Abstract
Gene Wiki Plus (GeneWiki+) and the Online Mendelian Inheritance in Man (OMIM) are publicly available resources for sharing information about disease-gene and gene-SNP associations in humans. While immensely useful to the scientific community, both resources are manually curated, thereby making the data entry and publication process time-consuming, and to some degree, error-prone. To this end, this study investigates Semantic Web technologies to validate existing and potentially discover new genotype-phenotype associations in GWP and OMIM. In particular, we demonstrate the applicability of SPARQL queries for identifying associations not explicitly stated for commonly occurring chronic diseases in GWP and OMIM, and report our preliminary findings for coverage, completeness, and validity of the associations. Our results highlight the benefits of Semantic Web querying technology to validate existing disease-gene associations as well as identify novel associations although further evaluation and analysis is required before such information can be applied and used effectively.
1. Introduction
Over the last decade, due to the advances in using high-throughput genotyping technologies, Genome-Wide Association Studies (GWAS) have successfully associated the most common form of genetic variant, the Single Nucleotide Polymorphism (SNP), with more than 40 common diseases and traits. These advances have led to a heavy influx of large amounts of association data interlinking genes, SNPs, proteins, diseases and drugs, and have enabled a network-based approach to understanding human disease that offer a platform for systematically identifying novel disease genes, drug targets and biomarkers for complex diseases. However, such datasets are highly heterogeneous in nature, both syntactically and semantically, thereby requiring the development of new approaches for biomedical data integration. Chen et al. [ 1 ] identified three basic requirements for developing such an approach:
A data model capable of capturing and modeling large-scale biomedical network data.
A data integration framework that can map and merge network data across multiple, heterogeneous data sources.
A collection of computational services and tools that can analyze, discover, and validate novel associations spanning genes, SNPs, drugs and diseases.
The Semantic Web, and in particular the Linked Data [ 2 ] paradigm, provides the requisite backbone for building applications that can address these requirements. Using Resource Description Framework (RDF) as the underlying data representation model, it is feasible to create large biomedical networks where multitude relevant biomedical entities can be interlinked into a semantic graph. This has been the mission of the W3C Linked Open Data (LOD) project, where as of December 2011, more than 300 datasets from multiple domains (e.g., genes, drugs, proteins, diseases, anatomy), with approximately 30 billion RDF triples have been connected via more than 500 million links. The creation of such a huge integrated-network dataset provides challenges as well as novel opportunities on which very expressive queries can be executed to answer important biomedical research questions.
To this end, the overall objective of the proposed study is to use the existing datasets within the LOD infrastructure to extract genotype-phenotyping associations for commonly occurring chronic diseases [ 3 ]. In particular, our goal is to demonstrate the applicability of SPARQL querying capabilities using two public knowledgebases, namely Online Mendelian Inheritance in Man (OMIM [ 4 ]) and Gene Wiki Plus (GeneWiki+ [ 5 ])—a part of Wikipedia, to extract disease-gene-SNP associations for six chronic diseases (Arthritis, Asthma, Cancer, Diabetes, Dementia and Obesity) and report our preliminary findings with respect to coverage, completeness and validity of the associations.
Our results indicate that the overlap for disease-gene associations between OMIM and GeneWiki+ is at best minimal. This could be attributed to various reasons, including a stale dataset provided by the current OMIM SPARQL endpoint. We also conclude that via SPARQL-based querying capabilities, our technique can deduce disease-gene associations that were not explicitly stated in the RDF graph, although further validation and verification is required before such deductive data can be applied effectively within applications for scientific and clinical research.
2. Background and Materials
2.1. Online Mendelian Inheritance in Man (OMIM) Knowledgebase
The Online Mendelian Inheritance in Man (OMIM [ 4 ]) is a comprehensive knowledgebase of human genes and genetic disorders that is distributed by the U.S. National Center for Biotechnology Information (NCBI) to support research in genomics. Updated on a daily basis, each entry in OMIM contains free text descriptions of genetic loci, inheritance patterns, allelic variants, biochemical and clinical features, as well as molecular and population genetics. Most OMIM entries also include information about accompanying phenotyping features within a clinical synopsis section (available as structured text). Additionally, OMIM entries are in general linked to other public databases such as Entrez Gene (NCBI LocusLink database), Nomenclature (HUGO Nomenclature Committee), RefSeq (NCBI reference sequences), GenBank (NCBI nucleotide sequence database), Protein (NCBI protein sequence database) and UniGene (NCBI UniGene project).
As of July 2012, OMIM contains approximately 21,000 detailed entries on human genes and genetic disorders, including around 5,300 phenotypes with a clinical synopsis. While this data can be downloaded as tab-delimited text files, an RDF-based representation and/or a SPARQL endpoint currently is not provided by OMIM or NCBI. To meet this requirement, the Bio2RDF project [ 7 ] has created a public SPARQL endpoint for OMIM data. For this study, the OMIM data accessible via the SPARQL endpoint was last updated on July 20 th , 2011.
2.2. Gene Wiki for Community Annotation
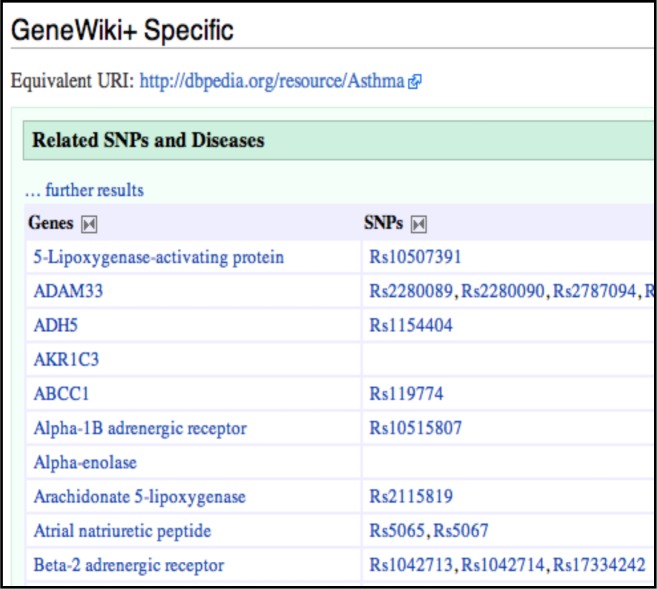
Gene Wiki [ 5 ] is a community intelligence and crowdsourcing project within Wikipedia where the goal is to build a gene-specific review article for every gene in the human genome, where each article is collaboratively written, continuously updated and community reviewed. Each gene article in Gene Wiki includes a free text summary as well as an “infobox” that contains links to public databases harvested from NCBI’s Entrez Gene, gene ontology annotations, and when available, protein structure identifiers. GeneWiki+ [ 8 ] is a semantic mirror of Gene Wiki and is built on top of the Semantic Mediawiki framework. Consequently, GeneWiki+ enables semantic queries for retrieval of large amounts of information which also includes additional public data sources, such as SNPedia [ 9 ]—a Semantic Wiki database of SNPs. For example, Figure 1 shows a snapshot of disease-gene-SNP association from GeneWiki+ for Asthma. For our study, we extracted disease-gene-SNP association data as RDF files from GeneWiki+ (February 12 th , 2012 data update). Creating a local dump of the RDF data files (approximately 80MB) allowed us to experiment with complex SPARQL queries that could not be executed within the GeneWiki+ environment.
Figure 1 .
Gene and SNP associations for Asthma, as obtained from GeneWiki+
3. Methods
The methods developed for this study can be divided into 2 main aspects: (1) extraction of disease-gene and disease-gene-SNP associations from OMIM and GeneWiki+, respectively, via SPARQL queries and (2) comparative analysis of query results.
3.1. Extracting disease-gene associations from OMIM and GeneWiki+
To extract the disease-gene associations from GeneWiki+, we first downloaded the February 12 th , 2012 snapshot of GeneWiki+ data as RDF files, and loaded the dataset in a local Virtuoso [ 10 ] server to create a SPARQL endpoint. The SPARQL query comprised of a regular expression filter for selecting the appropriate phenotype, and graph matching patterns to identify the genotype-phenotype associations. Note that, unlike OMIM, GeneWiki+ explicitly specifies “disease-SNP” associations (in addition to the “disease-gene” associations). Consequently, we utilized SPARQL's querying capabilities with the disease-SNP associations in GeneWiki+ to deduce novel disease-gene associations that were not explicitly stated in the RDF dump file for GeneWiki+. As an example, ILR6 is inferred as being associated with Asthma according to the query, although in GeneWiki+, this association is not explicitly stated. Additional results based on our SPARQ-based querying capabilities are discussed in Section 4.
To extract the disease-gene associations from OMIM, we devised a simple SPARQL query for execution at Bio2RDF’s OMIM endpoint ( http://omim.bio2rdf.org/sparql ). In essence, the query comprises a regular expression filter on the phenotype and disease descriptions, and extracts the appropriate RDF triples comprising the genotypephenotype associations. Additional details of the SPARQL queries and execution patterns are available via our project website: http://informatics.mayo.edu/LCD/index.php/DbSNP_OMIM_GWP .
3.2. Comparative Data Analysis between OMIM and GeneWiki+
Once the disease-gene associations from OMIM were extracted, we compared the results to GeneWiki+ to validate existing associations as well as potentially discover novel associations. For this task, we queried GeneWiki+ articles for the six chronic diseases namely, arthritis, asthma, cancer, diabetes, dementia and obesity, and identified the relationships to genes and SNPs. These six chronic diseases were chosen due to their prevalence in the general population as well as the availability of several published GWAS results [ 12 ].
To perform the comparative analysis, we took a “disease-centric view”. That is, for a given disease, we compared the genes identified from OMIM with that of GeneWiki+, and determined the extent of overlap in the data sets. We then selected one of the diseases, for example Asthma, and recorded the PubMed article references to provide a justification for a given disease-gene association. As mentioned above, due to the querying capabilities provided via SPARQL, our query resultset also contained associations that were not explicitly described in either knowledgebases. While this is a very unique feature in applying Semantic Web technologies, without appropriate validation and justification of the newly identified relationships, such information cannot be applied and used effectively. Hence, in our analysis, we lend special emphasis on verification of novel associations that have been deduced via the query inferencing process. Note that, for this study, we restricted the list of diseases to a set of six chronic diseases, although one can trivially apply our methods to other diseases.
4. Results
Table 1 shows the results of the analysis of disease-gene associations, where each gene is uniquely identified based on its Entrez Gene identifier. There are several aspects that can be deduced from these findings: (1) the total number of unique disease-gene associations in GeneWiki+ was significantly higher than OMIM. For example, for Cancer, only 83 (55+28) unique disease-gene pairs were extracted from OMIM, whereas 1060 (1032+28) pairs from GeneWiki+. Such a finding is contrary to our initial hypothesis that OMIM would potentially contain more unique disease-gene pairs curated than GeneWiki+. (2) The overlap between the numbers of genes extracted from GeneWiki+ and OMIM was significantly less (<5% on average) for all the six diseases. Dementia had the largest overlap (7.5%), and Cancer the lowest (1.6%). (3) Overall, the number of unique genes associated with Cancer (1115) was highest followed by Diabetes (263) and Arthritis (248). This is consistent with the number of genome-wide association studies conducted for Cancer, Diabetes and Arthritis [ 12 ].
Table 1 .
Comparing disease-gene pairs between OMIM and GeneWiki+
| Disease Type | Total Unique Genes | Genes Unique to GeneWiki+ | Genes Unique to OMIM | Genes common to both GeneWiki+ and OMIM |
|---|---|---|---|---|
| Arthritis | 248 | 232 | 9 | 7 |
| Asthma | 130 | 113 | 5 | 12 |
| Cancer | 1115 | 1032 | 55 | 28 |
| Dementia | 44 | 35 | 4 | 5 |
| Diabetes | 263 | 219 | 20 | 24 |
| Obesity | 162 | 145 | 4 | 13 |
Table 2 shows the results of the analysis for disease-gene-SNP associations for Asthma. By querying the disease-SNP associations for Asthma via SPARQL, we deduced 65 disease-gene associations from GeneWiki+ that were not explicitly stated (in the RDF dump files). For example, gene IL6R does not have an explicit association with Asthma in GeneWiki+. However, the SNP rs4129267 , which is a SNP within the IL6R gene, has an explicit association with Asthma. Consequently, our query deduced the association between IL6R and Asthma. We validated and confirmed that the association between IL6R and Asthma has been reported in a manuscript by Doganci et al. [ 11 ]. Similarly, the gene SH2B3 does not have an explicit association with Asthma, but the SNP rs3184504 within SH2B3 does, and hence the disease-gene association was deduced automatically. As indicated in Table 2 , we could not validate 63 disease-gene associations identified via our querying process using data from PubMed articles. Examples include TNF and NFKB2 —genes that were associated with Asthma by SPARQL queries, but there were no studies providing such evidence. Finally, as indicated in Table 1 , only 17 genes were associated with Asthma in OMIM (5 genes uniquely identified in OMIM, and 12 genes common between OMIM and GeneWiki+). In particular, for the 5 genes uniquely identified in OMIM, three were mentioned in the GeneWiki+ text, although those associations were not stated in the RDF files, which would indicate that the issue is related to data curation. Additionally, our analysis identified 28 genes that did not appear in the OMIM SPARQL query resultset for Asthma, but were in fact, associated with Asthma. As a concrete example, even though the gene DPP10 was not in the resultset in querying the OMIM SPARQL endpoint, a manuscript by Allen et al. [ 13 ] explicitly associates DPP10 with Asthma. Our manual validation of OMIM text descriptions revealed that all the 28 genes were in fact associated with Asthma, although such associations were not surfaced via the OMIM SPARQL endpoint.
Table 2 .
Disease-gene-SNP association results for Asthma
| Finding Type | Total Number |
|---|---|
| Gene-disease associations identified via SPARQL | 130 |
| Gene-disease associations identified via SPARQL (inference only) | 65 |
| Gene-disease associations validated via PubMed citations | 67 |
| Gene-disease associations without any validation | 63 |
5. Summary and Discussion
5.1. Discussion
One of the important goals in current biomedical research is the discovery of novel associations between diseases, genetic and environmental factors. Achieving this goal arguably requires coordination and integration of data from several biomedical resources and repositories. This reported study demonstrates the applicability of Semantic Web technologies for integration and querying of heterogeneous publicly available data sources. Its preliminary findings highlight several issues in terms of developing methods for Web-scale data integration and interpretation of query results. First, noticeably the data extracted from GeneWiki+ and OMIM had significantly less overlap. While this could in part be attributed, at least in part, to the near real-time synchronization of GeneWiki+ with content from Wikipedia compared to an outdated OMIM SPARQL endpoint provided by Bio2RDF (last updated on July 2011), the content gap underscores the necessity for rigorous validation of information extracted by in-silico approaches. Second, the content gap also illustrates the fact that deducing disease-gene associations via the SPARQL querying and graph pattern matching capabilities can lead to identification of association pairs that may or may not be biologically relevant. On one hand, this can potentially facilitate faster curation of disease-gene associations in public repositories, including GeneWiki+, but on the other hand, unless the associations are adequately supported by scientific literature, they are biologically questionable. Finally, as demonstrated via the SPARQL querying capabilities, our approach also introduces the possibility of discovering novel disease-gene associations that have been reported in GWAS but do not have an explicit relationship in centrally curated data sources, such as OMIM. With appropriate validation, one could naturally augment such findings within the public data sources.
5.2. Related Work
Over the last decade, a large number of computational methods and tools have been developed for in-silico identification and prioritization of disease-gene-SNP associations. Syndrome 2 Gene (S2G [ 14 ]) prioritizes candidate disease genes based on similarity to known disease genes underlying both the query disease and similar phenotypes via querying 18 different databases, including OMIM. It implements a host of algorithms that allow an efficient search for candidate genes on a genomic locus, using known genes whose defects cause phenotypically similar syndromes. The Phenotype-Genotype mapper (PGMapper [ 15 ]) maps candidate genes to phenotype-related terms in OMIM and PubMed databases. For a given phenotype keyword (or search term), it dynamically matches the phenotypic traits to genes or disease loci by integrating mapping information from Ensemble and gene function information from OMIM and PubMed. Gene 2 Disease (G2D [ 16 ]) is another toolkit that prioritizes positional candidate disease genes either by linking candidate genes directly to disease phenotypes using literature and text mining, or by using functional links between candidates in one locus and either known disease gene or those in a different candidate locus. It accepts as input an OMIM phenotype identifier or an Entrez Gene identifier to find genes associated with an inherited disease, with the assumption that the identified candidate genes will produce a similar variant of the disease of interest. Finally, ICSNPathway [ 17 ] provides a solution to bridge the gap between GWAS and disease mechanism study by generating hypotheses for SNP→gene→pathway(s). Essentially, it identifies candidate causal SNPs and their corresponding candidate casual pathways from GWAS by integrating linkage disequilibrium analysis, functional SNP annotations and pathway-based analysis.
Our work in this study, while similar in spirit to the aforementioned efforts, demonstrates the applicability of Semantic Web and Linked Open Data technologies for disease-gene-SNP association mining. Such an approach highlights three main advantages. First, by leveraging publicly available datasets and developing approaches for federated querying using open and Web-based protocols, our approach illustrates the potential of integrating and extracting information from multiple data sources dynamically without creating “local dumps”. This not only has the advantage of retrieving the most updated data, but also avoids the overhead to maintain and update a local copy of the original resource. Second, as more and more biomedical and clinical data sources become available as Linked Data, our methods can be scaled to incorporate new types of information with minimal re-engineering. And last but not the least, our approach opens the possibility of doing logical inferencing to detect inconsistencies on the derived data as well as identify novel associations that are currently implicit within disparate data sets.
5.3. Future Work and Conclusion
In this work, we demonstrate the applicability of Semantic Web and Linked Data technologies for integrating heterogeneous public repositories for identifying genotype-phenotype associations. The methods proposed along with the preliminary results highlight the promise of applying such technologies in mainstream biomedical sciences research. In terms of future work, we intend to incorporate pathway data from KEGG [ 18 ] as well as patient-specific information from Mayo Clinic’s patient electronic health record [ 19 ], which can be achieved by expanding our existing set of federated SPARQL queries without a complete reconfiguration of our existing system.
References
- 1. Chen H , et al. Semantic web for integrated network analysis in biomedicine . Briefings in Bioinformatics . 2009 ; 10 ( 2 ): 177 – 192 . doi: 10.1093/bib/bbp002. [DOI] [PubMed] [Google Scholar]
- 2. Bizer C , Heath T , Berners-Lee T . Linked Data - The Story So Far . International Journal on Semantic Web and Information Systems . 2009 ; 5 ( 3 ): 1 – 22 . [Google Scholar]
- 3. Hindorff L , et al. A Catalog of Published Genome-Wide Association Studies . 2011 January ; Available from: http://www.genome.gov/gwastudies .
- 4. Hamosh A , et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders . Nucleic Acids Research . 2005 ; 33 ( suppl 1 ): D514 – D517 . doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huss JW , et al. The Gene Wiki: community intelligence applied to human gene annotation . Nucleic Acids Research . 2010 ; 38 ( suppl 1 ): D633 – D639 . doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bizer C , et al. DBpedia - A crystallization point for the Web of Data . Web Semant. . 2009 ; 7 ( 3 ): 154 – 165 . [Google Scholar]
- 7. Belleau F , et al. Bio2RDF: Towards a mashup to build bioinformatics knowledge systems . Journal of Biomedical Informatics . 2008 ; 41 ( 5 ): 706 – 716 . doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 8. GeneWiki+: A Semantic Mirror of the Gene Wiki project on Wikipedia . 2011 December ; Available from: http://genewikiplus.org/
- 9. SNPedia: A Wiki Investigating Human Genetics . 2011 December ; Available from: http://www.snpedia.com/
- 10. Virtuoso Universal Server . [cited 2011 January 16, 2011]; Available from: http://virtuoso.openlinksw.com/
- 11. Doganci A , et al. The IL-6R α chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo . The Journal of Clinical Investigation . 2005 ; 115 ( 2 ): 313 Р325 . doi: 10.1172/JCI22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pearson T , Manolio T . How to Interpret a Genome-wide Association Study . Journal of the American Medical Association . 2008 ; 299 ( 11 ): 1335 – 1344 . doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 13. Allen M , et al. Positional cloning of a novel gene influencing asthma from Chromosome 2q14 . Nat Genet . 2003 ; 35 ( 3 ): 258 – 263 . doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 14. Gefen A , Cohen R , Birk OS . Syndrome to gene (S2G): in-silico identification of candidate genes for human diseases . Human Mutation . 2010 ; 31 ( 3 ): 229 – 236 . doi: 10.1002/humu.21171. [DOI] [PubMed] [Google Scholar]
- 15. Xiong Q , Qiu Y , Gu W . PGMapper: a web-based tool linking phenotype to genes . Bioinformatics . 2008 ; 24 ( 7 ): 1011 – 1013 . doi: 10.1093/bioinformatics/btn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Iratxeta C , Bork P , Andrade-Navarro MA . Update of the G2D tool for prioritization of gene candidates to inherited diseases . Nucleic Acids Research . 2007 ; 35 ( suppl 2 ): W212 – W216 . doi: 10.1093/nar/gkm223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang K , et al. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework . Nucleic Acids Research . 2011 ; 39 ( suppl 2 ): W437 – W443 . doi: 10.1093/nar/gkr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanehisa M , et al. From genomics to chemical genomics: new developments in KEGG . Nucleic Acids Research . 34 ( suppl 1 ): D354 – D357 . doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pathak J , Kiefer R , Chute C . Applying Linked Data Principles to Represent Patient’s Electronic Health Records at Mayo Clinic: A Case Report . 2nd ACM SIGHIT International Health Informatics Symposium . 2012 [Google Scholar]