Abstract
Identifying factors responsible for variation in drug response is essential for the effective use of targeted therapeutics. We profiled signaling pathway activity in a collection of breast cancer cell lines before and after stimulation with physiologically relevant ligands, which revealed the variability in network activity among cells of known genotype and molecular subtype. Despite the receptor-based classification of breast cancer subtypes, we found that the abundance and activity of signaling proteins in unstimulated cells (basal profile), as well as the activity of proteins in stimulated cells (signaling profile), varied within each subtype. Using a partial least squares regression approach, we constructed models that significantly predicted sensitivity to 23 targeted therapeutics. This analysis identified key proteins that could serve as biomarkers of drug sensitivity. For example, one model showed that the response to the growth factor receptor ligand heregulin effectively predicted the sensitivity of cells to drugs targeting the cell survival pathway mediated by PI3K (phosphoinositide 3-kinase) and Akt; whereas the abundance of Akt or the mutational status of the enzymes in the pathway did not. Thus, basal and signaling protein profiles may yield new biomarkers and enable the identification of appropriate therapies in cancers characterized by similar functional dysregulation of signaling networks.
INTRODUCTION
Large-scale sequencing of human tumors has identified an increasing number of genes encoding signaling proteins that are mutated, overexpressed, or deleted in cancer; examples include the genes encoding the kinase Akt, the lipid phosphatase PTEN, the epidermal growth factor (EGF) receptors ErbB2 and ErbB1, mitogen-activated protein kinases (MAPKs), and the proto-oncoprotein Raf (1) (see table S1 for a list of gene names and abbreviations used here). Many drugs targeting these proteins are in clinical use or development, but most drugs work in only a subset of patients (2-7). In a few cases, single genetic factors are highly predictive of drug response in cell lines and human tumors: Bcr-Abl translocation predicts sensitivity to imatinib in leukemia (8) and the BRAFV600E mutation predicts at least initial sensitivity to vemurafenib in melanoma (4, 5). However, the situation is usually more complex.
Early stage breast cancer is generally treated surgically, and adjuvant drugs are chosen on the basis of the morphology of the cancer and its molecular subtype, which is defined by the abundance of three receptors (9). The HER2amp subtype is defined by amplification of the receptor tyrosine kinase (RTK) ErbB2 (also known as Her2) and is typically scored using immunohistochemistry or by assaying for gene amplification. Overexpression of the estrogen receptor (ER) or progesterone receptor (PR) defines the HR+ (hormone receptor positive) subtype. In triple negative breast cancers (TNBCs), the abundance of all three receptors is low. HER2amp status serves as a biomarker for therapy with antibodies that target ErbB2, such as trastuzumab or pertuzumab (2, 3, 10-13), and HR+ status is a biomarker for therapy with hormone receptor antagonists, such as tamoxifen (14, 15). TNBCs are usually treated with cytotoxic chemotherapy (14, 16), sometimes in combination with ErbB1 inhibitors (17) and function-blocking antibodies targeting ErbB family members (18). However, breast cancer subtypes are heterogeneous (19-21), classical molecular subtypes (as described above) and those defined by whole-genome expression profiling are not identical (22), and even the best available biomarker, HER2amp status, correctly predicts response to trastuzumab in only a subset of patients (2, 3, 10, 11). The need for better biomarkers is particularly urgent for TNBCs, which appear to be genetically more heterogeneous than other breast cancer subtypes (23) and patients with these tumors have poor prognosis (24).
Projects like the Cancer Cell Line Encyclopedia aim to identify genomic features, such as gene amplification, mutation, deletion, or epigenetic modifications, that correlate with and are ultimately predictive of drug response (20, 21, 25-27). However, biochemical data on drug targets, such as abundance or phosphorylation status, are potentially more predictive of drug response than genomic features (28-30). Despite this and the availability of some systematic steady-state protein data (21, 22), few studies have tried to relate the biochemistry of signal transduction to drug response on a large scale (31).
Here, we investigated whether measurements of the basal and stimulated states of signal transduction networks are predictive of drug response as measured by the GI50 value, the concentration of drug that reduces cell number by 50% relative to a no-drug control when assayed a fixed time after drug exposure (3 days in the current work). We measured the abundance and basal phosphorylation state of nuclear and cell surface receptors and of downstream signaling kinases in a standardized NCI-ICBP43 cell line collection (21). The choice of which receptors to measure was a practical one: We focused on RTKs, because they are clearly implicated in breast cancer biology, and assayed all receptors for which we could verify the specificity and linearity of plate-based immune assays. Biological ligands present in the microenvironment alter drug sensitivity (32, 33) and thus some features of signal transduction may not be apparent by steady-state profiling. Therefore, we also measured the activities of downstream signaling kinases before and after exposing cells to a diverse set of growth factors and cytokines. We then evaluated how the resulting set of ~3 × 105 receptor and cell response measurements segregated with molecular subtype and whether they could predict the sensitivity of cells to a range of targeted anti-cancer drugs (20). We found that predictors could be constructed for more than half of the drugs we analyzed and that data on molecular subtype, signaling biochemistry, and genetic status could be combined to construct hybrid response biomarkers that are mechanistically plausible and may translate into the clinic.
RESULTS
Mapping protein profiles onto subtype
We profiled signaling in two ways in cell lines of the ICBP43 panel that could reliably be grown as adherent cells (table S2). The “basal profiles” set was defined as the data for the abundance and basal phosphorylation of 22 receptors and 3 downstream kinases (ERK, Akt and Src) (table S3) and the abundance of the ER and PR, and where possible we estimated steady-state levels in molecules per cell by ELISA calibrated with recombinant protein. All basal phosphorylation and protein abundance data were obtained by ELISA (see Materials and Methods). For the RTK phosphorylation data, we measured the amount of total phosphotyrosine rather than of individual phosphorylation sites, with the exception of IGF1R, for which we measured phosphorylation of Tyr1131. Note that such an approach ignores the fact that different phosphorylation events on the same protein often have different biological effects. To generate the “signaling profiles” set, we exposed the cells individually to 22 growth factors or cytokines (table S4) for 10, 30, or 90 min and monitored the response by immunofluorescence microscopy of key signaling kinases and transcription factors (table S5). Activation of NF-κB (nuclear factor kappaB) was based on nuclear translocation of its subunit p65; activation of all other signaling proteins was based on phosphorylation. Not every ligand was analyzed with every assay (fig. S1),
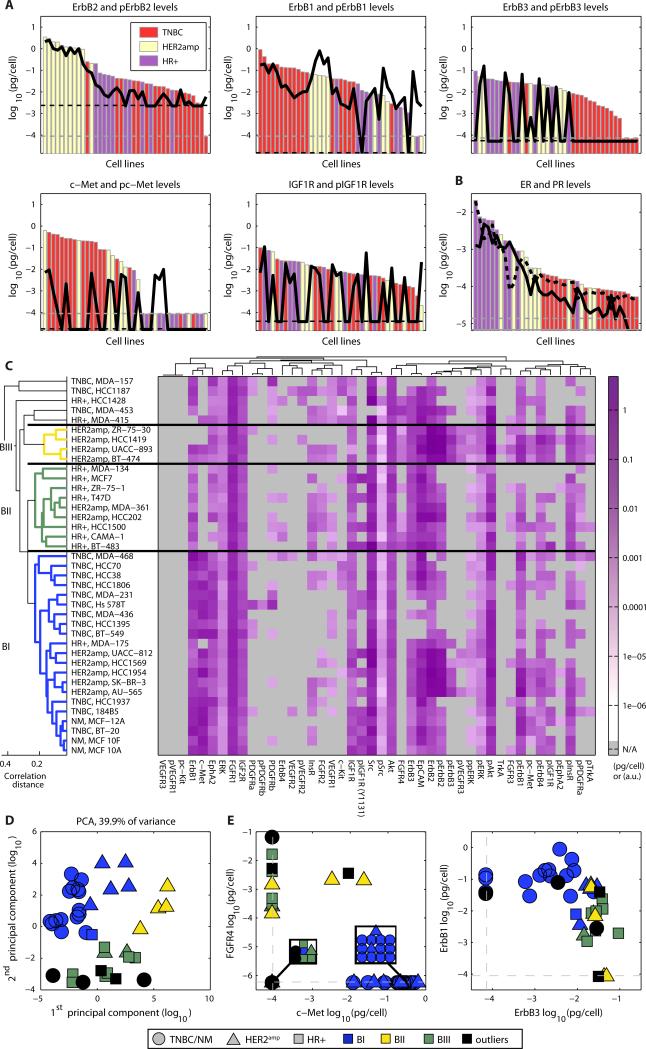
The basal profiling revealed that some RTKs, such as ErbB2, ErbB1, ErbB3, c-Met, and IGFR1 (insulin-like growth factor 1 receptor), were present in many of the cell lines (Fig. 1A); whereas others, such as PDGFR (platelet-derived growth factor receptor) and VEGFR (vascular endothelial growth factor receptor), were detectable in only a few lines (data S1). Absolute abundance varied from ~107 molecules per cell for ErbB2 to ~100 molecules per cell for c-Kit or IGF1R (data S1). The abundance of ErbB2 varied 10-fold among HER2amp cells (and ~103-fold across the collection), but the HR+ line with the greatest amount of ErbB2 had only ~3-fold less receptor than the HER2amp line with the lowest amount (Fig. 1A). The same was true for amounts of ER and PR, which varied 100-fold with intermingling of subtypes (Fig. 1B). More generally, we found that all of the broadly distributed receptors in our dataset exhibited high variability in abundance across all molecular subtypes. Thus, these results show that the dichotomous classification of breast cancers by receptor presence or absence belies the fact that actual receptor abundance varies in a graded fashion across subtypes.
Fig. 1. Abundance and phosphorylation status of receptor tyrosine kinases reveals heterogeneity in receptors within clinical subtypes in 39 breast cancer cell lines.
(A) Bars indicate the abundance of the receptor tyrosine kinases, colored by clinical subtypes (red, TNBC or nonmalignant; yellow, HER2amp; purple, HR+). The black line is the amount of phosphorylated receptor. Dashed lines show the detection threshold for total protein abundance (grey) and phosphoprotein abundance (black). (B) Abundance of the estrogen and progesterone receptors (ER, solid; PR dashed; sum of ER and PR as bars). Horizontal gray lines show the detection thresholds (ER, solid; PR dashed). (C) Clustering of cell lines according to their basal profile defines three main clusters (BI, blue; BII, green; BIII, yellow; outliers, black). Color intensity in the heat map indicates abundance of the indicated protein or phosphoprotein in pg/cell for protein abundance and arbitrary units for phosphorylation measurements (measurements below detection threshold are in gray). Black horizontal lines show the cluster separations. (D) Projection of cell lines on the first two principal axes of a PCA (fig. S3) of the basal measurements. (E) Projection of cell lines on the indicated key variables identified by PCA. Dashed lines show the detection thresholds for each measurement.
Among the receptors profiled, ErbB2 was unique in that basal phosphorylation was highly correlated with protein abundance (Fig. 1A; Spearman's R=0.86; p=3×10−12), whereas the abundance and basal phosphorylation status of other receptors exhibited little or no correlation. A likely explanation is that ErbB2 can self-associate when present in high abundance and autoactivate in the absence of ligand (34), whereas most other RTKs are only activated by ligand binding. The amount of phosphorylated ErbB2 (pErbB2) in HER2amp cells also correlated closely with amounts of pErbB1, pErbB3, and pErbB4 (fig. S2A) (34, 35) and with the phosphorylated forms of the insulin and insulin-like growth factor receptors (pInsR and pIGFR1; fig. S2A). Although pIGF1R can crossactivate ErbB2 in trastuzumab-resistant HER2amp cells (36), our data showed that high phosphorylation of IGF1R occurred in 8/11 HER2amp cell lines (Fig. 1A, fig. S2B). IGF1R (but not ErbB2) were also highly phosphorylated in a subset of TNBC cells (fig. S2B). High phosphorylation of IGF1R and InsR is linked to poor patient survival across all breast cancer subtypes (37) and IGF1R has been targeted therapeutically (38, 39).
Hierarchical clustering of basal profiles revealed three clusters with several outliers: cluster BI included most of the TNBC lines (12/15) and BII most of the HR+ lines (7/10); HER2amp lines were present in all three clusters (Fig. 1C). Non-malignant cell lines (NM) did not have increased amounts of ErbB2, ER, or PR and clustered closely with a subset of the TNBC lines in BI. Principle component analysis (PCA) revealed a similar distribution as found by hierarchical clustering with the TNBC lines tightly grouped in BI, HR+ lines in BII, and HER2amp lines widely distributed (Fig. 1D). Variables identified by PCA analysis (fig. S3) explained the differences between the clusters. In BI, ErbB1 and c-Met were highly abundant; whereas FGFR4 (fibroblast growth factor receptor 4) and ErbB3 were of low abundance in these cell lines; cell lines in BII exhibited the converse pattern (Fig. 1E).
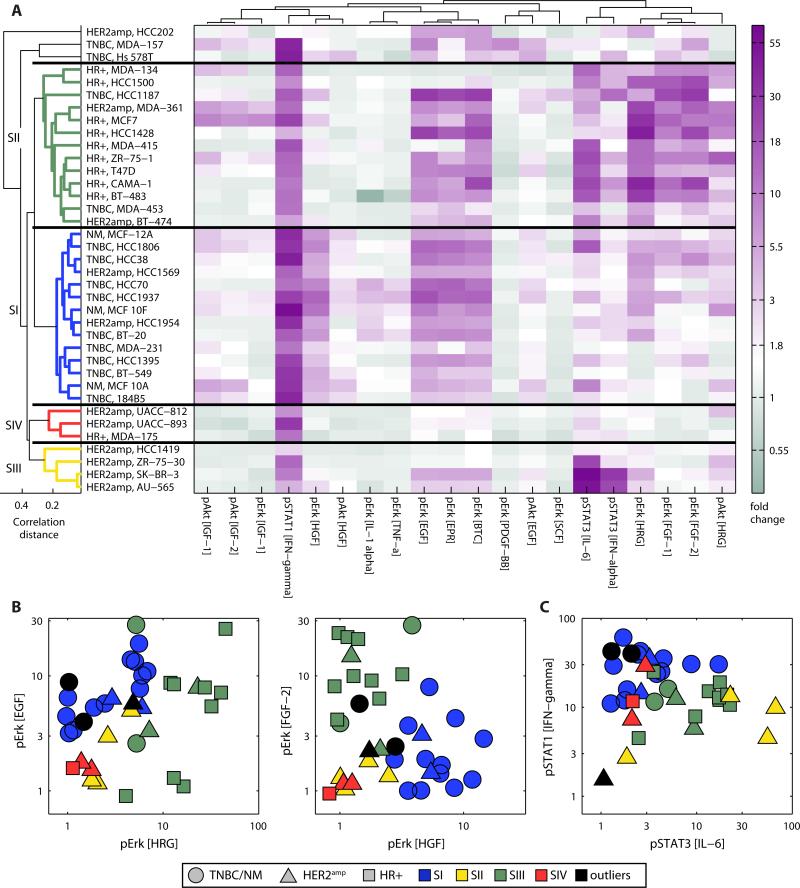
Analysis of the responses to growth factors or cytokines in 37 of the cell lines, which defined the signaling profiles, yielded four clusters, with TNBC cells falling together in SI and HR+ lines in SII (Fig. 2A, only the 20 most variable measurements are shown), consistent with their placement in BI and BII, respectively (Fig. 1C). When analyzing the degree to which phosphorylation of the extracellular signal-regulated kinase 1 and 2 (pERK) could be induced, we found that TNBC cells were generally more responsive to EGF and the c-Met ligand HGF, whereas HR+ cells responded more robustly to FGF-2 and HRG, a ligand that binds to ErbB2-ErbB3 heterodimers (Fig. 2B). To validate the signaling profiles collected by microscopy, we measured ligand responses using ELISA assays for pAkt and pERK, the linearity of which can be tested using recombinant protein standards. The profiles collected using the imaging and ELISA methods were highly correlated (fig. S4).
Fig. 2. Signaling response to extracellular ligands shows commonalities within breast cancer cell subtypes and identifies outliers.
(A) Heat map of variables representing the 20 most variable responses to growth factors or cytokines in 37 breast cancer cell lines identified by PCA. Color intensities indicate the fold change from vehicle-treated cells. Cell lines are clustered (SI, blue; SII, green; SIII, yellow; SIV, red; outliers, black) according to all ligand responses measured by HTM. (B) Projection of cell lines on the pERK response to the indicated receptor tyrosine kinase ligands at 100 ng/ml. (C) Projection of cell lines on the pSTAT3 response to IL-6 versus the pSTAT1 response to IFN-γ at 100ng/ml. Responses and ligands in (B and C) were selected from the most variable measurements identified by PCA (fig. S3).
Unsupervised clustering showed that HR+ and TNBC lines in the ICBP43 collection were distinguishable by the abundance and responsiveness of receptors that are not conventionally part of the subtype definition. These receptors include c-Met (for TNBC) and FGF receptors (for HR+), both of which are activated by components of the tumor microenvironment that are implicated in drug resistance (32, 33, 40) and are being intensively targeted by therapeutic antibodies and small molecule drugs (41, 42). From a molecular and diagnostic perspective the HER2amp subtype is generally regarded as the most distinctive, but unsupervised clustering divided HER2amp cells into multiple groups (Fig. 1C) based on 10-100 fold differences in the abundance of c-Met, FGFR4, and EGFR (Fig. 1E). Although many HER2amp cell lines responded weakly to most growth factors, perhaps because high pErbB2 causes constitutive downstream signaling, a subset of HER2amp cells was highly responsive to interleukin-6 (IL-6) (Fig. 2A, C). High amounts of IL-6 have been observed in some primary breast cancers and are associated with poor clinical outcomes (43-45). These results suggested that HER2amp cell lines exhibit substantial heterogeneity at the signaling and receptor level, which may possibly be a consequence of factors known or suspected to be involved in tumor progression and drug resistance (40, 46).
Predicting drug response from basal and signaling data
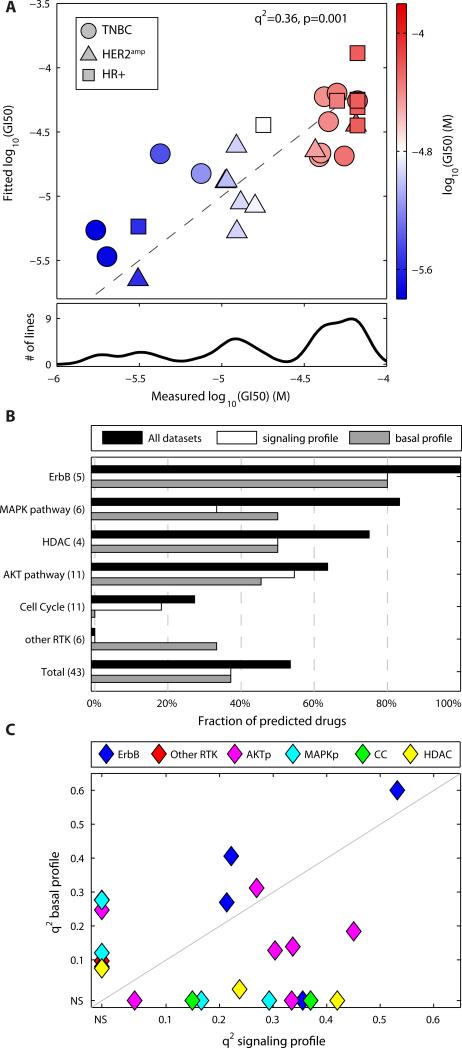
Previous attempts to correlate genomic features with drug response often start with a binary division into resistant and sensitive classes, with some exceptions (26). Although it is broadly true that subtype enriches for sensitivity to some drugs (HER2amp cells to ErbB2 inhibitors), in most cases GI50 varies continuously across the cell line panel (20). There is, for example, no principled way to divide the trimodal distribution of GI50 values for the ErbB1 inhibitor erlotinib into resistant and sensitive classes (Fig. 3A). Therefore, rather than create a classifier, we attempted to predict actual GI50 values directly using partial least squares regression (PLSR). The quality of each prediction was assessed by a q2 value derived from a leave-one-out cross-validation and the significance of prediction was assessed by the p-value of the correlation between predictions and measurements (see text S1, fig. S5). In the case of the ErbB1 inhibitor erlotinib (Fig. 3A), subtype was a poor response predictor (20), but we were able to build a PLSR model (q2=0.36 and a p~0.001) to correctly identify sensitive TNBC and HR+ lines (in blue in Fig. 3A) from a large set of insensitive lines. Although Erlotinib is not a standard treatment for breast cancer, it is part of 32 ongoing trials (47).
Fig. 3. Basal profiles and signaling profiles predict responses to targeted inhibitors.
(A) Projection of the fit of the PLSR model based on the signaling profile data predicting GI50 values for erlotinib responses against the measured response for each cell line (red, resistant; blue, sensitive). Lower plot shows the trimodal distribution of the measured GI50 values across the cell lines. (B) Fraction of drugs for each class (see Table 1) predicted by the totality of the data (black), the signaling profile based on the maximal response to the 15 growth factors (white), and the basal profile (gray). (C) Distribution of the q2 for drugs significantly predicted by the basal or signaling profile grouped by primary drug targets (AKTp, AKT pathway; MAPKp, MAPK pathway; CC, cell cycle). Non-significant predictions (NS) are set to zero. Colors represent different classes of drugs.
We built PLSR models to predict previously reported GI50 values for 43 targeted drugs (20) (data S2) using different partitions of our datasets that corresponded to the basal or signaling profiles (table S6). Overall, 23 models were statistically significant at a false discovery rate of 0.15 (Table 1; see data S3 for model coefficients). A majority of the drugs targeting ErbB, the MAPK pathway or phosphoinositide 3-kinase to Akt (PI3K/Akt) pathway, and histone deacetylases (HDACs) were predicted with statistical significance (Fig. 3B). The quality of predictions of the signaling profiles collected by ELISA (used for validation) and microscopy have a Pearson's correlation of R=0.52, p-value=3.0×10−4 (fig. S6, data S4). In general, models based on signaling profiles yielded higher q2 values than models based on basal profiles for drugs with intracellular targets, like the PI3K/Akt pathway (Fig. 3C).
Table 1. List of drugs for which GI50 values are significantly predicted using PLSR models.
Quality of the prediction (q2) for the drugs significantly predicted by any of the datasets used (see table S6). The Benjamini–Hochberg procedure with FDR=0.15 was used for significance and the p-value of the dataset with the highest q2 is reported.
| Drug name | Primary target | Drug class | highest q2 | corresponding p-value |
|---|---|---|---|---|
| Lapatinib | ERBB2 | ErbB | 0.600 | 6.83E-07 |
| Gefitinib | EGFR | ErbB | 0.464 | 5.98E-05 |
| TCS 2312 | CHEK1 | Cell Cycle | 0.455 | 2.15E-04 |
| GSK1059615 | PIK3CA | AKT pathway | 0.450 | 8.67E-05 |
| Oxamflatin | HDAC | HDAC | 0.420 | 1.83E-04 |
| TCS JNK 5a | MAPK9-10 | MAPK pathway | 0.419 | 1.80E-03 |
| Bosutinib | SRC | MAPK pathway | 0.406 | 9.49E-04 |
| Afatinib | ERBB2 | ErbB | 0.406 | 1.92E-03 |
| Rapamycin | MTOR | AKT pathway | 0.391 | 5.80E-04 |
| GSK1070916 | AURKB-C | Cell Cycle | 0.370 | 8.12E-05 |
| Erlotinib | EGFR | ErbB | 0.355 | 6.15E-04 |
| GSK2119563 | PIK3CA | AKT pathway | 0.337 | 8.93E-04 |
| A6730 SIGMA | AKT1-2 | AKT pathway | 0.312 | 1.75E-03 |
| GSK2126458 | pan-PIK3 | AKT pathway | 0.303 | 9.49E-04 |
| GSK1120212 | MAP2K1-2 | MAPK pathway | 0.293 | 1.07E-03 |
| AG1478 | EGFR | ErbB | 0.270 | 3.22E-03 |
| AZD6244 | MAP2K1-2 | MAPK pathway | 0.264 | 1.18E-02 |
| Temsirolimus | MTOR | AKT pathway | 0.247 | 8.85E-03 |
| PD 98059 | MAP2K1-2 | MAPK pathway | 0.241 | 8.73E-03 |
| LBH589 | HDAC | HDAC | 0.237 | 1.49E-02 |
| GSK461364 | PLK1 | Cell Cycle | 0.202 | 1.29E-02 |
| TGX-221 | PIK3CB | AKT pathway | 0.152 | 2.94E-03 |
| Vorinostat | HDAC | HDAC | 0.075 | 3.31E-03 |
Predicting sensitivity to PI3K/Akt inhibitors from the response to heregulin
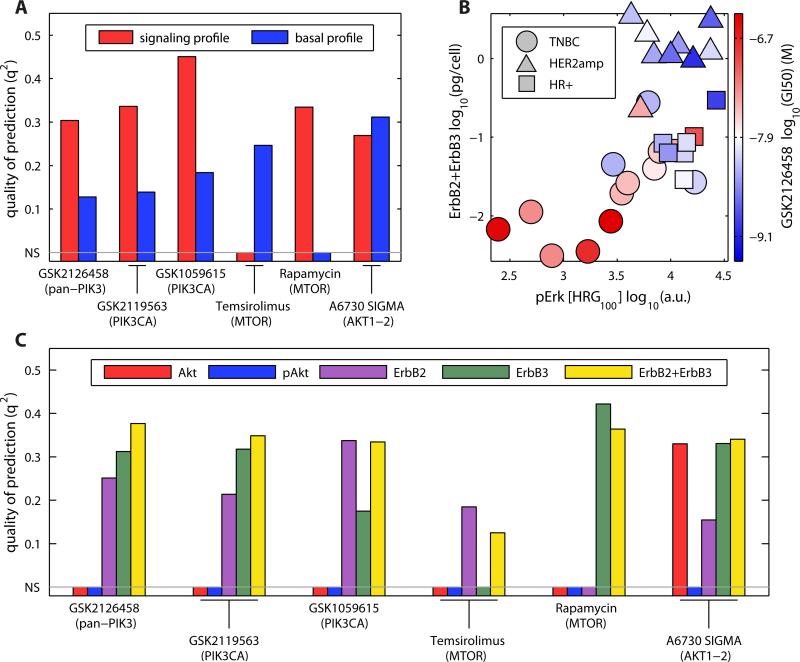
Drugs targeting proteins in the PI3K/Akt pathway are important investigational agents for breast cancer with 24 agents in ~150 trials (47). Furthermore, PIK3CA (encoding the PI3K p110α subunit) and PTEN (encoding a phosphatidylinositol phosphatase) are frequently mutated in breast cancer (1, 22). We found that mutational status was not a significant predictor of drug response for ICBP43 cell lines (table S7) and no clear correlation has been reported in the literature (48). We constructed statistically significant PLSR models for six drugs that target various enzymes in the PI3K/Akt pathway—three targeting PI3K, two targeting mechanistic target of rapamycin (mTOR), and one targeting Akt using either the signaling profile dataset or the basal profile dataset (Fig. 4A). Signaling profile data produced statistically significant PLSR models for five of the six drugs targeting the PI3K/Akt pathway. Although the sensitivity to some drugs was also predicted by basal profiles, models that used signaling data yielded substantially higher q2 values for four out of six drugs. Inspection of the models showed that the abundance of pERK in cells exposed to the ligand heregulin (HRG) had the highest coefficients, suggesting that responsiveness to HRG, but not other ligands that induce pAkt, is linked to sensitivity to PI3K/Akt inhibitors (fig. S7A). HRG responses were also the key variables in models built on the ligand responses collected by ELISA (fig. S7B).
Fig. 4. Response to heregulin and ErbB3 abundance are key predictors for response to drugs targeting the PI3K/Akt pathway.
(A) Quality of the predictions for the drugs targeting the PI3K/Akt pathway using their basal and signaling (based on maximal response) profiles. Non-significant predictions (NS) are set to zero. (B) Projection of cell lines according to the pERK response to heregulin and cumulative amounts of ErbB2 and ErbB3 overlaid with the GI50 value to GSK2126458 (red, resistant; blue, sensitive). (C) Quality of the prediction for drugs targeting the PI3K/Akt pathway using a linear model with a single variable or the variables ErbB3 and ErbB2.
Ligand response can be measured in cultured cells but not easily in tumors. We, therefore, looked for basal measurements that might serve as a surrogate for the HRG response. HRG binds to heterodimers of ErbB2 and ErbB3, and we observed that the sum of their abundances could substitute for the amount pERK in HRG-treated cells (Fig. 4B; illustrated for GSK2126458). Using ErbB2 and ErbB3 amounts we predicted GI50 values for six drugs with similar or better accuracy than was achieved with the original PLSR models (Fig. 4C). Importantly, we did not arrive at the [ErbB2 + ErbB3] predictor by systematically searching through the dataset for measures correlated with drug response; the danger of such a search is that it potentially uncovers random correlations between a large number of independent variables (biochemical measurements) and a dependent variable (GI50 value), particularly when the number of cell lines is small. Instead, we used PLSR to identify the most predictive model in a principled way and then generated a related, but easier to measure, predictor by combining information on top coefficients of the models and the prior knowledge that HRG functions through the ErbB2-ErbB3 complex. Although one might have expected total or phosphorylated amounts of Akt to serve as an effective surrogate, we found that neither measure of Akt was an effective predictor of the response (Fig. 4C), although cells carrying either mutations in the region of PIK3CA encoding the helical domain or in PTEN did have increased pAkt (p=0.029) compared with cell lines lacking either of these mutations (data S1), consistent with previous reports (28). The [ErbB2 + ErbB3] predictor is potentially assayable in clinical samples and appears to distinguish sensitive from insensitive TNBC cells, an area in which the clinical need is greatest.
Improving drug prediction by stratification of cancer cells
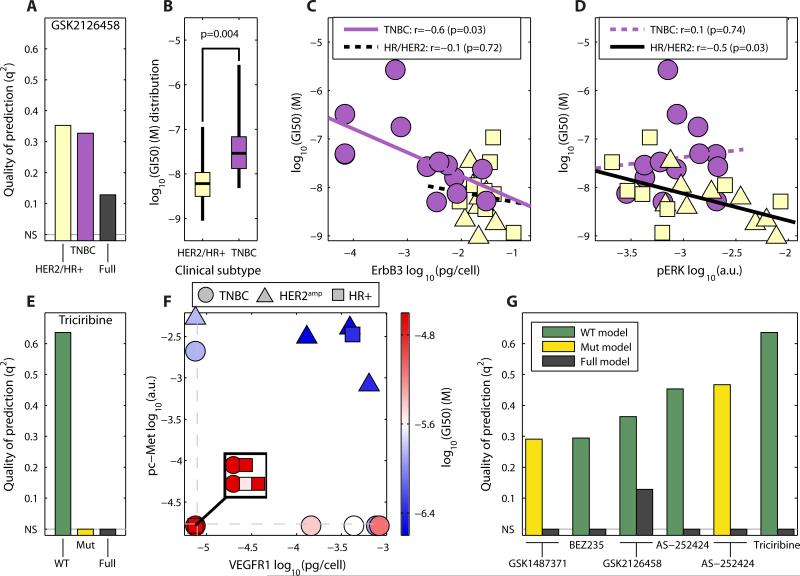
A second way in which to combine experimental data and prior knowledge is to build PLSR models for different subsets of cell lines based on their molecular subtype or mutational status. We applied this to the Pan-PI3K inhibitor GSK2126458, which is only poorly predicted by basal profiles when all cell lines are considered (q2~ 0.1; Fig. 5A). By dividing the cell lines according to their subtype, TNBC versus HR+ and HER2amp (HR+/HER2amp), we obtained a good prediction for both groups (q2~ 0.35; Fig. 5A). TNBC cell lines are generally resistant, but a few are as sensitive as HR+/HER2amp cells and the model captured this (Fig. 5B). Subtype-specific models predicted GI50 values for TNBC and HR+/HER2amp cells using different biochemical features: ErbB3 abundance alone could discriminate sensitive and insensitive TNBC cells (confirming one component of the [ErbB2 + ErbB3] predictor); whereas basal pERK was the key variable for HR+/HER2amp cells (Fig. 5C, D, data S5). The abundance of ErbB3 had no predictive value for the HR+/HER2amp cells because the amount of ErbB3 varied little among the cell lines of these subtypes (Fig. 1A).
Fig. 5. Inclusion of clinical and genomic information can improve predictions of drug response.
(A) Quality of the prediction for models stratified according to subtype (TNBC versus HR+/HER2amp) for the pan-PI3K inhibitor GSK2126458. Predictions by subset are shown according to a model built on just the indicated subset of cell lines (yellow and purple bars) or all of them (black bar). (B) Distribution of GI50 values for GSK2126458 by subtype. (C, D), Correlation of ErbB3 (C) and pERK (D) with GSK2126458 GI50 values for each of the three cancer subtypes. A dashed line shows a non-significant correlation. (E) Quality of the prediction of models stratified according to mutational status of PIK3CA and PTEN for triciribine. Full, all cell lines; WT cell lines wild-type for both PIK3CA and PTEN; Mut, cell lines with mutations in either or both of PIK3CA and PTEN. Non-significant predictions (NS) are set to zero. (F) Projection of the abundance of pc-MET and VEGFR1 overlaid with the triciribine GI50 values only for cell lines wild-type for PIK3CA and PTEN (red, resistant; blue, sensitive). Dashed lines show the detection threshold for each measurement. (G) Quality of the prediction of models stratified according to mutational status of PIK3CA and PTEN (WT versus Mut) for drugs targeting the PI3K/Akt pathway. Only those drugs for which stratification improves prediction are shown.
Whereas relatively few models were improved by stratifying by subtype, stratification by mutational status had a larger impact. In the case of triciribine, an inhibitor of Akt that also targets other kinases, we could not construct a significant PLSR model using biochemical data alone, but excluding cells carrying PIK3CA or PTEN mutations resulted in a highly predictive model (Fig. 5E, data S5). In the model for wild-type lines, cells sensitive to triciribine had high amounts of phosphorylated c-Met and high VEGFR1 and were distributed across all three clinical subtypes (Fig. 5F, blue). When we built models stratified by mutational status for the eleven drugs targeting the PI3K/Akt pathway in our dataset, prediction was improved for either wild-type or PIK3CA and PTEN mutant cell lines in six cases (Fig. 5G), resulting in significant predictions using either a general or stratified model for ten out of the eleven drugs. Because of varying sample sizes (not all drugs were tested on all cell lines), it is difficult to precisely quantify the degree of improvement, but our findings suggest that molecular determinants of drug response differ in wild-type and PIK3CA/PTEN mutant cells.
Identifying response outliers with modeling
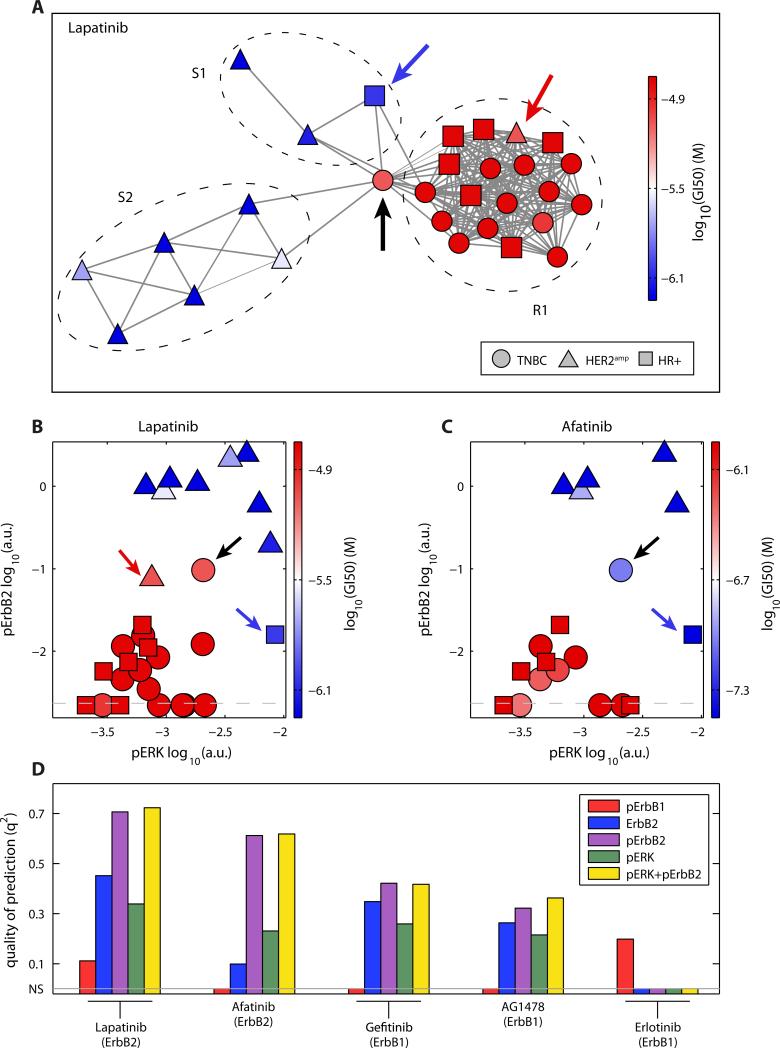
Drugs targeting ErbB are a special case in breast cancer because they are important clinically and well predicted by HER2amp status. However, the correlation is not perfect, which creates two challenges: to predict HER2amp lines that are resistant to ErbB-targeted drugs and to identify TNBC and HR+ lines that are sensitive. We built PLSR models for ErbB-targeted drugs using basal profiles and visualized cell lines on the landscape of model variables (Fig. 6A). This landscape was built by drawing edges between pairs of cell lines that lie within a threshold distance in the space of model variables. In the case of lapatinib, a standard of care drug for combination therapy (49) or in patients that relapse following trastuzumab-based therapy (50), we identified one resistant cluster (R1) and two sensitive clusters (S1 and S2) (Fig. 6A). The sole sensitive HR+ line fell into S1 (MDA-MB-175, blue arrow) and the sole resistant HER2amp line fell in R1 (MDA-MB-361, red arrow). The variables that differentiate R1 from S1 and S2 were pERK, which in MDA-MB-175 was significantly higher than in other HR+ lines (p=1.4×10−4), and pErbB2, which in MDA-MB-361 was lower than in other Her2amp lines (p=4.0×10−4). The abundance of pc-Met and pErbB3 distinguished S1 from S2 (fig. S8). The abundance of pERK and pErbB2 are markers of sensitivity to lapatinib (Fig. 6B) and were also the variables that identified the single TNBC line (MDA-MB-453) that is sensitive to afatinib, another approved ErbB2 inhibitor (Fig. 6C, black arrow). MDA-MB-453 lay at the intersection between the R1 cluster and the S1 and S2 clusters (black arrow, Fig. 6A) and it had a significantly higher pErbB2 abundance than the other TNBC lines (p=3.6×10−6). Thus, we found that pathway activity (represented by pErbB2 and pERK abundance) yielded higher q2 for models of ErbB-targeted drugs than did HER2amp status (Fig. 6D). These results showed that biochemical profiles can identify drug-sensitive outliers among HR+ and TNBC lines.
Fig. 6. The amounts of pErbB2 and pERK are key predictors of response to most drugs targeting ErbB1 or ErbB2.
(A) Network representation of the cell lines in high-dimensional space defined by the variables of the predictive models for lapatinib using the basal profile, with connections indicating similar amounts of the proteins or phosphoproteins. Color corresponds to GI50 values (red, resistant; blue, sensitive). (red arrow, MDA-MB-361 HER2amp; blue arrow, MDA-MB-175 HR+; black arrow MDA-MB-453 TNBC) (B,C) Projection of pErbB2 and pERK overlaid with the GI50 values for lapatinib (B) and afatinib (C). Color corresponds to GI50 values (red, resistant; blue, sensitive). Dashed lines show the detection threshold for pErbB2; arrows point the same cell lines as in (A). (GI50 value for MDA-MB-361 not available for afatinib). (D) Quality of the prediction for drugs targeting ErbB1 or ErbB2 using a linear model with a single variable or the variables pERK and pErbB2. Non-significant predictions (NS) are set to zero.
DISCUSSION
Here, we showed that biochemical profiles of basal and ligand-stimulated signaling pathways can be used to construct PLSR-based models that predict responsiveness to 23/43 targeted drugs examined by Heiser et al. (20). In contrast to many genomic classifiers of drug sensitivity in breast cancer, our models predicted continuous GI50 values, which rarely divide cleanly into resistant and sensitive classes. We found that signaling profiles rather than basal profiles yielded more predictive models for drugs targeting intracellular targets. This seems logical because signaling profiles report on network activity; however, larger datasets are necessary to assess the statistical significance of the differential predictive power of basal and signaling profiles.
Inspection of PLSR model variables revealed which features of signal transduction biochemistry were predictive of sensitivity to which drug. In many cases, the number of variables was small (5 to 12). Surprisingly, our data indicated that target abundance or target phosphorylation rarely correlated with the sensitivity of cells to a drug against that target, except in the case of ErbB2, which also had a positive correlation between abundance and basal phosphorylation. In this sense, ErbB2 is not a prototype for a protein-based biomarker, but rather a special and likely rare exception. More generally, we found that classification of breast cancer lines using a dichotomous high-low score for receptor abundance obscured the graded variation that is observed across cell lines. Unsupervised clustering divided lines into clusters primarily on the basis of the abundance or ligand responsiveness of receptors such as c-Met, FGFR4, and IL6R, none of which are in the subtype definitions of TNBC, HER2amp, or HR+, but which are implicated in drug resistance (51-53) or are the targets of investigational antibody-based therapeutics (54-58). Moreover, large-scale sequencing of HER2amp tumors also shows that the subtype comprises several distinguishable classes (22). Subdividing the HER2amp subtype could be beneficial clinically if future studies establish a correlation of genetic and biochemical profiles with preexisting or acquired resistance to trastuzumab, currently the most important drug for this type of tumor.
Breast cancers are frequently mutated at PIK3CA or PTEN loci (1, 22) and many drugs targeting the PI3K/Akt pathway are in active clinical development (47). Unfortunately, neither mutational status nor subtype is very predictive of sensitivity to these drugs. However, we could build predictive models for the majority of drugs targeting the PI3K/Akt pathway based on network profiling. Even among drugs that supposedly have the same molecular target, the models differed, which may be due to differences either in the isoform specificity or off-target activities of the individual drugs. Rather than mutational status of PIK3CA or PTEN, or the abundance of pAkt, we found that for a subset of PI3K/Akt-targeted drugs, responsiveness to the ligand HRG had significant predictive value, as did the abundance of ErbB2 and ErbB3, which are easier to measure in tumor samples. These results suggest that pathway activity or the potential for inducibility is an effective indicator of drug sensitivity. Because predictors are correlative, we cannot say precisely how responsiveness to the ErbB2-ErbB3 ligand HRG and sensitivity to PI3K/Akt inhibitors are linked mechanistically, but heregulin and ErbB3 are both implicated in breast cancer biology (59, 60). Additional work with clinical samples is required to determine whether such a biomarker will be practically useful.
In many cases, our ability to predict GI50 values from pathway data is improved by including information about the cell line's mutational status. Remarkably, prediction of sensitivity to drugs that target ErbB2 (such as lapatinib) or ErbB1 (such as gefitinib) is also improved by adding data on PIK3CA or PTEN mutational status. These findings highlight the value of including both genomic and biochemical data in predictors of drug sensitivity and demonstrate a need for better understanding of dysregulated Akt signaling in breast cancer cells. Our data also suggest that PIK3CA or PTEN mutational status may act as a cofounding factor rather than a predictor of responses to targeted drugs.
The experiments described here serve as proof of principle to study drug response across panels of cells lines based on systematic biochemical analysis of basal and induced cell states. The protein profiles in the current work contain only fifty to two hundred variables per cell line, many fewer than could be collected by whole-genome sequencing or mRNA expression profiling (although this may change with high-throughput mass spectrometry). Nonetheless our results suggest that even a limited number of protein measurements centered on pathways targeted by drugs can be highly informative. If we compare this work to the transcriptional and genetic profiling in Cancer Cell Line Encyclopedia (26) we find that our models have an equal or better Pearson's correlation for the predictions of all four drugs common to our work. We speculate that the wide range of genetic changes that occur in cancer are canalized into a simpler set of changes at the level of signal transduction networks. If so, systematic mechanistic analysis of signaling pathway status across diverse cancer genotypes should substantially simplify our understanding of tumorigenesis. Of course, genomic and transcriptional data are easier to collect from patients than are proteomic measurements, but we found that, in some cases, very specific proteomic data were predictive of drug response. Combining such proteomic data on signaling networks with genomic data already routinely collected in the clinic might yield hybrid clinical biomarkers that could improve therapy selection in cancer patients.
MATERIALS AND METHODS
Cell culture
All cells were obtained from the ATCC and grown according to recommendations, except for BT-474, MCF7, MDA-MB-415, and MDA-MB-436. All culture conditions are provided in table S2. All cells were free of Mycoplasma. Cells were plated either in 15-cm dishes or 96-well dishes to achieve 70-80% confluency at the time of lysis. Cells were grown for 24 h and then starved in serum-free media without additives for 18 h prior to exposure to ligands for signal profiling or lysis for basal profiling.
Extracellular ligands
All ligands used in this study are listed in table S4 and prepared as specified by the manufacturer. Ligands were dissolved according to recommendations from the manufacturer at 100 μg/ml, except for IFN-α which was supplied as a solution of 106 U/ml. For treatment of each cell line with final concentrations of 100 ng/ml and 1 ng/ml (103 U/ml and 101 U/ml for IFN-α), ligands were further diluted into 10x treatment solution with the same media used for serum starvation.
ELISA- basal profile lysate collection
Cells were plated in 9x 15 cm dishes, washed with 12 ml of PBS, and drained for 30 sec. Two plates were treated for approximately 5 min with 1.5 ml 0.05% Trypsin (Corning, 25-052-CI), until cells detached from the plates. Tryspin was stopped with 8.5 ml of 10% FBS in PBS and gently pipetted up and down until they were in single-cell suspension. Cells from each plate were counted in duplicate using a Cellometer Auto T4 (Nexcelcom). Seven plates were lysed with 1 ml of lysis buffer containing: Mammalian Protein Extraction Buffer (M-PER; Thermo Scientific, 78501) supplemented with protease inhibitor cocktail (Sigma-Aldrich, P2714), 1 mM sodium orthovanadate (Sigma-Aldrich, S6508), 5 mM sodium pyrophosphate (Sigma-Aldrich, 221368), 50 μM oxophenylarsine (EMD Biosciences, 521000), and 10 μM bpV (phen; EMD Biosciences, 203695). Lysed cells were scraped off the plate, collected in microcentrifuge tubes, and incubated on ice for 30 min. Membranes and cell debris were sedimented by centrifugation at 20,000g for 10min at 4°C, the supernatants were pooled, aliquoted, and subsequently stored at −80°C.
ELISA - ligand response lysate collection
Cells were plated in two 96-well plates and treated with growth factors for 10, 30, or 90 min. The cells were washed in 200 μl of cold PBS and lysed in 30 μl of the M-PER lysis buffer with the inhibitors listed above. Plates were sealed, rocked on ice for 15 min, and stored at −80°C.
ELISA measurements
All ELISA assays used for this study are listed with their targets in table S3. For protein measurements, 384 well, black, flat-bottomed, polystyrene, high-binding ELISA plates (Corning, 3577) were incubated overnight at room temperature with capture antibodies (table 3), and then blocked with 2% bovine serum albumin (BSA) in PBS for 1 hour. Plates were washed four times with 0.05% Tween-20 in PBS (PBS-T) then incubated with lysates and recombinant protein standards for 2 hours at room temperature. After each antibody incubation, plates were washed four times with PBS-T. ELISAs were incubated with primary and secondary antibodies for 2 hours and 1 hour, respectively. All ELISAs were visualized with SuperSignal ELISA Pico Chemiluminescent Substrate (Pierce, 37069) and luminescent signal was measured with an EnVision Plate Reader (Perkin Elmer).
Data from ELISA measurements was extracted using MATLAB Statistical Toolbox. Briefly, raw data were background subtracted (raw values from assay buffer was subtracted from raw values of each sample). A dilution series of recombinant protein (standard curve) was used to convert raw signal into known protein concentration (pg/ml). A linear best fit line was calculated based on the standard curve, then background subtracted values for each sample were converted into pg/ml using the linear equation (y = mx + b) of the standard curve. Regressed data were multiplied by the dilution factor for each sample, then mean and standard error of the mean values were calculated for at least four technical replicates for the basal profile measurements and two biological replicates for the ligand response measurements. Data above or below the range of detection was set to the upper or lower detection limit, respectively. For steady-state protein levels, ELISA measurements (pg/ml) were normalized to the number of cells in the lysate (cells/ml) to get pg protein per cell.
High-throughput microscopy – ligand response
Experiments were performed as previously described (61). In short, cells were plated in 6x 96 well plates and treated with growth factors and cytokines for 10, 30, or 90 min. The cells were fixed for 10 min at 25°C in 2% paraformaldehyde . Plates were washed with 200 μl PBS-T and stored at 4°C until assaying. Cells were permeabilized with 100 μl of methanol for 10 min at 25°C, washed with 200 μl PBS-T, and blocked with 40 μl of Odyssey blocking buffer (OBB; LICOR) for 1 hour at 25°C. Cells were treated with 40 μl of primary antibody diluted 1:400 in OBB, sealed, and incubate overnight at 4°C on rocking platform. Cells were washed twice with 200 μl of PBS-T and treated with 40 μl of secondary antibody diluted 1:2000 in OBB incubated for 1 hour at 25°C. Cells were washed in 200 μl of PBS-T, then 200 μl of PBS, and were stained with 40 μl of 250 ng/ml Hoechst 33342 (Invitrogen) and 1:1000 Whole Cell Stain (blue; Thermo Scientific) in PBS. Cells were washed two times with 200 μl of PBS and imaged in an imageWoRx high-throughput microscope (Applied Precision). Data was extracted using ImageRail and stored for processing in semantic data cubes (61). All antibodies used for high-throughput microscopy are listed with their targets in table S5.
Clustering parameters
The clustering in Figure 1C and 2A were made using the MATLAB statistical toolbox. The distance used in the algorithm is the correlation distance (that is 1-R where R is the Pearson's correlation) and the ‘average’ linkage was used to draw the clustering tree.
Prediction algorithm
Our prediction uses a linear model based on a partial least squares regression (PLSR) with variable selection based on variable importance projection (VIP) (62). All codes are written in MATLAB using the standard embedded function such as “simpls” for building the PLSR model. The detailed description of the algorithm can be found in the text S1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Chopra, C. Zechner, S. Rosenthal, J. Gray, and I. Overton for helpful discussions. Funding: This work was support by Stand Up to Cancer Project (AACR-SU2C-DT0409), NIH grant U54-HG006097, NIH grant CA112967, and a fellowship from the Swiss National Science Foundation (PBELP3_140652) to M.H. In-kind support for E.A.P., D.H.C., and B. S. was supplied by Merrimack Pharmaceuticals.
Footnotes
Author contributions: M.N., E.A.P., M.C., D.H.C., and L.Z. performed all experiments; M.H. performed all analysis; all authors wrote and edited the paper.
Data and materials availability: Supplementary materials and HMS LINCS website (http://lincs.hms.harvard.edu/explore/)
Competing interests: P.K.S. is a founder of Merrimack Pharmaceuticals and chair of its scientific advisory board; this relationship is actively managed by HMS in accordance with NIH regulations. B. S. is a shareholder in Merrimack Pharmaceuticals and a Board member of SilverCreek Pharmaceuticals.
REFERENCES AND NOTES
- 1.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M, Shak S, Stewart SJ, Press M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 3.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 4.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D'Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong RP, Baselga J. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin Oncol. 2003;21:2237. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr., Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 9.National Cancer Institute; 2013. [August 28th]. http://www.cancer.gov/ [Google Scholar]
- 10.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Lang I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Ruschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD. T. Herceptin Adjuvant Trial Study, Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 11.Smith I, Procter M, Gelber RD, Guillaume S, Feyereislova A, Dowsett M, Goldhirsch A, Untch M, Mariani G, Baselga J, Kaufmann M, Cameron D, Bell R, Bergh J, Coleman R, Wardley A, Harbeck N, Lopez RI, Mallmann P, Gelmon K, Wilcken N, Wist E, Sanchez Rovira P, Piccart-Gebhart MJ. HERA Study Team, 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369:29. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 12.Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, Miles D, Bianchi G, Cortes J, McNally VA, Ross GA, Fumoleau P, Gianni L. Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol. 2010;28:1138–44. doi: 10.1200/JCO.2009.24.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianni L, Pienkowski T, Im YM, Roman L, Tseng LM, Liu MC, Lluch A, Staroslawska E, de la Haba-Rodriguez J, Im SA, Pedrini JL, Poirier B, Morandi P, Semiglazov V, Srimuninnimit V, Bianchi G, Szado T, Ratnayake J, Ross G, Valagussa P. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 14.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 15.International Breast Cancer Study Group. Colleoni M, Gelber S, Goldhirsch A, Aebi S, Castiglione-Gertsch M, Price KN, Coates AS, Gelber RD. Tamoxifen after adjuvant chemotherapy for premenopausal women with lymph node-positive breast cancer: International Breast Cancer Study Group Trial 13-93. J Clin Oncol. 2006;24:1332. doi: 10.1200/JCO.2005.03.0783. [DOI] [PubMed] [Google Scholar]
- 16.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 17.Bernsdorf M, Ingvar C, Jörgensen L, Tuxen MK, Jakobsen EH, Saetersdal A, Kimper-Karl ML, Kroman N, Balslev E, Ejlertsen B. Effect of adding gefitinib to neoadjuvant chemotherapy in estrogen receptor negative early breast cancer in a randomized phase II trial. Breast Cancer Res Treat. 2011;126:463–70. doi: 10.1007/s10549-011-1352-2. [DOI] [PubMed] [Google Scholar]
- 18.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, Wolff AC, Hobday TJ, Ivanova A, Chiu WK, Ferraro M, Burrows E, Bernard PS, Hoadley KA, Perou CM, Winer EP. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30:2615–23. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo W-L, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer cell. 2006;10:529. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas, Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SWG, Morin GB, Watson P, Gelmon K, Chia S, Chin S-F, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra M. a., Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shastry M, Yardley DA. Updates in the treatment of basal/triple-negative breast cancer. Current opinion in obstetrics & gynecology. 2012;1 doi: 10.1097/GCO.0b013e32835c1633. [DOI] [PubMed] [Google Scholar]
- 25.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O'Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meric-Bernstam F, Akcakanat A, Chen H, Do KA, Sangai T, Adkins F, Gonzalez-Angulo AM, Rashid A, Crosby K, Dong M, Phan AT, Wolff RA, Gupta S, Mills GB, Yao J. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1777. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirzoeva OK, Das D, Heiser LM, Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM, Guan Y, Hu Z, Knight Z, Feiler HS, Gascard P, Parvin B, Spellman PT, Shokat KM, Wyrobek AJ, Bissell MJ, McCormick F, Kuo WL, Mills GB, Gray JW, Korn WM. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69:565–72. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, Ribas A, Li J, Moffat J, Sutherlin DP, Koeppen H, Merchant M, Neve R, Settleman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ignatoski KM, Lapointe AJ, Radany EH, Ethier SP. erbB-2 overexpression in human mammary epithelial cells confers growth factor independence. Endocrinology. 1999;140:3615. doi: 10.1210/endo.140.8.6939. [DOI] [PubMed] [Google Scholar]
- 35.Gamett DC, Pearson G, Cerione RA, Friedberg I. Secondary dimerization between members of the epidermal growth factor receptor family. J Biol Chem. 1997;272:12052. doi: 10.1074/jbc.272.18.12052. [DOI] [PubMed] [Google Scholar]
- 36.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 37.Law JH, Habibi G, Hu K, Masoudi H, Wang MY, Stratford AL, Park E, Gee JM, Finlay P, Jones HE, Nicholson RI, Carboni J, Gottardis M, Pollak M, Dunn SE. Phosphorylated insulin-like growth factor-i/insulin receptor is present in all breast cancer subtypes and is related to poor survival. Cancer Res. 2008;68:10238. doi: 10.1158/0008-5472.CAN-08-2755. [DOI] [PubMed] [Google Scholar]
- 38.Kurzrock R, Patnaik A, Aisner J, Warren T, Leong S, Benjamin R, Eckhardt SG, Eid JE, Greig G, Habben K, McCarthy CD, Gore L. A phase I study of weekly R1507, a human monoclonal antibody insulin-like growth factor-I receptor antagonist, in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:2458. doi: 10.1158/1078-0432.CCR-09-3220. [DOI] [PubMed] [Google Scholar]
- 39.Ramalingam SS, Spigel DR, Chen D, Steins MB, Engelman JA, Schneider CP, Novello S, Eberhardt WE, Crino L, Habben K, Liu L, Janne PA, Brownstein CM, Reck M. Randomized phase II study of erlotinib in combination with placebo or R1507, a monoclonal antibody to insulin-like growth factor-1 receptor, for advanced-stage non-small-cell lung cancer. J Clin Oncol. 2011;29:4574. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghav KP, Wang W, Liu S, Chavez-MacGregor M, Meng X, Hortobagyi GN, Mills GB, Meric-Bernstam F, Blumenschein GR, Jr., Gonzalez-Angulo AM. cMET and phospho-cMET protein levels in breast cancers and survival outcomes. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2269. doi: 10.1158/1078-0432.CCR-11-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nature reviews. Drug discovery. 2008;7:504. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 42.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1855. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 43.Bachelot T, Ray-Coquard I, Menetrier-Caux C, Rastkha M, Duc A, Blay JY. Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br J Cancer. 2003;88:1721. doi: 10.1038/sj.bjc.6600956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salgado R, Junius S, Benoy I, Van Dam P, Vermeulen P, Van Marck E, Huget P, Dirix LY. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. International journal of cancer. Journal international du cancer. 2003;103:642. doi: 10.1002/ijc.10833. [DOI] [PubMed] [Google Scholar]
- 45.Zhang GJ, Adachi I. Serum interleukin-6 levels correlate to tumor progression and prognosis in metastatic breast carcinoma. Anticancer research. 1999;19:1427. [PubMed] [Google Scholar]
- 46.Shattuck DL, Miller JK, Carraway KL, 3rd, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- 47. [August 28th 2013];ClinicalTrials. gov, http://clinicaltrials.gov.
- 48.Weigelt B, Downward J. Genomic Determinants of PI3K Pathway Inhibitor Response in Cancer. Frontiers in oncology. 2012;2:109. doi: 10.3389/fonc.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, Aura C, Gomez H, Dinh P, Fauria K, Van Dooren V, Aktan G, Goldhirsch A, Chang TW, Horvath Z, Coccia-Portugal M, Domont J, Tseng LM, Kunz G, Sohn JH, Semiglazov V, Lerzo G, Palacova M, Probachai V, Pusztai L, Untch M, Gelber RD, Piccart-Gebhart M, Neo AST. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A, Kaufman B, Skarlos D, Campone M, Davidson N, Berger M, Oliva C, Rubin SD, Stein S, Cameron D. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 51.Roidl A, Berger HJ, Kumar S, Bange J, Knyazev P, Ullrich A. Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin Cancer Res. 2009;15:2058–66. doi: 10.1158/1078-0432.CCR-08-0890. [DOI] [PubMed] [Google Scholar]
- 52.Moschetta M, Basile A, Ferrucci A, Frassanito MA, Rao L, Ria R, Solimando AG, Giuliani N, Boccarelli A, Fumarola F, Coluccia M, Rossini B, Ruggieri S, Nico B, Maiorano E, Ribatti D, Roccaro AM, Vacca A. Novel Targeting of Phospho-cMET Overcomes Drug Resistance and Induces Antitumor Activity in Multiple Myeloma. Clin Cancer Res. 2013;19:4371–4382. doi: 10.1158/1078-0432.CCR-13-0039. [DOI] [PubMed] [Google Scholar]
- 53.Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, Quraishi AA, Tawakkol N, D'Angelo R, Paulson AK, Chung S, Luther T, Paholak HJ, Liu S, Hassan KA, Zen Q, Clouthier SG, Wicha MS. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47:570–84. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith DC, Smith MR, Sweeney C, Elfiky AA, Logothetis C, Corn PG, Vogelzang NJ, Small EJ, Harzstark AL, Gordon MS, Vaishampayan UN, Haas NB, Spira AI, Lara Jr PN, Lin CC, Srinivas S, Sella A, Schöffski P, Scheffold C, Weitzman AL, Hussain M. Cabozantinib in patients with advanced prostate cancer: results of a phase II randomized discontinuation trial. J Clin Oncol. 2013;31:412–9. doi: 10.1200/JCO.2012.45.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choueiri TK, Vaishampayan U, Rosenberg JE, Logan TF, Harzstark AL, Bukowski RM, Rini BI, Srinivas S, Stein MN, Adams LM, Ottesen LH, Laubscher KH, Sherman L, McDermott DF, Haas NB, Flaherty KT, Ross R, Eisenberg P, Meltzer PS, Merino MJ, Bottaro DP, Linehan WM, Srinivasan R. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol. 2013;31:181–6. doi: 10.1200/JCO.2012.43.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.French DM, Lin BC, Wang M, Adams C, Shek T, Hötzel K, Bolon B, Ferrando R, Blackmore C, Schroeder K, Rodriguez LA, Hristopoulos M, Venook R, Ashkenazi A, Desnoyers LR. Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One. 2012;7:e36713. doi: 10.1371/journal.pone.0036713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato K, Tsuchiya M, Saldanha J, Koishihara Y, Ohsugi Y, Kishimoto T, Bendig MM. Reshaping a human antibody to inhibit the interleukin 6-dependent tumor cell growth. Cancer Res. 1993;53:851. [PubMed] [Google Scholar]
- 58.Voorhees PM, Manges RF, Sonneveld P, Jagannath S, Somlo G, Krishnan A, Lentzsch S, Frank RC, Zweegman S, Wijermans PW, Orlowski RZ, Kranenburg B, Hall B, Casneuf T, Qin X, van de Velde H, Xie H, Thomas SK. A phase 2 multicentre study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with relapsed or refractory multiple myeloma. Br J Haematol. 2013;161:357–66. doi: 10.1111/bjh.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hutcheson IR, Knowlden JM, Hiscox SE, Barrow D, Gee JM, Robertson JF, Ellis IO, Nicholson RI. Heregulin beta1 drives gefitinib-resistant growth and invasion in tamoxifen-resistant MCF-7 breast cancer cells. Breast cancer research : BCR. 2007;9:R50. doi: 10.1186/bcr1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garrett JT, Sutton CR, Kurupi R, Bialucha CU, Ettenberg SA, Collins SD, Sheng Q, Wallweber J, Defazio-Eli L, Arteaga CL. Combination of antibody that inhibits ligand-independent HER3 dimerization and a p110α inhibitor potently blocks PI3K signaling and growth of HER2+ breast cancers. Cancer Res. 2013 Aug 5; doi: 10.1158/0008-5472.CAN-13-1191. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Millard BL, Niepel M, Menden MP, Muhlich JL, Sorger PK. Adaptive informatics for multifactorial and high-content biological data. Nature Methods. 2011;8:487–493. doi: 10.1038/nmeth.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Enciso M, Tenenhaus M. Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Human genetics. 2003;112:581–92. doi: 10.1007/s00439-003-0921-9. [DOI] [PubMed] [Google Scholar]
- 63.Witten IH, Frank E, Hall MA. Data Mining. Third edition. Morgan Kaufmann Publishers; Burlington, MA.: chapter 5.3. [Google Scholar]
- 64.Varma A, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.