Abstract
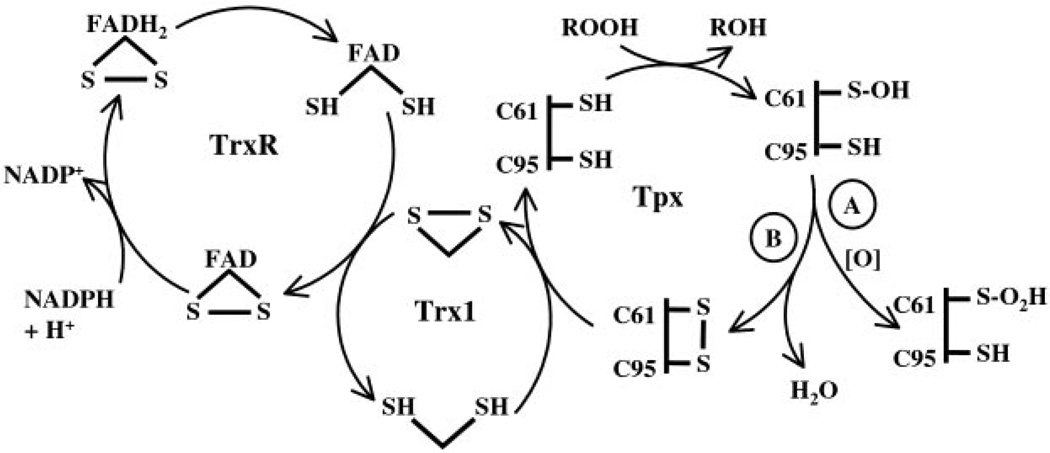
Escherichia coli thiol peroxidase (Tpx, p20, scavengase) is part of an oxidative stress defense system that uses reducing equivalents from thioredoxin (Trx1) and thioredoxin reductase to reduce alkyl hydroperoxides. Tpx contains three Cys residues, Cys95, Cys82, and Cys61, and the latter residue aligns with the N-terminal active site Cys of other peroxidases in the peroxiredoxin family. To identify the catalytically important Cys, we have cloned and purified Tpx and four mutants (C61S, C82S, C95S, and C82S,C95S). In rapid reaction kinetic experiments measuring steady-state turnover, C61S is inactive, C95S retains partial activity, and the C82S mutation only slightly affects reaction rates. Furthermore, a sulfenic acid intermediate at Cys61 generated by cu-mene hydroperoxide (CHP) treatment was detected in UV-visible spectra of 4-nitrobenzo-2-oxa-1,3-diazole-labeled C82S,C95S, confirming the identity of Cys61 as the peroxidatic center. In stopped-flow kinetic studies, Tpx and Trx1 form a Michaelis complex during turnover with a catalytic efficiency of 3.0 × 106 m−1 s−1, and the low Km(9.0 µm) of Tpx for CHP demonstrates substrate specificity toward alkyl hydroperoxides over H2O2 (Km > 1.7 mm). Rapid inactivation of Tpx due to Cys61 over-oxidation is observed during turnover with CHP and a lipid hydroperoxide, 15-hydroperoxyeicosatetraenoic acid, but not H2O2. Unlike most other 2-Cys peroxiredox-ins, which operate by an intersubunit disulfide mechanism, Tpx contains a redox-active intrasubunit disulfide bond yet is homodimeric in solution.
Oxidative stress defenses combat reactive oxygen species (1, 2) such as superoxide (), hydrogen peroxide (H2O2) and the hydroxyl radical (OH·) generated by the host immune response, environmental factors, and the incomplete reduction of oxygen to water during aerobic respiration, all of which are hazardous to proteins, DNA, and lipids (3, 4). Escherichia coli protects cellular components from oxidative damage by employing a variety of antioxidant defense enzymes, such as hydroperoxidases (catalases) I and II (gene products of katG and katE, respectively) that decompose H2O2 (5), and superoxide dismutases (manganese superoxide dismutase, sodA; iron super-oxide dismutase, sodB; copper-zinc superoxide dismutase, sodC) (6) that eliminate . Additional defenses in E. col against alkyl and lipid hydroperoxides are provided by multiple, non-heme peroxidases including 1) the peroxidatic component from the alkyl hydroperoxide reductase system (AhpC)1 (7); 2) a weakly active, thioredoxin (Trx)-dependent bacterioferritin-comigratory protein (BCP) (8); and 3) the periplasmic thiol peroxidase (Tpx, p20, scavengase) (9). In addition, a glutathione peroxidase homologue, the gene product of btuE (10), has been identified in E. coli, and preliminary investigations have indicated Trx-dependent peroxidatic activity against organic hydroperoxides (ROOH) and H2O2.2
AhpC, BCP, and Tpx are all members of the ubiquitous peroxiredoxin (Prx) family within the Trx superfamily of protein folds; however, the three E. coli Prx members are highly diverged from one another and are representative of three distinct Prx subfamilies (11–13). Generally, the Prx active site contains a disulfide bond composed of a conserved N-terminal Cys (Cys46 from Salmonella typhimurium AhpC) and the C-terminal Cys (Cys165′ from S. typhimurium AhpC) from the other subunit of the antiparallel dimer, resulting in two symmetrical active sites per dimer (14). Most Prxs contain two conserved Cys (2-Cys Prxs); however, in some homologues, only the N-terminal Cys is retained (the 1-Cys Prxs) (15). In many instances, the homodimers assemble into toroid-shaped decamers (15), a redox-dependent process in S. typhimurium AhpC whereby reduced decamers disassociate into dimers upon oxidation (16). To detoxify peroxides, the reduced N-terminal Cys attacks the peroxide -O–O- bond, with concomitant formation of a Cys sulfenic acid (Cys-SOH) intermediate, which then condenses with the C-terminal Cys to regenerate the stable disulfide bond at the active site (14, 17). For most bacterial Prxs, disulfide reduction is achieved by a specialized electron donor, AhpF (18), whereas many other Prx systems (both bacterial and eukaryotic) receive electrons from a reducing system composed of Trx and Trx reductase (TrxR) (13, 15).
Homologues of tpx are distributed throughout most or all eubacterial species, both Gram-negative and Gram-positive, and are found in pathogenic strains such as Haemophilus in-fluenzae, Streptococcus pneumoniae, and Helicobacter pylori (19), but biochemical and genetic analyses have been limited primarily to E. coli Tpx. In response to oxidative stress, E. coli up-regulates Tpx expression through an oxygen-responsive promoter element that is repressed by the transcriptional regulators ArcA and Fnr under anaerobic conditions (9, 20, 21). In addition, tpx deletion mutants, while still viable, were more susceptible to oxidative stress and displayed diminished colony sizes and numbers after peroxide exposure (22). In vitro studies have confirmed that Tpx forms a Trx-linked peroxidase system capable of reducing H2O2 and ROOH and protecting against glutamine synthetase inactivation by a mixed function oxidation system (9).
Tpx contains three Cys residues in its primary sequence, Cys61, Cys82, and Cys95. Of these, Cys61 aligns with the per-oxidatic, N-terminal Cys of other Prxs; whereas Cys95 does not align with the conserved C-terminal Cys of other 2-Cys Prxs, it is conserved among all Tpx homologues (19). Previous mutagenesis studies have presented conflicting information about which Cys residues are involved in peroxide attack and have indicated that both Cys61 and Cys95 are essential for activity, whereas loss of Cys82 only slightly attenuates activity (22, 23). In this report, we identify Cys61 as the peroxidatic Cys forming a Cys-SOH intermediate and firmly establish its intrasubunit linkage to Cys95 in the redox-active disulfide. Although Tpx monomers are not covalently linked, analytical ultracentrifugation studies reported herein demonstrate that the enzyme is a homodimer in solution.
EXPERIMENTAL PROCEDURES
Materials
SDS, ultrapure glycine, ultrapure urea, EDTA disodium salt, dithiothreitol (DTT), ammonium sulfate, β-mercaptoethanol, 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), Tris base, and other buffer reagents were purchased from Research Organics (Cleveland, OH). Bacteriological media components were from Difco. Ethanol was obtained from Warner Graham Company (Cockeysville, MD). Isopropyl β-d-thio-galactopyranoside and X-gal (5-bromo-4-chloro-3-inolyl-β-d-galactopyranoside) were from Inalc (Milan, Italy). Vent DNA polymerase was purchased from New England Biolabs (Beverly, MA). Restriction enzymes, T4 DNA ligase, calf intestinal phosphatase, Taq DNA polymerase, MgCl2 solutions, and restriction buffers were obtained from Promega (Madison, WI). Agarose medium EEO (electrophoresis grade), organic solvents (high pressure liquid chromatography (HPLC) grade), water (optima grade), sodium borate, acetic acid, sodium chloride, and H2O2 were from Fisher. Acrylamide/bis (40%) solution was purchased from Bio-Rad. Ampicillin powder, chloramphenicol, streptomycin sulfate, formic acid, calcium chloride, cumene hydroperoxide (CHP), dimethyl sulfoxide (Me2SO), protocatechuic acid, protocatechuate-3,4-dioxygenase, lipoxygenase, Saccharomyces cerevisiae glutathione reductase, E. coli glutaredoxin 1, and insulin were from Sigma. Ethyl hydroperoxide was from Polysciences, Inc. (Warrington, PA). Glutathione, 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD chloride), 4-vinylpyridine, and tert-butyl hydroperoxide were from Aldrich. NADPH and NADH were from Roche Molecular Biochemicals. l-1-(Tosylamino)-2-phenylethyl chloromethyl ketone-treated trypsin was obtained from Worthington. Pierce supplied the immobilized Tris[2-carboxyethyl] phosphine (TCEP) disulfide-reducing gel, Gel Code Blue stain, and trifluoroacetic acid ampules. Molecular Probes, Inc. (Eugene, OR) supplied the methyl methanethiolsulfonate and fluorescein-5-maleimide. Arachidonic acid was from NuCheck Prep (Elysian, MN). Stratagene (La Jolla, CA) supplied the Pfu Turbo polymerase, DpnI, and E. coli XL-1 Blue competent cells. All ultrafiltration was carried out using Apollo 7-ml high performance centrifugal concentrators (Orbital Biosciences, Topsfield, MA) unless otherwise noted.
Cloning of tpx into an Expression Vector
Genomic DNA prepared from a 400-ml overnight culture of an E. coli K-12 strain (XL-1 Blue) was used as the PCR template (24). The gene of interest was amplified with the following PCR primers: forward primer, 5′-GCGAATTCAGGAGGAAGAATAGATGTCACAAACCGTACATTTCCAGGGC-3′ and reverse primer 5′-GCCTGCAGTTATGCTTTCAGTACAGCC-3′ (engineered restriction sites underlined) synthesized in the DNA Synthesis Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. PCR mixtures (50 µl) contained 200 µm dNTPs, 2 units of Vent DNA polymerase, 20 pmol each of forward and reverse oligonucleotides, 1 mm MgCl2, and 0.5 µg of genomic DNA. The reaction was carried out in a Mini Cycler (MJ Research, Waltham, MA) as follows: 95 °C at 30 s, 55 °C for 45 s, and 72 °C for 1.5 min (35 cycles) and then 72 °C for 15 min. The 507-bp tpx PCR product purified with the QIA-quick PCR Cleanup kit (Qiagen, Studio City, CA) was then ligated into the pCR2.1 TA cloning vector (Invitrogen) after Taq DNA polymerase/ dNTP treatment. Plasmid DNA purified from the E. coli host using the Wizard Miniprep Kit (Promega) was screened for the presence of insert by digestion with the appropriate restriction enzymes (EcoRI and PstI). The DNA fragment containing tpx was excised from agarose gels and purified using the Gene Clean II Kit (Bio 101, Inc., Vista, CA). The fragment was ligated using T4 DNA ligase into the pPROK1 expression vector (Clontech, Palo Alto, CA; expression under control of the tac promoter) that had been similarly digested, isolated from an agarose gel, and pretreated with calf intestinal phosphatase to generate pPROK1/tpx.
Site-directed Mutagenesis of Tpx
The four mutant E. coli Tpx enzymes, C61S, C82S, C95S, and C82S,C95S were created by following the protocol outlined in the QuikChange site-directed mutagenesis kit (Stratagene) using primers complementary to the coding and noncoding template sequence (pPROK1/tpx) containing a double-base mismatch. To generate the C61S mutation, the forward primer 5′-GATACCGGTGTTTCGGCCGCATCAGTACG-3′ and a complementary reverse primer were used (underlined letters indicate the base pair mismatch). To generate the C95S mutation, the forward primer 5′-CGCCCAGTCTCGTTTCTCGGGCGCAGAAGG-3′ and a complementary reverse primer were employed, and C82S was constructed using the forward primer 5′-CACCGTTGTGCTGTCTATCTCTGCCGATCTGCC-3′ and its complementary reverse primer. C82S,C95S was created using C95S plasmid as template with the forward and reverse primers for the C82S mutation. Reaction mixtures (50 µl) contained 10–50 ng of template DNA (pPROK1/tpx or pPROK1/tpxC95S), 125 ng of each primer, 200 µm dNTPs, and 1 µl of Pfu Turbo polymerase. Twelve cycles of 95 °C for 30 s and 55 °C for 13 min were carried out in a Mini Cycler followed by 55 °C for 10 min to finish extending products. To digest methylated template, each reaction mixture was treated with 1 µl of DpnI at 37 °C for 1 h.
Bacterial Strains and Culture Procedures
Ligated DNA (pPROK1/tpx) and mutagenesis products were transformed into XL-1 Blue cells. Single colonies were selected on Luria-Bertani (LB) plates containing ampicillin (50 µg/ml), and those containing the recombinant DNA were evaluated for protein expression by SDS-PAGE after induction with 0.4 mm isopropyl β-d-thiogalactopyranoside. Isolated plasmid DNA for each construct was sequenced throughout the coding region by automated DNA sequencing at the Comprehensive Cancer Center of Wake Forest University. Bacterial stocks containing each plasmid with the sub-cloned gene were prepared from a single colony and stored at −80 °C in LB broth containing 15% (v/v) glycerol. Culture procedures were generally the same as reported earlier (25).
Purification of Recombinant and Mutant Tpx Proteins
A modification of a previous Tpx purification protocol was used for this study (9). All procedures were carried out in a standard buffer (pH 7.0) consisting of 25 mm potassium phosphate with 1.0 mm EDTA. Briefly, 100 ml of XL-1 Blue E. coli harboring the pPROK1/tpx plasmid were added to 10 liters of LB medium containing 0.5 g of ampicillin supplemented with 0.2% glucose in a BioFlo 2000 fermentor (New Brunswick Scientific, Edison, NJ). Isopropyl β-d-thiogalactopyranoside (0.4 mm) was added at A600 = 0.9, and bacteria were harvested by centrifugation 16 h after induction. Pelleted bacteria were disrupted with a Bead Beater (Bio-Spec Products, Bartlesville, OK), and cell extracts treated with streptomycin sulfate to precipitate nucleic acids were subjected to 30 and 75% (NH4)2SO4 treatments to precipitate proteins (25). The protein mixture resuspended in standard buffer containing 10% (NH4)2SO4 was applied to a 24 × 2.5-cm Phenyl-Sepharose 6 Fast Flow Column (Amersham Biosciences), washed with 10% (NH4)2SO4 buffer, and eluted with deionized H2O. Protein fractions were evaluated for contamination of overexpressed Tpx by SDS-PAGE, and the purest fractions were pooled. After dialysis against 5 mm potassium phosphate buffer (pH 7.0), the protein was loaded onto a Q-Sepharose column (Amersham Biosciences) pre-equilibrated in 5 mm potassium phosphate and eluted with a linear gradient from 5 to 30 mm potassium phosphate (1 liter total volume). Again, fractions were analyzed for Tpx by SDS-PAGE, and the pure fractions were pooled, concentrated, and aliquotted for storage at −20 °C.
Purifications of mutant Tpx proteins were carried out in the same manner as for wild type Tpx with a few exceptions. The pooled C61S mutant Tpx protein from the Phenyl-Sepharose column was loaded onto a 24 × 2.5-cm DEAE-cellulose column (DE52; Whatman, Kent, UK) equilibrated with 30 mm potassium phosphate, and eluted with a linear gradient from 30 to 80 mm potassium phosphate (1 liter total volume). To completely purify C61S, fractions containing the mutant protein were reapplied to the Phenyl-Sepharose and DEAE-cellulose columns. Initial attempts at purifying the C95S mutant under the same conditions as wild type resulted in aggregation of the mutant protein, as observed by multiple bands during SDS-PAGE of the subsequent fractions, even after many rounds of purification over the two columns. DTNB titration of the isolated protein even after DTT treatment gave less than one thiol (data not shown), suggesting that Cys61 had become irreversibly overoxidized to a sulfinic (Cys-SO2H) or sulfonic (Cys-SO3H) acid. The addition of 2 mm DTT to all buffers prior to bacterial disruption and over the course of the purification of C95S gave pure protein after one round of purification on the two columns. C82S was purified in buffers containing 2 mm DTT according to the same protocol as C95S; however, pure C82S,C95S was obtained using a slightly altered protocol. 20% instead of 10% (NH4)2SO4 was used to treat C82S,C95S prior to its application to a Phenyl-Sepharose column equilibrated with 20% (NH4)2SO4, and C82S,C95S was eluted using a gradient of 20% (NH4)2SO4 in 25 mm potassium phosphate with 2 mm DTT to 0% (NH4)2SO4 in deionized water with 2 mm DTT. Elution from the Q Sepharose column was achieved with a gradient of 20–120 mm potassium phosphate in 2 mm DTT. Prior to assay, each mutant was subjected to ultrafiltration and washed with standard buffer to remove DTT and then immediately incubated with equal volumes of TCEP gel.
Other Protein Purifications
Purifications of E. coli TrxR (26) and E. coli Trx1 (27) were carried out as described previously. S. typhimurium AhpF and S. typhimurium AhpC were purified as reported previously (25).
Fluorescein-5-Maleimide Labeling of Proteins
Wild type, C61S, C82S, C95S, and C81S,C95S Tpx (100 µg each), prereduced with DTT that was removed by ultrafiltration, were reacted with fluorescein-5-maleimide (10 equivalents) under denaturing (4 m urea) and reducing (100 eq of TCEP) conditions for 16 h at 4 °C in standard buffer. Protein samples (5 µg) were separated on 18%, 20-cm-long SDS-polyacrylamide gels. Densitometry to assess the protein contents of fluorescein-labeled bands was conducted using the Quantity One quantitation software from Bio-Rad, and image files were generated with a ChemiImager 5500 digital imaging system from Alpha Innotech Corp. (San Leandro, CA) and a near UV light source and UV filter. For quantitation, six amounts of wild type Tpx (0.03–5 µg) were electrophoresed along with each of the mutant samples. Electrophoresis samples also included one with 5 µg of C61S, to which 0.25 µg of wild type Tpx was intentionally added, and one into which 1.7 µg each of wild type Tpx and C61S and C82S,C95S mutants were loaded to assess the quality of the separation.
The labeled proteins were also submitted for electrospray mass spectrometry, as described below, following ultrafiltration into water to remove buffer using Apollo concentrators (Orbital Biosciences).
Analytical Ultracentrifugation Analyses
To determine the oligomeric state of wild type and mutant proteins, samples of different concentrations were analyzed by sedimentation equilibrium at various speeds on an Optima XL-A analytical ultracentrifuge (Beckman Instruments, Palo Alto, CA) outfitted with absorbance optics. Tpx, C61S, and C95S (purified in DTT) were exchanged into a buffer of 25 mm potassium phosphate, 1 mm EDTA, and 0.15 m NaCl, pH 7.05, via ultrafiltration. Three different concentrations of each protein (39.5, 184.2, and 342.1 µm) in 110 µl were loaded into three of the six sectors of each cell, and buffer (125 µl) was loaded into the remaining sectors as a reference. Data were obtained at 11,000, 16,000, and 20,000 rpm at 20 °C following equilibration for 8, 10, and 12 h at each speed. Data with absorbances higher than 1.4 were excluded from data analysis. The partial specific volumes for Tpx and the mutants were calculated from the amino acid composition to be 0.7433 cm3 g−1 and 0.7434 cm3 g−1, respectively (28). Multiple data sets were globally fit to the equation for a single ideal species using WinNonLin2 (29). The buffer density of 1.00773 g/cm3 was determined using a DA-310 M precision density meter (Mettler Toledo, Hightstown, NJ) at 20 °C.
Mass Spectrometric Analyses
Protein samples were extensively dialyzed in deionized water (6 liters) in a Slide-A-Lyzer cassette (Pierce) overnight prior to analysis by electrospray ionization mass spectrometry (ESI-MS; Micromass, Manchester, UK) precalibrated with horse heart myoglobin. The protein sample (1 µm) in 1% formic acid was injected at a flow rate of 300 µl/h, and positively charged ions in the m/z range of 800–1800 were analyzed using MassLynx software (version 3.5; Micromass).
Spectral Experiments
Most spectral assays were conducted on a single wavelength, thermostatted (25 °C) Gilford 220 updated recording spectrophotometer (Oberlin, OH) with a Beckman DU monochrometer (Fullerton, CA) unless otherwise noted. Microbiuret assays for proteins to determine extinction coefficients, disulfide assays with 2-nitro-5-thiosulfobenzoate, and thiol quantification with DTNB were conducted as described previously (25, 30). To further quantify Tpx proteins by absorbance, the following experimentally determined extinction coefficients at 280 nm were used: Tpx and C82S,C95S, 3800 ± 300 m−1 cm−1; C95S, 3500 ± 200 m−1 cm−1; C82S, 4200 ± 600 m−1 cm−1; C61S, 5300 ± 700 m−1 cm−1. Other extinction coefficients used were as follows: E. coli TrxR, 11,300 m−1 cm−1 (454 nm) (31); E. coli Trx1, 13,700 m−1 cm−1 (280 nm) (32); S. typhimurium AhpC, 24,300 m−1 cm−1 (280 nm) (25); S. typhimurium AhpF, 13,100 m−1 cm−1 (450 nm) (25); E. coli glutaredoxin 1, 12,400 m−1 cm−1 (280 nm) (33); NADPH, 6200 m−1 cm−1 (340 nm); NADH, 6220 m−1 cm−1 (340 nm); 2-nitro-5-thiobenzoate (TNB2−), 14,150 m−1 cm−1 (412 nm) (34).
Steady-state Kinetic Analysis of Tpx and Mutants
Michaelis constants (Km) and turnover numbers (kcat) with CHP as the substrate for Tpx were obtained as described previously for H. pylori TrxR (35). Briefly, reactions were conducted on an Applied Photophysics DX-17 MV stopped-flow spectrophotometer (Surrey, UK) at 25 °C, and activity was followed by monitoring the decrease in fluorescence or absorbance of NADPH over time. Use of the stopped-flow spectrophotometer for mixing and data acquisition helped alleviate nonlinearity and reproducibility problems inherent in the manual mixing methods (14). Reaction mixtures in one syringe contained NADPH (150 µm), Trx1 (2–80 µm), TrxR (0.25–10 µm, in a molar ratio of 1:8 TrxR to Trx1, so that the electron transfer to Trx1 is not rate-limiting), and Tpx (0.2 µm) in peroxidase buffer consisting of 50 mm potassium phosphate (pH 7.0), 0.1 m (NH4)2SO4, and 0.5 mm EDTA. CHP (1–50 µm) was prediluted into Me2SO (in a ratio of 1:50 with the solvent) and mixed with peroxidase buffer in the other syringe. In all cases, concentrations given are final concentrations achieved after mixing of the contents of the two different syringes. Under the conditions given, Tpx was supplied in rate-limiting concentrations, and due to the saturable interaction between Tpx and Trx1, the reaction was linearly dependent on Trx1, provided that concentrations supplied were below the Km. Assays were designed so that Trx1 levels were at least 10-fold higher than those of Tpx to maintain the peroxidase as the limiting enzyme. Flavin-dependent oxidase activity did not contribute significantly to NADPH oxidation (0.05 s−1) and was not subtracted from initial rates. After conversion to concentration units/min, the primary rate data were fit to a rectangular hyperbolic curve function in Sigma Plot (Jandel Scientific, San Rafael, CA). The data obtained for each substrate concentration were plotted according to the Hanes-Woolf representation of the Michaelis-Menten equation to give intersecting lines at the y axis, indicating a substituted (ping-pong) mechanism and were further evaluated using the Hanes-Woolf equation for a two-substrate, substituted reaction as previously described (35). Because of the much larger Km of Tpx for H2O2, larger amounts of substrates were present in the reaction mixtures, and the sensitivity required previously in the fluorescence assay was no longer necessary. Therefore, all H2O2 data were collected in absorbance mode on the stopped flow spectrophotometer with the same protein assay mixtures as above, except that the second syringe contained varying H2O2 concentrations (0.25–10 mm). Synthesis of 15-hydroperoxyeicosatetraenoic acid (15-HPETE) was carried out as described by O’Flaherty et al. (36).
Sulfenic Acid Trapping Experiments
To observe the formation of Cys-SOH as a reaction intermediate, experiments with C61S, C95S, and C82S,C95S mutants labeled with NBD chloride were carried out as described previously (17, 37), with a few exceptions. Each mutant (171 µm in 100 µl, enough protein to give an absorbance of 0.5 at 280 nm) was preincubated with DTT for 1 h and then desalted by ultrafiltration. C61S and C95S proteins in standard buffer were made anaerobic by repeated flushing with argon gas and vacuum in alternating cycles for 30 min. Anaerobic solutions of NBD chloride (50 mm in Me2SO) and CHP (110 mm in Me2SO) were prepared by bubbling argon through the preparations for 15 min. Under anaerobic conditions, the C61S and the C95S mutants were oxidized by H2O2 (1 eq), and then the proteins were incubated with NBD chloride (10 eq) for 5 min. Excess NBD chloride was removed by ultracentrifugation, and the absorbance properties of the protein samples were analyzed (200–600 nm) on a Beckman DU 7500 diode array spectrophotometer (Fullerton, CA). Labeling of prereduced C165S AhpC from S. typhimurium (48.3 µm) with NBD chloride was used as a positive control. Due to its sensitivity toward oxidation, C82S,C95S was stored with equal volumes of TCEP gel after removal of DTT and treated with CHP rather than H2O2 prior to NBD chloride labeling.
Tryptic Peptide Separations by HPLC
To prepare proteins for digestion, prereduced Tpx (10 nmol) was treated with or without H2O2 (1 eq) and then incubated in MES (125 mm, pH 6.5) buffer with EDTA (1 mm) prior to reaction with 4-vinylpyridine and subsequent denaturation in urea (1.6 m) as described (30). The addition of 4-vinylpyridine to block free Cys was necessary to prevent adventitious disulfide bond formation or migration following denaturation. Following dialysis and solvent removal, exhaustive tryptic digestion of Tpx, C61S, or C95S in either oxidized or reduced forms was carried out as described (30). Tryptic maps were generated by injecting samples into a Rainin Dynamax HPLC system equipped with an AquaPore RP-300 C8 column (4.6 × 100 mm) and were eluted using a 90-min gradient consisting of 5–60% Solvent B (Solvent A was 0.1% trifluoroacetic acid in deionized, ultrapure H2O; Solvent B was 70% acetonitrile with 0.08% trifluoroacetic acid in H2O). Peaks were detected at 215 and 254 nm on a Dynamax UV-DII dual wavelength detector. To isolate the disulfide-containing fragments, peaks at 68 (P1) and 70 min (P2) from oxidized Tpx were isolated and then further purified on a shallower gradient. Acetonitrile and trifluoroacetic acid were then removed by vacuum centrifugation, and solutions were brought to a final volume of 100 µl with deionized H2O prior to analysis by ESI-MS.
RESULTS
Characterization of Purified Tpx and Mutants
Homogeneous, recombinant wild type Tpx and mutant proteins were isolated using a modification of the previous purification protocol (9), and all mutants were purified in the presence of 2 mm DTT to prevent overoxidation of free Cys residues (14). Mutant enzymes were difficult to prepare under oxidizing conditions due to irreversible protein dimerization, and C95S purified without DTT displayed significantly less activity than C95S purified under reducing conditions. DTNB titrations of denatured, prereduced wild type Tpx and mutants confirmed the expected thiol content for each (Table I). Additionally, ESI-MS of Tpx indicated a mass of 17,700 atomic mass units, 135 atomic mass units less than the mass of 17,835 atomic mass units predicted by the tpx open reading frame, indicating the loss of the initiating Met from E. coli Tpx. Each single mutant exhibited a mass of 17,686 atomic mass units, corresponding to a 16 atomic mass units loss due to the Cys to Ser mutation, whereas the 17,672-atomic mass units mass for C82S,C95S confirmed the double Cys to Ser mutation. The Cys to Ser mutations are conservative, and spectral and circular dichroism scans conducted in the far ultraviolet region revealed no gross structural perturbations among the mutants compared with the wild type protein (data not shown).
Table I. DTNB analysis of free thiols on wild type and mutant Tpx.
For details, see “Experimental Procedures.”
| Prereduced | Oxidizeda | |||
|---|---|---|---|---|
| Native | Denaturedb | Native | Denaturedb | |
| Tpx | 1.90 ± 0.03 | 2.60 ± 0.09 | 0.47 ± 0.07 | 0.84 ± 0.09 |
| C61S | 1.2 ± 0.1 | 2.0 ± 0.3 | 0.98 ± 0.10 | 1.82 ± 0.04 |
| C82S | 1.70 ± 0.01 | 2.00 ± 0.04 | 0.02 ± 0.01 | 0.17 ± 0.08 |
| C95S | 1.1 ± 0.1 | 1.7 ± 0.2 | 0.19 ± 0.03 | 0.43 ± 0.03 |
| C82S,C95S | 0.78 ± 0.01 | 0.85 ± 0.04 | 0.02 ± 0.02 | 0.07 ± 0.02 |
Prereduced + 1 eq. of H2O2.
In 4 m guanidine HCl.
As mutant proteins were isolated from E. coli expressing low amounts of wild type Tpx, labeling experiments were conducted using fluorescein-5-maleimide to shift protein molecular weights according to the number of cysteine residues (an increase in 427 g/mol per fluorescein-labeled cysteine residue) and to enhance quantitation by densitometry. Using long gels and 18% acrylamide, singly, doubly, and triply labeled proteins were nicely separated (data not shown). Including data from kinetic studies described below, which give an upper limit of 0.4% for the degree of contamination of the C61S mutant by wild type Tpx (if all activity is due to the presence of wild type), densitometry of the gel samples taking into account this information indicates a wild type Tpx contamination level of less than 5%. Verification of the single (C82S,C95S), double (C61S, C82S, and C95S mutants), and triple (wild type Tpx) labeling of these proteins by fluorescein was obtained by mass spectrometry. Furthermore, purification protocols for C61S and C82S,C95S mutants were significantly different from that for the wild type enzyme, making contamination by the wild type enzyme less likely in these cases.
Oligomeric State of Tpx
Earlier studies using a gel filtration column standardized with molecular weight markers suggested that oxidized Tpx was a monomer of 16.8 katomic mass units (9). In addition, the 2-atomic mass units difference for Tpx ESI-MS masses obtained under reducing (17,698.4 ± 0.62 atomic mass units) and nonreducing (17,696.7 ± 0.23 atomic mass units) conditions and the nonreducing SDS-PAGE analysis (see below) exclude the possibility of a covalent dimer for oxidized Tpx. Nonetheless, most other 2-Cys Prx proteins examined thus far, except for human PrxV (38), exist as covalent dimers when oxidized and noncovalent dimers or higher order oligomers when reduced (15). The oligomeric state of Tpx in solution was, therefore, examined by analytical ultracentrifugation sedimentation equilibrium experiments. Data analyses of three separate data sets each (40–340 µm) gave weight-average molecular weights for Tpx of 29,500 and 32,000 for the oxidized and reduced enzyme, respectively, indicating that two Tpx monomers self-associate in solution independent of redox state. Tpx dimerization is not concentration-dependent above 40 µm, because similar molecular weights are obtained at different speeds and all three concentrations. Detection of dimeric molecular weights for both C61S and C95S under the same experimental conditions indicates that these mutations do not perturb self-association.
Most 2-Cys Prxs are linked by an intersubunit disulfide bond at the active site in their oxidized state (25, 39). However, previous SDS-polyacrylamide gel studies of wild type Tpx revealed only a 19-kDa band under reducing and nonreducing conditions, suggesting the formation of an intrasubunit, rather than intersubunit, disulfide bond upon oxidation (9, 19). In the present study, a second species (~36 kDa) was observed on SDS-PAGE conducted under nonreducing conditions only when a thiol-specific blocking agent, methyl methanethiolsulfonate, was excluded from sample preparations prior to denaturation (data not shown), indicating that any covalent dimerization of Tpx is artifactual. Furthermore, during SDS-PAGE analysis under nonreducing conditions, only a very small downward shift in protein migration is observed upon oxidation (from an apparent molecular mass of 18.5 to 18.2 kDa, data not shown), consistent with intrasubunit disulfide bond formation as analyzed further using tryptic mapping techniques.
Identification of the Intrasubunit Disulfide Linkage
To assess the redox state of Cys residues and identify the location of the putative intrasubunit disulfide linkage, a series of biochemical assays were conducted on oxidized and reduced wild type Tpx. Reduced Tpx contained no disulfide bonds (as quantified by 2-nitro-5-thiosulfobenzoate assays) and 1.90 ± 0.03 or 2.60 ± 0.09 free thiols/monomer under native or denaturing conditions, respectively (Table I), indicating the presence of one buried, slow reacting Cys thiol. After the addition of 1 eq of peroxide to prereduced Tpx, the two accessible thiol groups were lost (Table I), and one disulfide bond (0.90 ± 0.01) per monomer was gained.
To identify the disulfide-forming Cys residues, tryptic maps of oxidized and reduced, pyridylethylated Tpx were generated by HPLC and compared at 215 and 254 nm (Fig. 1). Instead of two strong peaks at 254 nm in the reduced, pyridylethylated chromatograms, four strong peaks were observed around 54, 56, 58, and 74 min, which allowed for the identification of Cys-containing peptides. After comparing the oxidized and reduced tryptic maps at 215 nm, two new peaks representing the disulfide-containing peptides were detected in the oxidized map at 68 (P1) and 70 min (P2), representing the disulfide-containing peaks (Fig. 1). P1 and P2 were reisolated on a shallower gradient and then reduced and pyridylethylated and reanalyzed by HPLC. P1 and P2 each gave two new peaks corresponding to two of the 254-nm absorbing peaks observed in the original chromatogram of the reduced, pyridylethylated protein (data not shown). Masses of P1 (3830 atomic mass units) and P2 (3702 atomic mass units) as determined by ESI-MS correspond to Cys61- and Cys95-containing peptides and verify that Cys61 and Cys95 compose the active site intra-subunit disulfide bond (Fig. 1). The presence of adjacent tryptic digest sites upstream of Cys61 resulted in two disulfide-containing peaks due to the partial digestion after Arg47.
FIG. 1. HPLC separation of tryptic peptides from either reduced or oxidized pyridylethylated Tpx and tryptic digest sites.
Exhaustive digests of reduced (A and C) or oxidized (B and D) Tpx (5 nmol) that was pyridylethylated during denaturation prior to incubation with trypsin were fractionated on an AquaPore RP-300 C8 column. Gradient conditions are given under “Experimental Procedures,” and peptides were detected at 215 nm (A and B) and 254 nm (C and D). The closed arrows in each map indicate the following peaks: a pyridylethylated fragment containing Cys82 (74 min) (A and C) and disulfide-containing peaks P1 (68 min) and P2 (70 min) (B and D). The open arrows indicate fragments resulting from reduction of P1 or P2. E, a portion of the sequence of Tpx from E. coli (residues 45–113, accession number AAC74406) is shown with the tryptic sites (underlined) flanking the three Cys (boldface type) numbered from the initiating Met. The two Cys61 fragments resulting from the flanking tryptic sites are highlighted with different lines for P1 (dotted) and P2 (solid), and the total ESI-MS mass of each disulfide-containing peak (including the Cys95-containing fragment indicated by the dashed line) is given above the site of initial hydrolysis by trypsin.
Kinetic Characterization of Tpx and Inactivation during Catalysis
The Trx1-linked peroxidase activity of Tpx and its differential activity with ROOH and H2O2 were reported earlier (9, 19). In this study, the true kcat and Km values of Tpx for Trx1, CHP, and H2O2 were determined from Hanes-Woolf plots of the initial rate data (Table II). All lines in the Hanes-Woolf primary plots intersect the origin, indicating a bisubstrate, ping-pong (substituted) reaction mechanism for Tpx, which can be depicted as a sequence of consecutive reactions,
Trx1red + Tpxox ↔ Trx1ox + Tpxred
Tpxred + ROOH → Tpxox + ROH + H2O
Reactions 1 and 2
where ROH is the corresponding alcohol. These redox reactions are analogous to those observed for other peroxidases from H. pylori and Crithidia fasciculata (35, 40) that are recycled by Trx or a Trx homologue, but unlike those systems that display an infinite Km for Trx, Tpx interacts with Trx1 in a saturable manner with a Km of 22.5–25.5 µm (Table II).
Table II.
Steady-state kinetic parameters of Tpx
| Peroxide | kcat | Km | kcat/Km (Trx1) | kcat/Km (peroxide) | |
|---|---|---|---|---|---|
| Trx1 | Peroxide | ||||
| s−1 | µm | m−1 s−1 | m−1 s−1 | ||
| H2O2 | 76.0 ± 8.1 | 25.5 ± 2.9 | 1730 ± 360 | 3.0 × 106 | 4.4 × 104 |
| CHP | 70.1 ± 7.1 | 22.5 ± 4.7 | 9.1 ± 1.8 | 3.1 × 106 | 7.7 × 106 |
Tpx steady-state reactions followed for longer than 5 s exhibited rapidly declining NADPH oxidation rates over time for CHP, but not H2O2, that were dependent on the concentration of peroxide. This observed inactivation of Tpx required a threshold level of CHP, whereby at high levels of CHP (>150 µm), but not at low CHP levels (<100 µm), activity diminished rapidly and nonlinearly and resulted in incomplete consumption of peroxide (Fig. 2). Activity could not be regained with the addition of new substrates; however, supplementation of reactions with fresh Tpx restored NADPH oxidation, which again diminished rapidly due to inactivation of the newly added Tpx (data not shown). Inactivated reaction mixtures (i.e. 200 µm CHP and levels of NADPH, TrxR, Trx1, and Tpx matching those used for the steady-state assays) that had been incubated with DTT and subsequently washed could not regenerate activity in the presence of fresh NADPH and CHP. These data suggest that inactivation is due to an irreversible overoxidation process that is most likely occurring at the active site Cys-SOH, a transient species that can be readily and irreversibly overoxidized by excess peroxide to Cys-SO2H or Cys-SO3H (14, 41). Prereduced Tpx incubated overnight in excess CHP retained full activity, indicating that inactivation requires Tpx turnover. ESI-MS verification of active site Cys overoxidation was obtained by injecting freshly inactivated reaction mixtures and immediately determining Tpx mass after turnover. After subtracting the contribution of TrxR and Trx1 ions from the reaction mixture spectra, the mass of Tpx was found to be 17,732 atomic mass units, an increase in 32 atomic mass units, which is consistent with the addition of two oxygen atoms. Together, these data strongly suggest that during turnover, Tpx’s active site Cys is converted to Cys-SO2H in an overoxidation process relying on a threshold amount of peroxide and that the Cys-SOH formed at the active site follows one of two pathways with each catalytic cycle (Fig. 3).
FIG. 2. Steady-state kinetic analysis of Tpx with various concentrations of CHP.
All three protein components (TrxR (1.5 µm), Trx1 (10 µm), and Tpx (1 µm)) were incubated in 150 µm NADPH for 5 min in one syringe and then were mixed with varying amounts of CHP (0 µm (closed circles), 100 µm (open triangles), 200 µm (closed squares), and 400 µm (open diamonds)) in another syringe (final concentrations after mixing; see “Experimental Procedures”). Reaction progress was monitored at 340 nm on a stopped flow spectrophotometer at 25 °C, and rates were extrapolated from the linear portion of the curve (0–2 s) using linear regression analysis. At 100 µm CHP, Tpx was not inactivated and gave linear absorbance changes for the duration of the reaction and full peroxide consumption (open triangles). At higher CHP concentrations, Tpx activity diminished rapidly and nonlinearly without complete consumption of CHP or NADPH (closed squares and open diamonds). Stopped-flow data were collected every 50 ms, but only data from every 1.5 s are represented by the symbols.
FIG. 3. Scheme of TrxR/Trx1 reduction and electron transfer pathways for Tpx during catalysis and inactivation.
During Tpx turnover, a small proportion of Tpx becomes overoxidized and is removed from the reaction cycle (path A). The remaining Tpx reforms the redox-active disulfide to continue the catalytic cycle (path B). Eventually, after multiple turnovers in the presence of ≥150 µm CHP, the enzyme primarily converts to the R-SO2H species, few disulfide-containing species remain, and activity declines, leading to irreversible inactivation. This scheme depicts the overall flow of electrons but not necessarily the precise redox forms of TrxR involved in turnover.
Bisubstrate kinetic analyses have confirmed that Tpx reacts differentially with soluble peroxides as evidenced by the large difference in Km for H2O2 and CHP (1750 and 9.1 µm, respectively) (Table II). Upon examination of the Km(app) values for other soluble peroxides (in reactions conducted at [Trx1] = 10 µm), we find that whereas H2O2 and CHP maintain their status as the worst and best substrates, respectively (Km(app) values of 648 versus 3.9 µm), Tpx has a lower Km(app) for ethyl hydroperoxide than for t-butyl hydroperoxide (30.7 versus 66.6 µm). Examination of Tpx reactivity with a physiological alkyl hydroperoxide, a relatively insoluble fatty acid hydroperoxide, 15-HPETE, resulted in a complex pattern of [15-HPETE]-dependent enzyme activity and inactivation. At low concentrations (2–6 µm), activity increased steadily with peroxide concentrations, but at higher [15-HPETE] (10–30 µm), activity was significantly decreased (Fig. 4). Incubation of the Tpx reaction mixture (i.e. NADPH, TrxR, Trx1, and Tpx) with increasing amounts of 15-HPETE followed by the addition of a noninactivating amount of CHP (50 µm) resulted in loss of Tpx activity with higher concentrations of 15-HPETE (10–30 µm) (data not shown), suggesting that Tpx is also inactivated by this substrate. The resulting, nonhyperbolic curve representing [15-HPETE]-dependent activity (Fig. 4) is not amenable to typical Michaelis curve fits and is further complicated by the limited solubility of 15-HPETE in aqueous buffers. Therefore, apparent second order rate constants were calculated from the slope of a line drawn through the points for 0–4 µm to give a kcat(app)/Km(app) of 2.6 × 105 m−1 s−1. These studies verify that while Tpx is sensitive to overoxidation by fatty acid hydroperoxides (and other soluble alkyl hydroperoxides such as CHP), significant activity at low concentrations of these substrates is achieved with rates comparable with those obtained for other soluble peroxides. Similar trends of inactivation with increasing concentrations of fatty acid hydroperoxide were obtained during studies of human PrxII with linoleic acid hydroperoxide (42), although the peroxide levels required for overoxidation in this case were much higher (250 µm) than those required for Tpx inactivation.
FIG. 4. Steady-state assay of Tpx as a function of 15-HPETE concentration.
Reaction mixtures in one syringe containing NADPH (150 µm), Trx1 (10 µm), TrxR (1.5 µm), and Tpx (1 µm) were mixed with varying amounts of 15-HPETE (0–30 µm) in another syringe on the stopped-flow spectrophotometer at 25 °C (final concentrations after mixing). Each rate is the average of three experiments and was obtained by linear regression analysis of the linear portion of the reaction progression (0–1 s) prior to inactivation. Standard peroxidase buffer was used in all cases as described under “Experimental Procedures.”
Tpx Specificity toward Trx1 Reduction
We examined the specificity of the interaction between Tpx and Trx1 by testing whether or not other E. coli enzymatic disulfide/dithiol reductants could serve as reductase systems. Because Trx2 (trxC) is up-regulated in response to oxidative stress (43, 44) and exhibits similar disulfide reductase capabilities as Trx1 (44, 45), we postulated that Trx2 may be able to substitute for Trx1. However, replacement of Trx2 in the steady-state Tpx assay did not result in NADPH oxidation (data not shown). Previous work also indicated that Tpx was active in the presence of high levels of glutathione (10–20 mm) (9); however, replacing the TrxR/Trx1 reductase system with glutathione reductase and glutathione (up to 40 mm) did not support Tpx turnover in our stopped-flow assays (data not shown). Additionally, neither E. coli glutaredoxin 1 nor E. coli AhpF were capable of reducing Tpx, indicating that Tpx peroxidase activity specifically requires Trx1.
Cys61 Is the Peroxidatic Center in Tpx
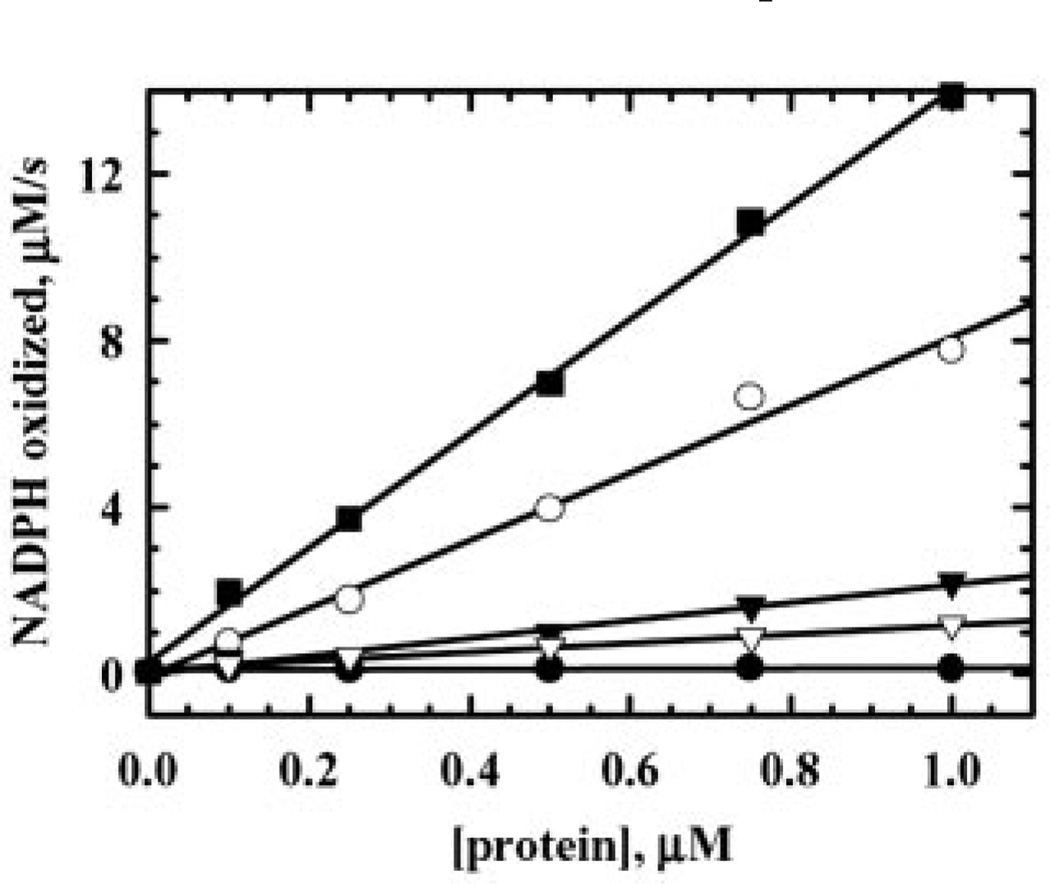
To address which of the three Cys residues in Tpx are involved in the direct reduction of peroxide, Cys to Ser mutants were analyzed under steady-state assay conditions using a stopped-flow spectrophotometer. Unlike in previous studies, where both C61S and C95S were inactive (22, 23), we detected activity with C95S (~20% of wild type activity with 10 µm Trx1) but not with C61S in our assays (Fig. 5), indicating that Cys61 is the essential peroxidatic Cys of the disulfide pair (the activity of C61S is about 0.4% that of wild type Tpx and may be entirely due to a very small amount of wild type contamination in the C61S protein). C95S is also more sensitive to inactivation than wild type Tpx, as illustrated by less robust reaction rates that are more quickly diminished even in the presence of low concentrations of peroxide (data not shown). The loss of Cys82 does not have a large effect on activity (~72% activity compared with wild type Tpx); interestingly, removal of both Cys82 and Cys95 in the double mutant, C82S,C95S, dramatically decreases activity below that of C95S to about 10% of wild type activity (Fig. 5), suggesting a modest, stabilizing effect of Cys82 in the absence of Cys95. When the single Cys Tpx mutants were assayed with varying amounts of Trx1 (0–80 µm) at one concentration of CHP (50 µm), we found that C82S retained the same low Km(app) for Trx1 as wild type Tpx (14.8 versus 12.3 µm), whereas C95S had a much higher Km(app) for Trx1 (50.7 µm). Our labeling and densitometry data verifying less than 5% contamination of the mutants by wild type Tpx, taken together with the value of 20% for the rate of C95S compared with wild type Tpx (at 10 µm Trx1) and the unique kinetic properties of this mutant (higher Km(app) for Trx1 and more rapid inactivation during turnover), clearly indicate that C95S does retain activity, although removal of part of the Tpx redox-active disulfide center adversely affects its normal catalytic activity with Trx1 and exacerbates its inactivation by peroxides.
FIG. 5. Steady-state kinetic analysis of wild type and mutant Tpx proteins.
Tpx reaction mixtures in one syringe containing NADPH (150 µm), Trx1 (10 µm), TrxR (1.5 µm), and varying amounts (0–1 µm) of Tpx (closed squares), C82S (open circles), C95S (closed triangles), C82S,C95S (open triangles), or C61S (closed circles) were mixed with CHP (50 µm) on the stopped-flow spectrophotometer in standard peroxidase buffer (final concentrations after mixing; see “Experimental Procedures”). Each rate is the average of three experiments conducted at 25 °C and was obtained by linear regression analysis of the linear portion of the reaction (0–1 s) prior to inactivation.
Identification of the Sulfenic Acid Form of Cys61
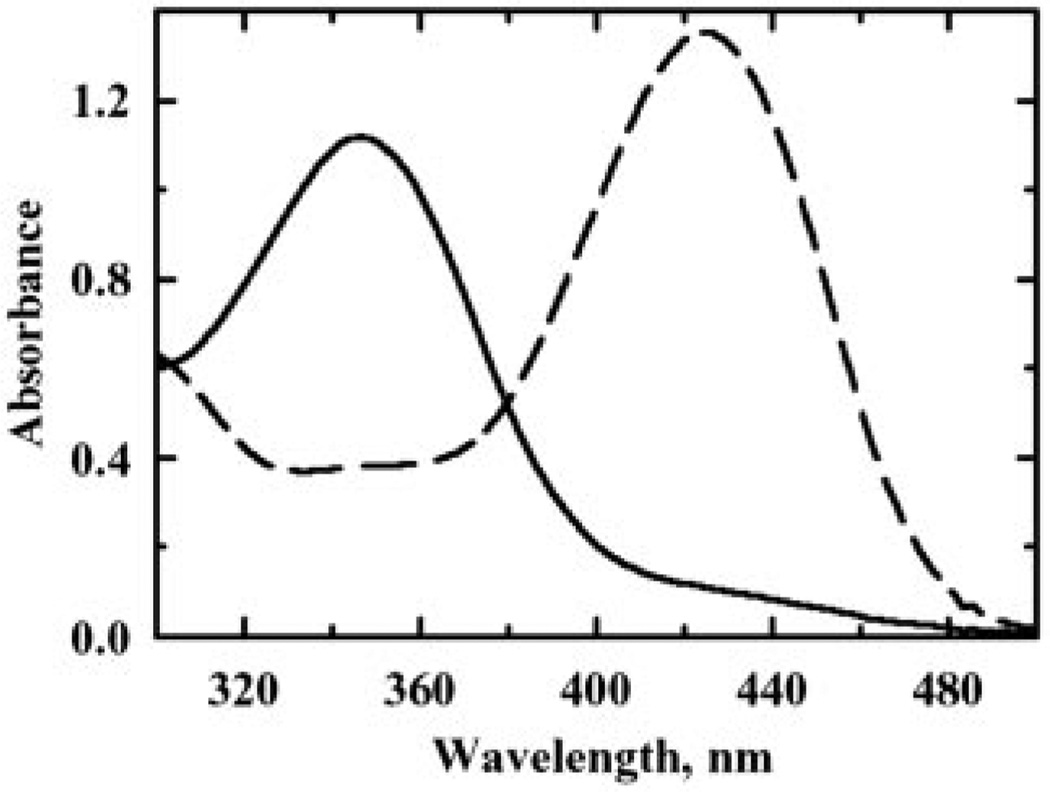
NBD chloride labeling and x-ray crystallography have directly demonstrated the presence of a R-SOH intermediate on the peroxidatic Cys of several Prx homologues (17, 46). During catalysis, the labile Cys-SOH is quickly attacked by the other half-cysteine of 2-Cys Prxs to reform the redox active disulfide; therefore, to stabilize and trap Cys-SOH, the C-terminal Cys of the active site disulfide pair must be removed. Because C95S and C82S, but not C61S, display a reduced thiol titer upon oxidation (Table I) and Cys61 is essential for activity (Fig. 5), SOH formation on Cys61 was hypothesized. Detection of SOH in C95S was hampered by artifactual enzyme dimerization and apparent NBD migration to Cys82, resulting in a low Cys-SOH signal (data not shown). Therefore, NBD chloride labeling to detect the putative Cys-SOH intermediate was conducted using a Tpx double mutant, C82S,C95S, which still maintained slight activity during steady-state assays (Fig. 5). In accordance with SOH formation following peroxide treatment, the thiol titer of C82S,C95S also decreased in a manner similar to that of C95S (Table I). In the resulting ultraviolet-visible wavelength spectral scan, oxidized, NBD-labeled C82S,C95S exhibited an absorbance maximum at 347 nm (Fig. 6), a signal characteristic of NBD adducts of R-SOH (R-S(O)-NBD) (17, 37), which clearly signifies the detection and trapping of approximately stoichiometric amounts of SOH at Cys61, the only Cys in this mutant protein. The absorbance maximum at 420 nm for reduced C82S,C95S (Fig. 6), a signal diagnostic for NBD-modified thiols (R-S-NBD), was much attenuated in the oxidized spectra, confirming the loss of the thiolate species (RS−) upon reaction with peroxide. C61S with or without peroxide pretreatment exhibits only the 420-nm peak after reaction with NBD chloride, confirming the lack of SOH formation on Cys82 or Cys95 (data not shown).
FIG. 6. Spectrophotometric analysis of NBD-labeled Tpx mutants.
Prereduced C82S,C95S treated with 1 eq of cumene hydroperoxide (solid line) or no peroxide (dashed line) was modified with NBD chloride (10×) for 5 min. To remove excess reagent, treated samples were washed with 5 ml of buffer by ultrafiltration, and then labeled proteins were analyzed from 200 to 600 nm.
DISCUSSION
Although Tpx was excluded initially from the Prx family (9) due to its low sequence identity (17% compared with E. coli AhpC), the high z score (>70) for the alignment of Tpx with 2-Cys mammalian Prx homologues in fold and function assignment system (FFAS) analysis (47) confirms homology of Tpx with the Prxs. Not surprisingly then, Tpx shares many features of the Prx catalytic mechanism, including a reliance on the conserved N-terminal Cys for peroxide reduction (15). Of Tpx’s three Cys residues (Cys61, Cys82, and Cys95), Cys61 was shown in our mutagenesis studies to be essential for attack of the peroxide’s -O–O- bond (Fig. 4) and to form a Cys-SOH intermediate upon oxidation (Fig. 6). Our assay data, collected using stopped-flow spectrophotometry, captured initial reaction rates before overoxidation of Cys61, and it is likely that prior inability to detect mutant C95S activity (22, 23) was due to the rapid overoxidation of Cys61 during purification in the absence of DTT and/or during the manual mixing procedures used in assay methods. Tpx does not form the hallmark intersubunit disulfide linkage of typical 2-Cys Prxs, and instead Cys61 links with Cys95 in an unusual intramolecular disulfide bond, a reaction intermediate common to atypical 2-Cys Prxs such as human PrxV (15). Whereas Cys95 does not align with the C-terminal Cys of other 2-Cys Prxs, the participation of Cys95 as a disulfide partner signals that Cys95 is functionally equivalent to the “resolving” Cys of other 2-Cys Prxs (15).
Investigations of Prx interactions with their reducing systems have revealed two different kinetic patterns where 1) the formation of enzyme substrate complexes is an observable phenomenon (25, 26, 48, 49) or 2) reduction of the Prx is a bimolecular process, whereby infinite values for Km and Vmax characterize activity (13, 35). The specific reduction of Tpx by Trx1 occurs with a Km of 25 µm and a catalytic efficiency of ~3 × 106 m−1 s−1 (Table II). This rate is about 10-fold faster than rates of ~105 m−1 s−1 achieved generally by other Prx systems (13) and much faster than the weak peroxidase activity displayed by BCP (~104 m−1 s−1, calculated from apparent maximal velocity (Vmax) and Km values obtained under non-steady-state conditions for linoleic acid hydroperoxide (8)). On the basis of catalytic efficiency, Tpx is the most potent reductant of alkyl hydroperoxides in E. coli when compared with the other two Prx family members, AhpC and BCP. Whereas some Prxs interact with Trx in a nonsaturable manner, it is unknown if this type of activity arises as a result of differential affinity for Trx binding or because the Km for the reducing substrate is higher than the concentrations tested.
Earlier reports of the greater catalytic efficiency of Tpx with t-butyl hydroperoxide over H2O2 (9) were confirmed in our own kinetic studies with CHP and H2O2 as substrates (giving true Km values of 9.1 and 1750 µm, respectively) (Table II). As a result, Tpx does not seem to exhibit the requirements for a minimal peroxide binding site as observed for the Trx-dependent Prx from H. pylori (35). In light of this, the nature of the Tpx peroxidatic binding site is likely to be complex, and due to its high affinity for CHP, hydrophobic interactions may be the predominating forces that influence substrate interaction with the enzyme.
Previously, it was shown that the primary role of AhpC was to keep concentrations of H2O2 in exponentially growing E. coli quite low (10−8 to 10−6 m) (7), and the role of AhpC in organic hydroperoxide detoxification was questioned because bacteria do not synthesize the polyunsaturated fatty acids required for lipid peroxidation. However, the ability of Tpx and other Prxs, including yeast type II thioredoxin peroxidase (50), to preferentially decompose organic hydroperoxides over H2O2 suggests that Tpx’s central role in vivo involves the reduction of complex ROOH, whereas AhpC mainly reduces H2O2. Several lines of evidence also highlight the importance of Prxs in bacterial ROOH resistance, including the isolation of mutants with increased resistance to organic solvents linked directly to a mutation in E. coli AhpC (51). Other bacterial peroxidases, such as Ohr from Xanthomonas campestris, are specifically up-regulated by organic hydroperoxides (52, 53), implying that defense against these peroxides is requisite for bacterial survival. It is also quite possible that during pathogenesis, bacteria take up polyunsaturated fatty acids from the host (54), creating the potential for bacterial lipid hydroperoxide formation. Whereas the exact nature and concentration of organic hydroperoxides that Prxs are exposed to in vivo is unknown, the existence of Prxs with selectivity for these peroxides suggests the possibility of more complex peroxides as Prx substrates.
Inactivation has been observed for many different Prx homologues during activity assays (14, 55–58), and, when investigated, irreversible overoxidation at the active site Cys is responsible for the loss of activity (57), with the terminal species being sulfinic acid in some cases (15). If Tpx inactivation requires a threshold level of peroxide that is substantially higher than in vivo concentrations, then the observed in vitro overoxidation for Tpx is due to the use of very high substrate concentrations in steady-state assays. This seems probable, considering that the endogenous in vivo concentrations of H2O2 are reportedly quite low in the absence of exogenous sources (59). Exposure to peroxide at endogenous concentrations of 2 µm or more is growth-inhibitory (7), and in these cases, peroxidase activity may no longer prevent oxidative damage to cellular components. Whereas the in vivo concentrations of other ROOH in bacteria are not known, it is likely that they would not exceed H2O2 levels. Therefore, subjecting Tpx during assays to exceptionally high peroxide levels may artificially promote overoxidation, a future consideration for all Prx steady-state analyses done without the benefits of stopped-flow reaction techniques.
To explain the existence of multiple Prxs, the differential cellular localization of Tpx and AhpC has been cited. Tpx has been characterized as a periplasmic protein (9, 60), despite the lack of a signal sequence for periplasmic transport,3 whereas AhpC has been localized to the cytoplasm (60). E. coli proteomics studies by Link et al. (60) showed that during the growth phase in minimal media and without induction by H2O2, Tpx (~1.6 µm) is 3.5-fold less abundant than AhpC and 2.5-fold more abundant than BCP. The relatively high expression of all three Prxs indicates a sustained requirement for peroxide detoxification during growth, and in these cases, a periplasmic peroxidase metabolizing peroxides prior to cytosolic entry would be quite beneficial. Although Trx1 is relatively abundant (0.3% of the cellular protein) (61), its cytoplasmic location (62) would restrict access of Tpx to its reductant and decrease the catalytic efficiency of the Tpx system. The designation of Tpx as a periplasmic protein should then be treated with caution until more careful localization studies are completed.
Detailed kinetic analyses of Tpx reaction rates have allowed us to clarify the roles of the three Cys in E. coli Tpx herein and demonstrate the essentiality of Cys61. The shared reliance on Cys-SOH formation and disulfide bond formation points to mechanistic similarities between Tpx and other members of the Prx family. Further kinetic and mechanistic studies on Prx family members may begin to delineate the functional roles of multiple Prxs in the same organism, the presence of which most likely signifies considerable selection pressure from oxidative stress and the need to combat reactive oxygen species.
Acknowledgments
We thank Dr. Daniel Ritz for the generous gift of E. coli Trx2, Dr. Joe O’Flaherty for assistance with 15-HPETE production, Dr. C. M. Reynolds for the cloning and initial purification of wild type E. coli Tpx, and Mark Morris for HPLC assistance and peptide analysis. We are particularly grateful to Mike Samuel at Wake Forest University Medical Center and Dr. Rodney Baker at the University of Mississippi School of Medicine for assistance with mass spectrometry analyses.
Footnotes
Project support was provided by National Institutes of Health Grant R01 GM50389 and by an Established Investigatorship from the American Heart Association (to L. B. P).
The abbreviations used are: AhpC, alkyl hydroperoxide reductase peroxidase component; AhpF, alkyl hydroperoxide reductase flavoprotein oxidoreductase component; Trx, thioredoxin; BCP, bacterioferritincomigratory protein; Tpx, thiol peroxidase; Prx, peroxiredoxin; Cys-SOH, cysteine sulfenic acid; TrxR, thioredoxin reductase; DTT, dithiothreitol; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); CHP, cumene hydroperoxide; NBD chloride, 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole; Cys-SO2H, cysteine sulfinic acid; Cys-SO3H, cysteine sulfonic acid; 15-HPETE, 15-hydroperoxyeicosatetraenoic acid; ESI-MS, electrospray ionization mass spectrometry; HPLC, high pressure liquid chromatography; MES, 2-(N-morpholino)-ethane sulfonic acid; TCEP, Tris[2- carboxyethyl]phosphine; Me2SO, dimethyl sulfoxide.
L. M. S. Baker and L. B. Poole, unpublished observations.
REFERENCES
- 1.Storz G, Tartaglia LA, Farr SB, Ames BN. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 2.Carmel-Harel O, Storz G. Annu. Rev. Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 3.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. 3rd Ed. Oxford: Oxford University Press; 1999. pp. 188–276. [Google Scholar]
- 4.Storz G, Imlay JA. Curr. Opin. Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 5.Lynch AS, Lin ECC. In: E. coli and Salmonella: Cellular and Molecular Biology VI. 2nd Ed. Neidhardt FC, editor. Washington, D. C.: American Society for Microbiology Press; 1996. pp. 1526–1538. [Google Scholar]
- 6.Carlioz A, Touati D. EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa Seaver L, Imlay JA. J. Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong W, Cha MK, Kim IH. J. Biol. Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 9.Cha MK, Kim HK, Kim IH. J. Biol. Chem. 1995;270:28635–28641. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 10.Rioux CR, Kadner RJ. Mol. Gen. Genet. 1989;217:301–308. doi: 10.1007/BF02464897. [DOI] [PubMed] [Google Scholar]
- 11.Poole LB. In: Signal Transduction by Reactive Oxygen and Nitrogen Species: Pathways and Chemical Principles. Torres M, Fukuto JM, Forman HJ, editors. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. pp. 80–101. [Google Scholar]
- 12.Jin D-Y, Jeang K-T. In: Antioxidant and Redox Regulation of Genes. Sen CK, Sies H, Bauerle P, editors. San Diego: Academic Press, Inc; 2000. pp. 381–407. [Google Scholar]
- 13.Hofmann B, Hecht HJ, Flohé L. Biol. Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 14.Ellis HR, Poole LB. Biochemistry. 1997;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- 15.Wood ZA, Schröder E, Harris JR, Poole LB. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 16.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 17.Ellis HR, Poole LB. Biochemistry. 1997;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 18.Poole LB, Reynolds CM, Wood ZA, Karplus PA, Ellis HR, Li Calzi M. Eur. J. Biochem. 2000;267:6126–6133. doi: 10.1046/j.1432-1327.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 19.Wan X-Y, Zhou Y, Yan Z-Y, Wang H-L, Hou Y-D, Jin D-Y. FEBS Lett. 1997;407:32–36. doi: 10.1016/s0014-5793(97)00302-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim HK, Kim SJ, Lee JW, Cha MK, Kim IH. Biochem. Biophys. Res. Commun. 1996;221:641–646. doi: 10.1006/bbrc.1996.0649. [DOI] [PubMed] [Google Scholar]
- 21.Kim SJ, Han YH, Kim IH, Kim HK. IUBMB Life. 1999;48:215–218. doi: 10.1080/713803496. [DOI] [PubMed] [Google Scholar]
- 22.Cha MK, Kim HK, Kim IH. J. Bacteriol. 1996;178:5610–5614. doi: 10.1128/jb.178.19.5610-5614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Wan XY, Wang HL, Yan ZY, Hou YD, Jin DY. Biochem. Biophys. Res. Commun. 1997;233:848–852. doi: 10.1006/bbrc.1997.6564. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel FM, Brent RB, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York: John Wiley & Sons, Inc; 1999. pp. 212–213. [Google Scholar]
- 25.Poole LB, Ellis HR. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 26.Poole LB, Godzik A, Nayeem A, Schmitt JD. Biochemistry. 2000;39:6602–6615. doi: 10.1021/bi000405w. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds CM, Poole LB. Biochemistry. 2000;39:8859–8869. doi: 10.1021/bi000826d. [DOI] [PubMed] [Google Scholar]
- 28.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical Ultracentrifugation in Biochemistry and Polymer Science. Harding SE, Rowe AJ, Horton CJ, editors. Cambridge, UK: The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 29.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole LB. Biochemistry. 1996;35:65–75. doi: 10.1021/bi951888k. [DOI] [PubMed] [Google Scholar]
- 31.Williams CH, Jr, Zanetti G, Arscott LD, McAllister JK. J. Biol. Chem. 1967;242:5226–5231. [PubMed] [Google Scholar]
- 32.Holmgren A, Reichard P. Eur. J. Biochem. 1967;2:187–196. doi: 10.1111/j.1432-1033.1967.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren A. Methods Enzymol. 1985;113:525–540. doi: 10.1016/s0076-6879(85)13071-5. [DOI] [PubMed] [Google Scholar]
- 34.Riddles PW, Blakeley RL, Zerner B. Anal. Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 35.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. J. Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Flaherty JT, Wykle RL, Thomas MJ, McCall CE. Res. Commun. Chem. Pathol. Pharmacol. 1984;43:3–23. [PubMed] [Google Scholar]
- 37.Poole LB, Ellis HR. Methods Enzymol. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- 38.Declercq JP, Evrard C, Clippe A, Stricht DV, Bernard A, Knoops B. J. Mol. Biol. 2001;311:751–759. doi: 10.1006/jmbi.2001.4853. [DOI] [PubMed] [Google Scholar]
- 39.Chae HZ, Uhm TB, Rhee SG. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7022–7026. doi: 10.1073/pnas.91.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nogoceke E, Gommel DU, Kiess M, Kalisz HM, Flohé L. Biol. Chem. 1997;378:827–836. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 41.Claiborne A, Miller H, Parsonage D, Ross RP. FASEB J. 1993;7:1483–1490. doi: 10.1096/fasebj.7.15.8262333. [DOI] [PubMed] [Google Scholar]
- 42.Cha MK, Yun CH, Kim IH. Biochemistry. 2000;39:6944–6950. doi: 10.1021/bi000034j. [DOI] [PubMed] [Google Scholar]
- 43.Prieto-Alamo MJ, Jurado J, Gallardo-Madueno R, Monje-Casas F, Holmgren A, Pueyo C. J. Biol. Chem. 2000;275:13398–13405. doi: 10.1074/jbc.275.18.13398. [DOI] [PubMed] [Google Scholar]
- 44.Ritz D, Patel H, Doan B, Zheng M, Åslund F, Storz G, Beckwith J. J. Biol. Chem. 2000;275:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- 45.Miranda-Vizuete A, Damdimopoulos AE, Gustafsson J, Spyrou G. J. Biol. Chem. 1997;272:30841–30847. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- 46.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 47.Rychlewski L, Jaroszewski L, Li W, Godzik A. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niimura Y, Poole LB, Massey V. J. Biol. Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 49.Poole LB, Higuchi M, Shimada M, Calzi ML, Kamio Y. Free Radic. Biol. Med. 2000;28:108–120. doi: 10.1016/s0891-5849(99)00218-x. [DOI] [PubMed] [Google Scholar]
- 50.Jeong JS, Kwon SJ, Kang SW, Rhee SG, Kim K. Biochemistry. 1999;38:776–783. doi: 10.1021/bi9817818. [DOI] [PubMed] [Google Scholar]
- 51.Ferranté AA, Augliera J, Lewis K, Klibanov AM. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7617–7621. doi: 10.1073/pnas.92.17.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. J. Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sukchawalit R, Loprasert S, Atichartpongkul S, Mongkolsuk S. J. Bacteriol. 2001;183:4405–4412. doi: 10.1128/JB.183.15.4405-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuenen JG, Rittenberg SC. J. Bacteriol. 1975;121:1145–1157. doi: 10.1128/jb.121.3.1145-1157.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chae HZ, Chung SJ, Rhee SG. J. Biol. Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 56.Koo KH, Lee S, Jeong SY, Kim ET, Kim HJ, Kim K, Song K, Chae HZ. Arch. Biochem. Biophys. 2002;397:312–318. doi: 10.1006/abbi.2001.2700. [DOI] [PubMed] [Google Scholar]
- 57.Yang K-S, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. J. Biol. Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 58.Flohé L, Budde H, Bruns K, Castro H, Clos J, Hofmann B, Kansal-Kalavar S, Krumme D, Menge U, Plank-Schumacher K, Sztajer H, Wissing J, Wylegalla C, Hecht HJ. Arch. Biochem. Biophys. 2002;397:324–335. doi: 10.1006/abbi.2001.2688. [DOI] [PubMed] [Google Scholar]
- 59.Costa Seaver L, Imlay JA. J. Bacteriol. 2001;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Link AJ, Robison K, Church GM. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- 61.Potamitou A, Holmgren A, Vlamis-Gardikas A. J. Biol. Chem. 2002;277:18561–18567. doi: 10.1074/jbc.M201225200. [DOI] [PubMed] [Google Scholar]
- 62.Lunn CA, Pigiet V. In: Thioredoxin and Glutaredoxin Systems: Structure and Function. Holmgren A, Bränden C, Jörnball H, Sjoberg B, editors. New York: Raven Press; 1986. pp. 165–177. [Google Scholar]