and after matching
| Variable | Before Matching | Similarity network | PSM | ||||
|---|---|---|---|---|---|---|---|
| Cilostazol (n= 223) | Control (n= 5534) | p-value * | Control (n= 223) | p-value | Control (n= 223) | p-value * | |
| Demographics | |||||||
| Age (at indication onset), mean (sd) | 71.22 (11.02) | 70.43 (12.46) | 0.30 | 72.05 (10.62) | 0.41 | 70.87 (11.51) | 0.75 |
| Gender (female), n (%) | 37.22 | 45.94 | <0.01 | 36.65 | 0.84 | 35.87 | 0.77 |
| Race, (%) | |||||||
| Asian | 8.52 | 7.41 | 0.56 | 6.33 | 0.37 | 10.31 | 0.51 |
| Black | 2.69 | 3.71 | 0.36 | 3.61 | 0.59 | 0.90 | 0.16 |
| Unknown | 22.87 | 26.17 | 0.25 | 22.17 | 0.82 | 20.63 | 0.57 |
| White | 65.47 | 62.22 | 0.32 | 67.87 | 0.55 | 67.27 | 0.69 |
| Comorbidities | |||||||
| Coronary artery disease, n (%) | 5.38 | 6.47 | 0.48 | 4.98 | 0.83 | 6.28 | 0.70 |
| Congestive heart failure, n (%) | 25.56 | 22.84 | 0.36 | 20.36 | 0.21 | 30.49 | 0.36 |
| Hypertension, n (%) | 10.76 | 11.31 | 0.80 | 9.50 | 0.75 | 10.31 | 0.88 |
| Co-prescriptions | |||||||
| Beta blocking agents, n (%) | 75.34 | 60.77 | <0.001 | 69.68 | 0.20 | 74.89 | 0.91 |
| ACE inhibitors, plain, n (%) | 78.03 | 69.57 | <0.01 | 67.87 | 0.01 | 78.92 | 0.81 |
| Platelet aggregation inhibitors excl. heparin, n (%) | 91.93 | 79.00 | <0.001 | 89.59 | 0.41 | 95.51 | 0.07 |
| Vasodilators, n (%) | 32.29 | 26.36 | 0.06 | 31.67 | 0.84 | 37.22 | 0.29 |
| History of | |||||||
| Cardiac arrhythmia, n (%) | 32.29 | 32.17 | 0.97 | 23.08 | 0.03 | 33.18 | 0.84 |
| Stroke, n (%) | 17.94 | 18.31 | 0.89 | 15.84 | 0.61 | 21.52 | 0.34 |
| Myocardial infarction, n (%) | 17.94 | 15.87 | 0.43 | 13.58 | 0.24 | 19.73 | 0.64 |
| Vascular surgical procedures, n (%) | 74.44 | 47.71 | < 0.001 | 65.61 | 0.05 | 74.44 | 1 |
| Bypass procedures, n (%) | 41.70 | 26.56 | <0.001 | 36.20 | 0.24 | 40.36 | 0.75 |
P-values are based on X 2 -test for categorical, and two-sample t-test for continuous variables
We profiled the safety of cilostazol with respect to MACE, MALE and cardiac arrhythmia outcomes. We compared differences in outcomes (see Figure 1B ) between the cilostazol group and control groups generated via PSM (green) and network-based patient-patient similarity clustering (blue) using logistic regression. The results show that in most outcome variables, PSM and network-based approach generate equivalent control groups.
Outcome analysis:
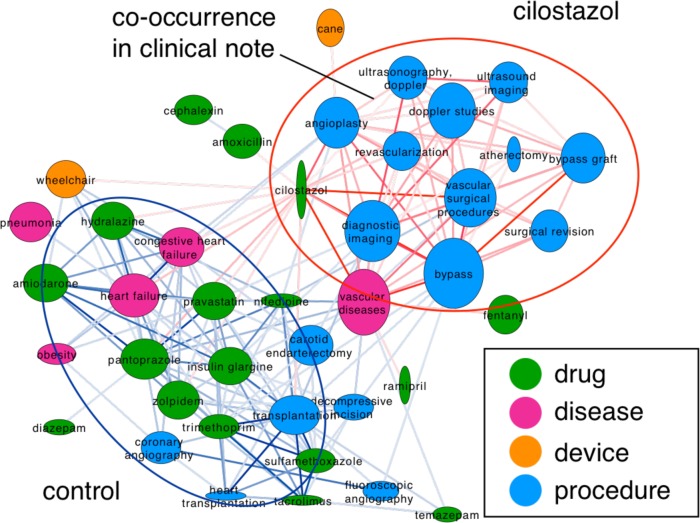
To compare the outcomes in cilostazol patients with the controls using a network-based approach, we focused on those nodes and edges that are over- or under-represented in the cilostazol group. Figure 2 depicts the resulting network, nodes represent the clinical concepts, the edges their association strength. Nodes are colored according to their semantic group; node height corresponds to frequency in the cilostazol cohort, and node width to the frequency in the control. Cilostazol is therefore a thin and tall node. We colored the edges according to the over-representation in the two groups of patients. Red edges are over-represented in the cilostazol patients and blue edges in the control group. The thickness of the edges represents the statistical significance of the over-representation. Two main clusters are visible, in the upper right part there is a cluster of concepts over-represented in the cilostazol group, and in the lower left part are concepts over-represented in the control group. The cilostazol cluster contains different vascular surgical procedures (angioplasty, revascularization and bypass procedures) and some imaging procedures (ultrasonography, doppler and ultrasound imaging) used to diagnose PAD and to monitor the vascular surgical procedures 25 ; thus it is not surprising to find a strong association between these concepts in the clinical notes. In contrast, the control cluster is more heterogeneous and contains concepts related to severe heart failure, which is a contraindication for Cilostazol use.
Figure 2:
Concept-concept network, nodes represent clinical concepts and edges their associations in the clinical notes.
Discussion
One key aspect of our work is the use of temporal ordering of clinical notes to group the annotations into concepts used for control group construction and concepts useful for outcome analysis. In particular, we used the annotations before the treatment (e.g., cilostazol) to construct a patient-patient similarity network from which we selected a matched control group. By analyzing the balance in expert-selected variables before and after matching, we showed that our network-based approach achieves similar performance compared to standard PSM. Moreover, we profiled differences in outcome between the cilostazol and the control groups and showed that both matching approaches, network-based and PSM produce similar results, confirming that both control groups are equivalent. Here, the main advantage of the network-based approach is that no expert-knowledge is required to identify potential confounders. In addition, we can use all concepts found in the patient’s clinical notes to compute similarity between the patients in their phenotypic profiles. Others have also shown that a combination of features (in particular diseases, medications, findings and medical procedures) achieve better performance in classifying patients compared with restricting the features to one semantic group (such as diseases) 26 .
We used a concept-concept association network to profile differences in outcome. In contrast to standard logistic regression analysis, which is restricted to those concepts considered meaningful by a medical expert, the network-based approach does not restrict the concept space. Using the networks as a tool for visualizing associations found in the clinical notes, we can analyze large numbers of concepts and their associations simultaneously as well as uncover the same results as regular methods would; as evidenced by the agreement with standard logistic regression analysis ( Figure 1B ) and confirmed by a medical expert. In contrast to standard clustering approaches, the use of networks allows for user-friendly visualization of the data, and the constructed networks can easily be enriched with additional information to guide the analysis. For example, the patient-patient similarity networks could be layered with the information about which concepts contributed most to the association between two patients, such that the researcher understands why a control is matched to a cilostazol patient, adding transparency to the analysis.
Limitations and future work:
Automatically generated annotations of clinical notes can contain errors. However, with accuracies ranging from 86% for recognizing diseases, in particular 92% for identifying PAD, 93% for drugs and 97% for negations 19 , the annotation derived features are “good enough” for such a study. In contrast to logistic regression the concept-concept network does not provide a single score (such as an odds ratio) for each outcome variable. In future work, we want to use the concept-concept network to identify those variables worth being included in a standard outcome analysis and see if this selection agrees with expert choices. Moreover, we plan to compare different similarity metrics 26 , and to investigate the use of bipartite networks similarly as previously described for analyzing associations of patients and SNPs 27 or cytokines 28 .
Conclusion
Network-based approaches are applicable for constructing control groups as well as for analyzing clinical outcomes as shown by our case study on the safety of cilostazol in PAD patients. Our results show similar performance of the network-based approaches compared to standard methods used in comparative effectiveness research 29 . One clear advantage of this approach is that the analysis is not limited to expert-selected variables, allowing the analysis of large numbers of variables and their associations simultaneously. In addition, visualizing associations between patients and concepts as networks allows for transparent interpretation.
Acknowledgments
ABM, PJL, RH and NHS acknowledge support from the NIH grant U54 HG004028 for the National Center for Biomedical Ontology.
References
- 1. Friedman C , Shagina L , Lussier Y , Hripcsak G . Automated encoding of clinical documents based on natural language processing . J. Am. Med. Inform. Assoc. . 2004 Sep-Oct; 11 ( 5 ): 392 – 402 . doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Savova GK , Masanz JJ , Ogren PV , et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications . J. Am. Med. Inform. Assoc. . 2010 ; 17 ( 5 ): 507 – 513 . doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shah NH , Bhatia N , Jonquet C , Rubin D , Chiang AP , Musen MA . Comparison of concept recognizers for building the Open Biomedical Annotator . BMC Bioinformatics . 2009 ; 10 ( Suppl 9 ): S14 . doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coloma PM , Schuemie MJ , Trifiro G , et al. Combining electronic healthcare databases in Europe to allow for large-scale drug safety monitoring: the EU-ADR Project . Pharmacoepidemiol Drug Saf . 2011 ; 20 ( 1 ): 1 – 11 . doi: 10.1002/pds.2053. [DOI] [PubMed] [Google Scholar]
- 5. Haerian K , Varn D , Vaidya S , Ena L , Chase HS , Friedman C . Detection of pharmacovigilance-related adverse events using electronic health records and automated methods . Clin. Pharmacol. Ther. . 2012 ; 92 ( 2 ): 228 – 234 . doi: 10.1038/clpt.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lependu P , Iyer SV , Fairon C , Shah NH . Annotation Analysis for Testing Drug Safety Signals using Unstructured Clinical Notes . J Biomed Semantics . 2012 ; 3 ( Suppl 1 ): S5 . doi: 10.1186/2041-1480-3-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lependu P , Liu Y , Iyer S , Udell MR , Shah NH . Analyzing patterns of drug use in clinical notes for patient safety . AMIA Summits Transl Sci Proc . 2012 ; 2012 : 63 – 70 . [PMC free article] [PubMed] [Google Scholar]
- 8. Hripcsak G , Albers DJ , Perotte A . Exploiting time in electronic health record correlations . J. Am. Med. Inform. Assoc. . 2011 Dec ; 18 ( Suppl 1 ): i109 – 115 . doi: 10.1136/amiajnl-2011-000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y , Salmasian H , Harpaz R , Chase H , Friedman C . Determining the reasons for medication prescriptions in the EHR using knowledge and natural language processing . AMIA. Annu. Symp. Proc. . 2011 ; 2011 : 768 – 776 . [PMC free article] [PubMed] [Google Scholar]
- 10. Lim J , Hao T , Shaw C , et al. A Protein-Protein Interaction Network for Human Inherited Ataxias and Disorders of Purkinje Cell Degeneration . Cell . 2006 ; 125 ( 4 ): 801 – 814 . doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 11. Przulj N , Wigle DA , Jurisica I . Functional topology in a network of protein interactions . Bioinformatics . 2004 ; 20 ( 3 ): 340 – 348 . doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
- 12. Hanauer DA , Rhodes DR , Chinnaiyan AM . Exploring Clinical Associations Using ‘-Omics’ Based Enrichment Analyses . PLoS ONE . 2009 ; 4 ( 4 ): e5203 . doi: 10.1371/journal.pone.0005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roque FS , Jensen PB , Schmock H , et al. Using electronic patient records to discover disease correlations and stratify patient cohorts . PLoS Comp. Biol. . 2011 Aug ; 7 ( 8 ): e1002141 . doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park J , Lee D-S , Christakis NA , Barabasi AL . The impact of cellular networks on disease comorbidity . Mol Syst Biol . 2009 ; 5 doi: 10.1038/msb.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bodenreider O , McCray AT . Exploring semantic groups through visual approaches . J. Biomed. Inf . 2003 ; 36 ( 6 ): 414 – 432 . doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu R , Musen MA , Shah NH . A Comprehensive Analysis of Five Million UMLS Metathesaurus Terms Using Eighteen Million MEDLINE Citations . AMIA. Annu. Symp. Proc. . 2010 ; 2010 : 907 – 911 . [PMC free article] [PubMed] [Google Scholar]
- 17. Wu ST , Liu H , Li D , et al. Unified Medical Language System term occurrences in clinical notes: a large-scale corpus analysis . J Am Med Inform Assoc . 2012 doi: 10.1136/amiajnl-2011-000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gautam KP , Jonquet C , Xu R , Musen M , Shah NH . The Lexicon Builder Web service: Building Custom Lexicons from two hundred Biomedical Ontologies . AMIA Annu. Symp. Pro. . 2010 ; 2010 : 587 . [PMC free article] [PubMed] [Google Scholar]
- 19. Chapman WW , Chu D , Dowling JN . ConText: An Algorithm for Identifying Contextual Features from Clinical Text . Paper presented at: Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing ; 2007 . [Google Scholar]
- 20. Chapman WW , Bridewell W , Hanbury P , Cooper GF , Buchanan BG . A simple algorithm for identifying negated findings and diseases in discharge summaries . J Biomed Inform . 2001 Oct ; 34 ( 5 ): 301 – 310 . doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 21. LePendu P , Iyer SV , Bauer-Mehren A , Harpaz R , Mortensen JM , Shah NH . Pharmacovigilance using free-text clinical notes . Under revision (Nature CPT) . doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sekhon JS . Multivariate and Propensity Score Matching Software with Automated Balance Optimization The Matching Package for R . Journal of Statistical Software . 2011 ; 42 ( 7 ) [Google Scholar]
- 23. Shannon P , Markiel A , Ozier O , et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks . Genome Res . 2003 ; 13 ( 11 ): 2498 – 2504 . doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chi YW , Lavie CJ , Milani RV , White CJ . Safety and efficacy of cilostazol in the management of intermittent claudication . Vasc Health Risk Manag . 2008 ; 4 ( 6 ): 1197 – 1203 . doi: 10.2147/vhrm.s3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lumsden AB , Davies MG , Peden EK . Medical and endovascular management of critical limb ischemia . J Endovasc Ther . 2009 Apr ; 16 ( 2 Suppl 2 ): II31 – 62 . doi: 10.1583/08-2657.1. [DOI] [PubMed] [Google Scholar]
- 26. Cao H , Melton GB , Markatou M , Hripcsak G . Use abstracted patient-specific features to assist an information-theoretic measurement to assess similarity between medical cases . J Biomed Inform . 2008 Dec ; 41 ( 6 ): 882 – 888 . doi: 10.1016/j.jbi.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhavnani SK , Bellala G , Victor S , Bassler KE , Visweswaran S . The role of complementary bipartite visual analytical representations in the analysis of SNPs: a case study in ancestral informative markers . J. Am. Med. Inform. Assoc . 2012 ; 19 ( e1 ): e5 – e12 . doi: 10.1136/amiajnl-2011-000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhavnani SK , Victor S , Calhoun WJ , et al. How cytokines co-occur across asthma patients: from bipartite network analysis to a molecular-based classification . J. Biomed. Inf. . 2011 Dec ; 44 ( Suppl 1 ): S24 – 30 . doi: 10.1016/j.jbi.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sox HC , Goodman SN . The methods of comparative effectiveness research . Annu. Rev. PublicHealth. . 2012 ; 33 : 425 – 445 . doi: 10.1146/annurev-publhealth-031811-124610. [DOI] [PubMed] [Google Scholar]