Abstract
Elevated expression of p130Cas (Crk-associated substrate)/BCAR1 (breast cancer antiestrogen resistance 1) in human breast tumors is a marker of poor prognosis and poor overall survival. p130Cas is a downstream target of the tyrosine kinase c-Src. Signaling mediated by p130Cas through its phosphorylated substrate domain (SD) and interaction with effector molecules directly promotes tumor progression. We previously developed a constitutively phosphorylated p130Cas SD molecule, Src*/SD (formerly referred to as Src*/CasSD), which acts as decoy molecule and attenuates the transformed phenotype in v-crk-transformed murine fibroblasts and human breast cancer cells. To test the function of this molecule in vivo, we established mouse mammary tumor virus (MMTV)-long terminal repeat-Src*/SD transgenic mice in which mammary gland development and tumor formation were analyzed. Transgenic expression of the Src*/SD molecule under the MMTV-long terminal repeat promoter did not interfere with normal mammary gland development or induce tumors in mice observed for up to 11 months. To evaluate the effects of the Src*/SD molecule on tumor development in vivo, we utilized the MMTV-polyoma middle T-antigen (PyMT) murine breast cancer model that depends on c-Src. PyMT mice crossed with Src*/SD mice displayed accelerated tumor formation. The earlier onset of tumors can be explained by the interaction of the Src* domain with PyMT and targeting the fused phosphorylated SD to the membrane. At membrane compartments, it might integrate membrane-associated active signaling complexes leading to increased proliferation measured by phospho-Histone H3 staining. Although these results were unexpected, they emphasize the importance of preventing the membrane association of Src*/SD when employed as decoy molecule.
Introduction
p130Cas (Crk-associated substrate) is the founding member of the Cas family (1) and was first identified as a major tyrosine-phosphorylated protein in cells transformed by v-crk and v-src (2). It functions as scaffold for large multiprotein complexes that integrate the response to various stimuli including growth factor and hormone signaling and integrin engagement (3,4). Elevated p130Cas levels are associated with poor prognosis in breast cancer patients and correlate with increased rate of relapse and poor response to tamoxifen (5). Increased p130Cas/BCAR1 expression was detected in tumor cells isolated from pleural effusions of breast cancer patients (6). Recent in vivo studies showed substantial mammary epithelial cell hyperplasia during development and pregnancy, and delayed involution in mouse mammary tumor virus (MMTV)-p130Cas transgenic mice (7).
The p130Cas molecule is characterized by multiple protein–protein interaction domains including an N-terminal SH3 domain, a central substrate domain (SD) containing multiple tyrosine phosphorylation sites, a serine-rich region and a C-terminal domain containing a bi-partite Src-binding motif (8). Inducible phosphorylation of the p130Cas SD on tyrosine residues is critical for its function. These phosphorylation events lead to coupling with the small adapter protein Crk via an SH2 domain interaction resulting in a molecular switch that promotes cell migration (9). Therefore, preventing the interaction of p130Cas with proteins that bind to its phosphorylated SD might be a useful treatment strategy for breast cancers with elevated p130Cas levels. We previously developed a dominant negative p130Cas molecule Src*/SD (formerly referred to as Src*/CasSD)—composed of an attenuated c-Src kinase domain fused to the p130Cas SD (10,11). Attenuation was achieved by mutating tyrosine 416 in the c-Src kinase domain to phenylalanine. Although this Src kinase mutant is inactive against exogenous substrates (12,13), it constitutively phosphorylates the p130Cas SD in the Src*/SD chimera independent of upstream signals. Src*/SD acts as a decoy for downstream binding partners thereby competing with endogenous p130Cas (10). In tamoxifen-resistant breast cancer cells (TAM-R) (14) characterized by elevated levels of breast cancer antiestrogen resistance 1 (BCAR1) messenger RNA, total and phosphorylated p130Cas protein and c-Src kinase activity, it attenuates the transformed phenotype and resensitizes these cells to tamoxifen (11,15,16).
To determine if this decoy approach alters tumor progression, we assessed here the in vivo effects of Src*/SD expression in the mammary gland of transgenic mice and in the Src-dependent MMTV-polyoma middle T-antigen (PyMT) breast cancer mouse model. PyMT is a membrane-anchored viral protein that assembles a large multiprotein complex at various membrane compartments (17). It is able to convert cultured rat cells to a fully transformed, tumorigenic phenotype (18). PyMT transgenic mice develop mammary gland tumors with short latency and tumor progression resembles that of human disease (19). Tumorigenicity in this model depends on the activity of the c-Src kinase (20).
We demonstrated that transgenic Src*/SD mice develop normal mammary glands and do not develop tumors. Expression of the Src*/SD molecule in mammary tumors induced by PyMT significantly accelerates tumor development. We have attributed this unexpected outcome to binding of Src*/SD to the PyMT protein and its targeting to membrane compartments. Thus, these in vivo data suggest that future decoy approaches to inhibit p130Cas signaling need to consider the subcellular location of the potential inhibitor.
Materials and methods
Generation of MMTV-Src*/SD transgenic mice
Animal experiments were performed adhering to ethical standards and approved by the institutional animal care and use committee at Boston University Medical Center. Mice of the FVB/N (FVB) strain were utilized to generate MMTV-Src*/SD (Src*/SD) transgenic animals. The transgene was amplified by PCR of the hemagglutinin epitope (HA)-tag-Src*/SD fragment from the pJ3H-Src*/SD plasmid (10) with the primers: MMTV5ʹHindIII: CCCAAGCTTATGTACCCATACGATGTTCCA (forward) and MMTV3ʹEcoRI: CGGAATTCTCATCGCTCGGCTGGTGG (reverse) and subcloned between the HindIII and EcoRI restriction sites downstream of the MMTV-long terminal repeat promoter and upstream of Sv40 polyadenylation signal of the MMTV-Sv40-Bssk vector kindly provided by K.Ravid (Boston University School of Medicine, Boston, MA). The transgenic mice were generated by the transgenic core facility of Boston University. The animals were screened for the transgene by PCR analysis using tail DNA and/or Southern blot analysis. For PCR analysis, primers P1 and P2 were used to amplify a 534bp fragment of the Src*/SD construct. P1 (forward): 5ʹ-TCGCTGCATGACCTCATGTGC-3ʹ and P2 (reverse): 5ʹ-TGG AGGGACTTGGTAGATGTC-3ʹ. Forward primer: 5ʹ-TTCCGCAAGTTCA CCTACC-3ʹ and reverse primer: 5ʹ-CGGGCCGGCCATGCTTTACG-3ʹ were used to amplify a 361bp fragment of the housekeeping gene S15 to check for DNA quality.
Animal models and assays for tumor progression
Two founder mice were identified that expressed Src*/SD in the expected tissues (L 67 and L 63, high and low expressing, respectively). MMTV/PyMT (PyMT) transgenic mice with an FVB strain background were purchased from the Jackson Laboratory. Genotyping for the PyMT transgene was performed according to the protocol provided by the Jackson Laboratory. Homozygous Src*/SD female mice (L 67 and L 63) were mated with male mice hemizygous for the PyMT transgene to obtain PyMT × MMTV-Src*/SD (PyMT × Src*/SD) mice. Double transgenic mice were identified by PCR analyses of tail DNA samples. Female mice were examined for mammary tumors by palpation twice a week. Tumor diameters were measured with calipers and tumor volume was estimated as described previously (21). Mice were then euthanized at different stages of mammary tumorigenesis, and their mammary glands and tumors were collected for histological analyses. Mammary gland development in MMTV-Src*/SD female mice was assessed in 6-, and 10-week-old virgin and involuting mammary glands. Mammary gland whole-mount analysis was performed on 5 mice per time point.
Mammary gland whole-mount analysis
The inguinal mammary fat pads were spread on microscope slides, fixed in Carnoy’s fixative overnight, hydrated and stained with carmine alum stain (Sigma–Aldrich, St Louis, MO) overnight. Subsequently, the samples were dehydrated, treated with xylene to remove the fat and cover slips were mounted with ‘Protocol’ Mount Medium (Fisher Scientific, Pittsburg, PA) and observed under a Olympus SZX16 stereo microscope (Center Valley, PA). Pictures were taken with a QImaging camera.
Generation of expression constructs
Src*/SD [SD-wild-type (WT)] containing the attenuated (*) Src kinase domain fused to amino acids 157–516 of rat p130Cas was described previously (10). Src*/SDh was generated by amplification of the complete human p130Cas SD (amino acid 109–419) using full-length BCAR1 complementary DNA (cDNA) (22) as template and Pfu polymerase. Following primers were used: CasA- forward (including ClaI site) ATA ATCGATCCCCAGCCAGACAGCGTC and CasA-reverse (containing NotI site) ATAGCGGCCGCTCA ACGTTCA GCTGGGGGAG. Subsequently, the amplified SD cDNA was subcloned using ClaI/NotI into the doxycycline (Dox)-inducible expression vector pC4-R(TO) containing the 5ʹ located cDNA for Src*. Src*/SD-CAAX was generated by C-terminal fusion of the H-Ras CAAX lipid anchor (GCMSCKCVLS) (23) to Src*/SDh by insertion mutagenesis using the QuikChange™ XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Src*/SDh served as template and primers were as follows: 5ʹ-TGCCTCCCCCAGCTGAACGT GGCTG CATGAGCTGCAAGTGTGTGCTCTCCTGAGCGGCCGCTCGACTTTTA-3ʹ (forward) and 5ʹ-TAAAAGTCGA GCGGCCGC TCA GGAGAGCACACACTTG CAGCTCATGCAGCCACGTTCAGCTGGGGGAGGCA-3ʹ (reverse). Constructs were validated by sequencing.
Cell lines, culture conditions and retroviral transductions
PyMT breast tumor cells were the kind gift from R.Hazan (Albert Einstein College of Medicine, New York, NY) (24). PyMT cells and 3Y1 rat fibroblasts were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% and 5% fetal bovine serum (FBS), respectively, 1mM sodium pyruvate, 2mM l-glutamine, 100U/ml penicillin and 100U/ml streptomycin. TAM-R cells were maintained as described previously (11,15). Dox-inducible retroviral expression constructs of Src*/SD, SrcKM/SD and Src* were generated as described previously (11). PyMT cells constitutively expressing Src*/SD and empty vector (EV) were established by transduction with retroviral pCXbsr expression plasmids (11). Retroviral transduction of PyMT and 3Y1 cells was carried out as described previously (21).
Reverse transcription–polymerase chain reaction assay
The Ultraspec-II RNA Isolation System (Biotecx Laboratories, Houston, TX) was used to extract RNA from tissue according to the manufacturer’s instructions. RNA was treated with RNAse-free DNAse. Subsequently, 5 μg of total RNA were transcribed into cDNA using random primers and the Superscript II Kit (Invitrogen, Carlsbad, CA). The Src*/SD was amplified by PCR (35 cycles) using 2 μl of cDNA as template and P1 and P2 primers described previously. A PCR with control primers for the S15 gene was performed as described above.
Antibodies
β-Actin (#A-5316; Sigma–Aldrich), phospho-extracellular signal-regulated kinase (ERK) 1/2 Thr202/Thr204 (#9106; Cell Signaling, Danvers, MA), HA (12CA5; Roche Applied Science, Nutley, NJ), Ki-67 (cloneSP6; Thermo Scientific, Waltham, MA), phospho-Histone H3 (pHistone-H3; #06-570; Millipore, Billerica, MA), PyMT (pAb762; S. Dilworth, Imperial College London) and tubulin (#CP06-100UG; Calbiochem, San Diego, CA) were used. CasB antibodies (directed against the SD of p130Cas) were established as described previously (25). ERK1 (sc-93), ERK2 (sc-154), HA (Y-11, sc-805), IgG control (sc-2343), Myc (9E10, sc-40), pan-Cadherin (CH-19, sc-59876), phospho-tyrosine (pTyr; PY-99, sc-7020) and PyMT (sc-53481) were from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 488 goat anti-mouse IgG, Alexa Fluor 594 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rabbit IgG were from Life Technologies (Molecular Probes, Grand Island, NY).
Histochemical analysis
Tumor samples from mice were fixed in 10% formalin, processed and embedded in paraffin by the Pathology Laboratory Services Core at Boston University School of Medicine. Immunohistochemical staining was performed on 5 μm sections using the standard ABC method (Vector Laboratories, Burlingame, CA) and counterstained with hematoxylin.
Immunofluorescence analysis and confocal microscopy
Tumor sections were incubated with pHistone-H3 rabbit polyclonal antibodies at room temperature for 1h followed by incubation with a secondary antibody labeled with Alexa Fluor 488 at room temperature for 1h. Nuclei were labeled with 4ʹ6-diamidino-2-phenylindole (DAPI). For each slide, pHistone-H3 positively stained cells and DAPI-stained cells were counted in seven ×20 fields per tumor (each field contained ~500–800 cells) and the results were averaged. The results are presented as mean and standard deviation from at least four tumors.
Constitutively expressing PyMT-Src*/SD and PyMT-EV cells were cultured in 8-well chamber slides. After fixation with 3% paraformaldehyde for 20min, cells were immunostained with PyMT (pAb762) and HA antibodies followed by Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit IgG. Imaging of tumor sections was performed with an Axiovert 200M (Carl Zeiss, Thornwood, NY) equipped with a charge-coupled device ORCA-ER camera (Hamamatsu), and PyMT cells were imaged with a confocal laser scanning microscope LSM510 (Carl Zeiss). The proprietary software was used for analysis.
Immunoblotting analysis
Mouse mammary gland tissues or tumor tissues were flash frozen in liquid nitrogen. Frozen tissue was disrupted in RIPA lysis buffer [50mM Tris–HCl (pH 8.0), 150mM NaCl, 1% Nonidet P-40 (NP-40), 0.5% sodium deoxycholate and 0.1% sodium dodecyl sulfate], containing protease inhibitors (Roche), using Dounce homogenization on ice. Whole-cell extracts (WCE) of PyMT cells were prepared in RIPA buffer as described previously (26). Lysates were cleared by centrifugation at 16 000g at 4°C for 30min. Tissue lysates and WCE were analyzed by immunoblotting (IB) as described in ref. 27.
Cell fractionation
The protocol was adapted from Patwardhan et al. (28). PyMT, TAM-R and 3Y1 cells were washed with cold STE buffer [150mM NaCl, 50mM Tris (pH 7.2) and 1mM ethylenediaminetetraacetic acid], resuspended in hypotonic buffer [10mM Tris–HCl (pH 7.2) and 0.2mM MgCl2) supplemented with protease inhibitors and incubated on ice for 10min, followed by homogenization in a 1.5ml of Dounce homogenizer (30 strokes). The homogenate was adjusted to 250mM sucrose and 1mM ethylenediaminetetraacetic acid and centrifuged in a Sorvall Ultracentrifuge WX100 (Thermo Scientific) for 45min at 145 000g. The pellet (P) fraction, containing the cellular membranes, was resuspended in 1× RIPA buffer supplemented with protease inhibitors and the soluble/cytoplasmic fraction (S) was supplemented with a 0.2 vol of 5× RIPA buffer. Samples were incubated for 30min on ice and cleared by centrifugation at 16 000g at 4°C for 20min. Equal volumes of the P and S fractions were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and analyzed by IB as described previously.
Immunoprecipitation analysis
Cells or tissues were incubated in lysis buffer [25mM N-2-hydroxyethylpiperazine-Nʹ-2-ethanesulfonic acid–KOH (pH 7.2), 150mM KCl, 2mM ethylenediaminetetraacetic acid and 1% Triton X-100] supplemented with protease inhibitors. The lysate was centrifuged for 10min at 16 000g at 4°C and subsequently precleared with protein A/G-agarose beads (Santa Cruz Biotechnology) for 2h at 4°C followed by overnight incubation with 2 μg of immunoprecipitation (IP) antibodies at 4°C. Protein A/G-agarose beads were added and incubated for 1h at 4°C. Precipitates were washed three times with lysis buffer and resuspended in 2× sample loading buffer and analyzed by IB.
Matrigel assay
PyMT transductants were subjected to matrigel assay as described previously (29). Colonies were analyzed using an Olympus phase contrast microscope at day 10. Rough and smooth colonies in three random fields were counted from each triplicate sample and values are presented as average ± standard deviation. Data were generated from three independent experiments.
Phase contrast microscopy
Cells were imaged by using an Olympus IMT-2 inverted phase contrast microscope, and images were recorded with a Nikon Coolpix 4300 charge-coupled device camera attached to the microscope.
Statistical analysis
Comparisons of palpability of tumors were made by analyzing Kaplan–Meier survival curves, and the log-rank test was used to assess for differences in tumor curves. The number of samples per group (n) is specified in Figure 2A. All P-values were two-tailed, and statistical significance was accepted at P ≤ 0.05. P-values in Figures 2B, 3 and 4 were calculated using unpaired Student’s t-test.
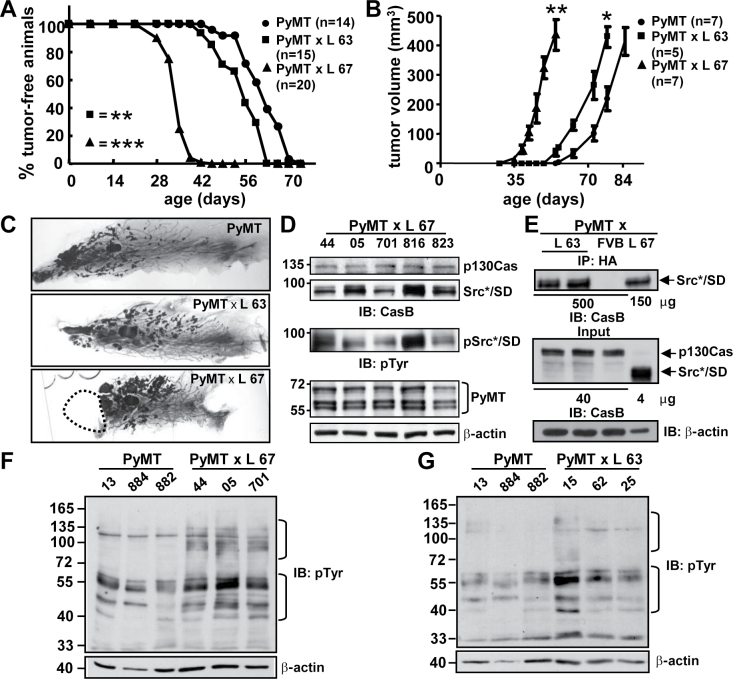
Fig. 2.
Tumor occurrence and Erk1/2 activity in PyMT and PyMT × Src*/SD compound animals. (A) Comparison of the kinetics of tumor formation in PyMT and PyMT × Src*/SD (L 67 and L 63) animals. n, number of mice examined. **P ≤ 0.05, ***P ≤ 0.01. P-values were calculated using log-rank test. (B) Tumor volumes were determined by caliper measurements as described previously in Materials and methods. Bars, standard error. PyMT × L 67 tumor volume versus PyMT tumor volume at day 55 (7.8 weeks), **P ≤ 0.01. PyMT × L 63 tumor volume versus PyMT tumor volume at day 79 (11.3 weeks), *P ≤ 0.05. (P-values were calculated using unpaired Student’s t-test). (C) Whole-mount mammary gland preparations of PyMT and PyMT × Src*/SD animals (L 63 and L 67). The area labeled with dashed line marks location where tumor was removed prior to whole-mount preparation. (D–F) The tumors were excised when they reached ~1cm in the largest dimension. (D) Transgene expression analysis in whole-tumor tissue lysates (20 μg) of individual mice of PyMT × Src*/SD animals (L 67) by IB with CasB, pTyr and PyMT antibodies. β-actin antibodies was used as loading control. (E) Analysis of Src*/SD expression by IP in low-expressing PyMT × Src*/SD tumors (L 63), FVB (negative control) and L 67 (positive control). Upper panel, whole-tissue lysates (500 and 150 μg as indicated) were immunoprecipitated with anti-HA antibodies. The precipitated proteins were identified by IB with CasB antibodies. Input lanes 40 and 4 μg (middle panel) and β-actin (lower panel), for comparison. (F and G) Whole-tissue lysates of PyMT, PyMT × L 67 (F) and PyMT × L 63 (G) were monitored for total tyrosine phosphorylation with pTyr and β-actin antibodies.
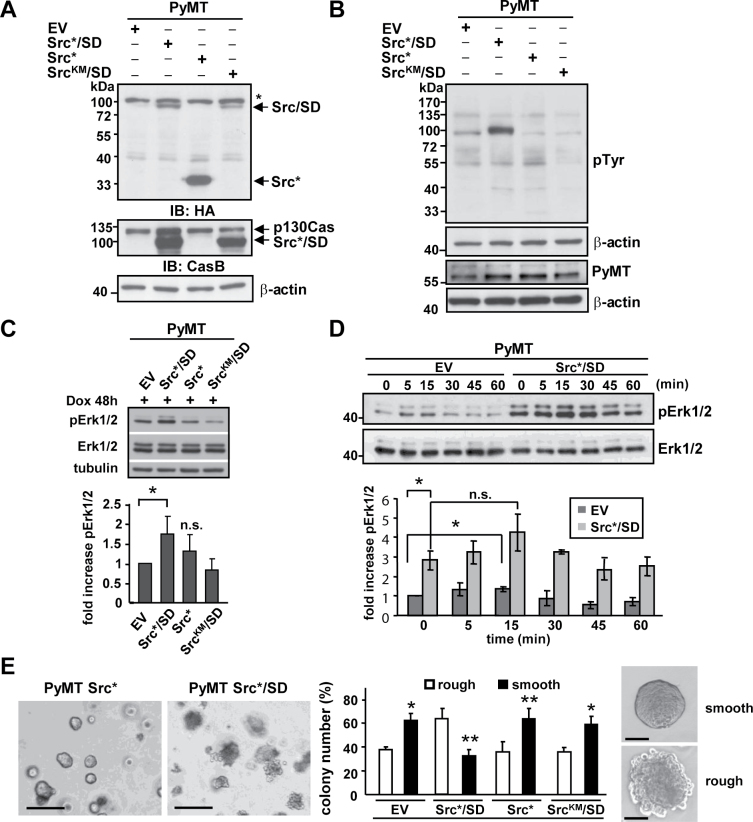
Fig. 4.
Src*/SD expression in PyMT-derived mammary tumor cells increases Erk1/2 activity. (A–E) Expression of Src*/SD and control plasmids (Src*, SrcKM/SD and EV) was induced in PyMT breast cancer cells with Dox (2 μg/ml) for 48h (A–D) and 10 days (E). (A) Expression of the individual constructs was determined by IB with HA and CasB antibodies. *, non-specific band. β-actin antibodies were used as loading control. (B) WCE was monitored for tyrosine phosphorylation of total cellular proteins with pTyr and β-actin antibodies, and for the expression of PyMT with PyMT and β-actin antibodies. (C) Upper panel, phosphorylation of Erk1/2 was analyzed by IB in WCE (20 μg) of PyMT cells expressing the indicated constructs. Total ERK1/2 and tubulin antibodies were used as controls. Lower panel, the fold increase in pErk1/2 was determined by densitometric analysis of three independent experiments. n.s., not significant; *P ≤ 0.05. (D) Dox-inducible PyMT-Src*/SD and PyMT-EV stable cell populations were starved in medium containing 0.5% FBS for 24h and subsequently stimulated with 20% FBS for the indicated times. Upper panel, WCE (20 μg) were analyzed by IB with pERK1/2 and ERK1/2 antibodies. Lower panel, fold increase in pErk1/2 was determined as described previously. Data represent ±standard deviation of duplicate of three independent experiments. (E) Cell populations were subjected to matrigel assay, and colony formation was observed for 10 days. Scale bar: left panel, 300 μm; right panel 50 μm. Colony appearance for Src*/SD and EV cells is depicted. Relative numbers of colonies with rough versus smooth boundaries are given for each cell population. Black bars, colonies with smooth boundaries; white bars, colonies with rough boundaries. Data represent ±standard deviation of duplicate of three independent experiments. *P < 0.05. **P < 0.01.
Results
Src*/SD does not alter mammary ductal outgrowth
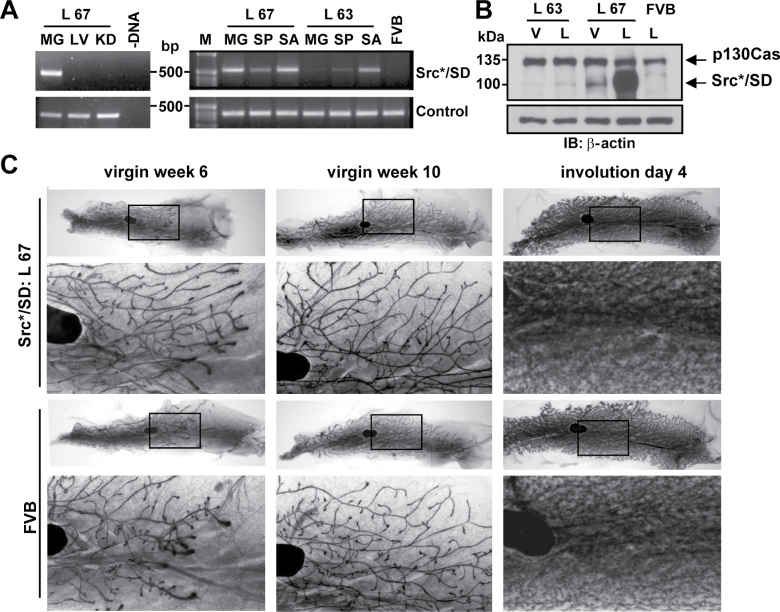
To develop a strategy for inhibiting oncogenic p130Cas signaling in vivo, the dominant interfering p130Cas molecule Src*/SD (10,11) and its potential side effects were investigated in a mouse model. Transgenic mice expressing Src*/SD under the hormonally regulated MMTV-long terminal repeat promoter were generated. Six founder mice carrying the transgene were identified by Southern blot analysis (data not shown). Transgene messenger RNA levels were determined by reverse transcription–PCR and IB analysis with CasB antibodies in mammary gland tissue of virgin and lactating mice (Figure 1A and B). Liver, kidney, salivary gland and spleen preparations were included as controls because the MMTV promoter is also active in other tissues than the mammary gland (30). Two lines (L 67 and L 63), displaying high and low expression levels, respectively, were chosen for further analysis. Hemizygous and homozygous Src*/SD mice appeared healthy, exhibited normal behavior, were fertile and the females were able to lactate and nurse their young. L 67 and L 63 mice were observed for up to 11 months of age and neither homozygous nor hemizygous, developed tumors.
Fig. 1.
Characterization of Src*/SD transgenic mice. (A) Messenger RNA expression analysis of the transgene in L 67 and L 63 by reverse transcription–PCR (top panel). MG, mammary gland (lactating); LV, liver; KD, kidney; SP, spleen; SA, salivary gland; FVB, FVB control mouse; –DNA, PCR control; control, ribosomal subunit S15 (bottom panel). DNA molecular weight markers (M) are indicated. (B) Protein expression in mammary gland tissue of virgin (V) and lactating (L) animals of L 67, L 63 and WT FVB mice analyzed by IB of whole-tissue lysates (20 μg) with CasB. β-actin is used as a loading control (bottom panel). (C) Mammary gland development in transgenic Src*/SD versus WT FVB animals. Whole-mount preparations of gland number 4 of virgin animals at 6 and 10 weeks and at day 4 of involution are presented. Insets are presented at higher magnification for each gland. Black region represents a lymph node.
The effects of the Src*/SD transgene on 6- and 10-week-old virgin mouse mammary gland ductal outgrowth, architecture and involution were examined using whole-mount analysis. Preparations of the virgin and involuting glands of FVB control mice and Src*/SD transgenic animals (L 67, Figure 1C; L 63, data not shown) displayed normal ductal architecture, including ductal elongation and development of the branching structure. At 10 weeks, the ducts filled the entire mammary fat pad, as expected. Together, these analyses revealed no differences in ductal architecture among the groups.
Mammary tumorigenesis is accelerated in female PyMT × Src*/SD mice
We next assessed the decoy function of the Src*/SD molecule in the c-Src-dependent PyMT transgenic mouse model, that was chosen, because c-Src kinase activity correlates with phosphorylated p130Cas in aggressive breast cancer cells (31). Moreover, tamoxifen resistance conferred by p130Cas has been linked to the physical interaction of c-Src and p130Cas (32). PyMT transgenic mice developed mammary gland tumors within 8–12 weeks (19). PyMT mice were crossed with the two Src*/SD transgenic mouse lines (L 67 and L 63, high and low expressing, respectively; Figure 1A and B). Mammary glands were palpated twice a week starting at 3 weeks of age until the development of mammary gland tumors was detected and the onset of palpable tumors for each mouse strain was plotted (Figure 2A). In the PyMT control group, palpable mammary tumors were detected the earliest at the age of 46 days and by 70 days in all mice. Fifty percent of these mice developed palpable breast tumors at 62 days. In the PyMT × Src*/SD (L 67) group, palpable mammary tumors developed as early as 28 days. By 35 days, 50% of the mice developed palpable tumors, and at 42 days, tumors were detectable in all of the mice. Low expression of Src*/SD in PyMT × Src*/SD (L 63) mice accelerated tumor induction only slightly (50% by 53 days). The log-rank test showed that the curves for L 67 and L 63 are significantly different with values of P ≤ 0.0001 and P ≤ 0.0072 (L 67 and L 63, respectively). Determination of the growth rate demonstrated that tumors grew significantly faster in the PyMT × Src*/SD (L 67) cohort as it took only 18 days (±2.7) from the time the tumors were palpable to reaching the size of 1cm compared with 23 (±5.9) and 24.5 days (±6.3) for the PyMT × Src*/SD (L 63) and PyMT cohorts, respectively.
Representative whole-mount preparations of mammary glands of PyMT and compound animals at day 52 are shown in Figure 2C. Lesion areas in PyMT × Src*/SD L 63 and L 67 animals were increased at this age compared with PyMT mice. Of note, L 67 mice showed multiple large lesion areas and developed tumor masses in the glands that had to be removed before whole-mount preparation was performed.
To examine whether the differences in the onset of tumor development in L 63 and L 67 correlate with the levels of Src*/SD, the expression was analyzed in tumor tissues. Tumors from L 67 and L 63 mice were harvested when they reached ~1cm in the largest dimension and subjected to IP and/or IB analysis with PyMT, Src*/SD, pTyr and HA-tag antibodies. PyMT was expressed at equal levels in the mammary gland tumors of both Src*/SD lines (Figure 2D and L 63 data not shown). Expression and phosphorylation of the Src*/SD protein were confirmed by IB in L 67 (Figure 2D). Because Src*/SD was expressed at very low levels in L 63 mammary gland tissue, IPs on tumor tissue lysates were performed. These assays revealed much higher levels of Src*/SD protein in L 67 tumors compared with L 63 tumors (Figure 2E, different protein amounts were used in the IPs as indicated), which reflects the transgene expression differences in L 67 versus L 63 single transgenic animals.
To further evaluate the mechanisms leading to the earlier onset of tumor development, the tyrosine phosphorylation profile on tumor samples of ~1cm within the largest dimension was analyzed by IB (Figure 2F and G). Compared with PyMT tumors, the L 67 and L 63 lesions displayed increased tyrosine phosphorylation of proteins ranging in size from 140 to 90 and 65 to 40kDa, which was more prominent in L 67.
These observations suggest that Src*/SD expression leads to activation of signaling pathways in the PyMT breast tumor model, which may contribute to the significantly earlier development of tumors compared with the PyMT animals.
Src*/SD promotes the development of more aggressive tumors at early stages of PyMT-mediated tumor development
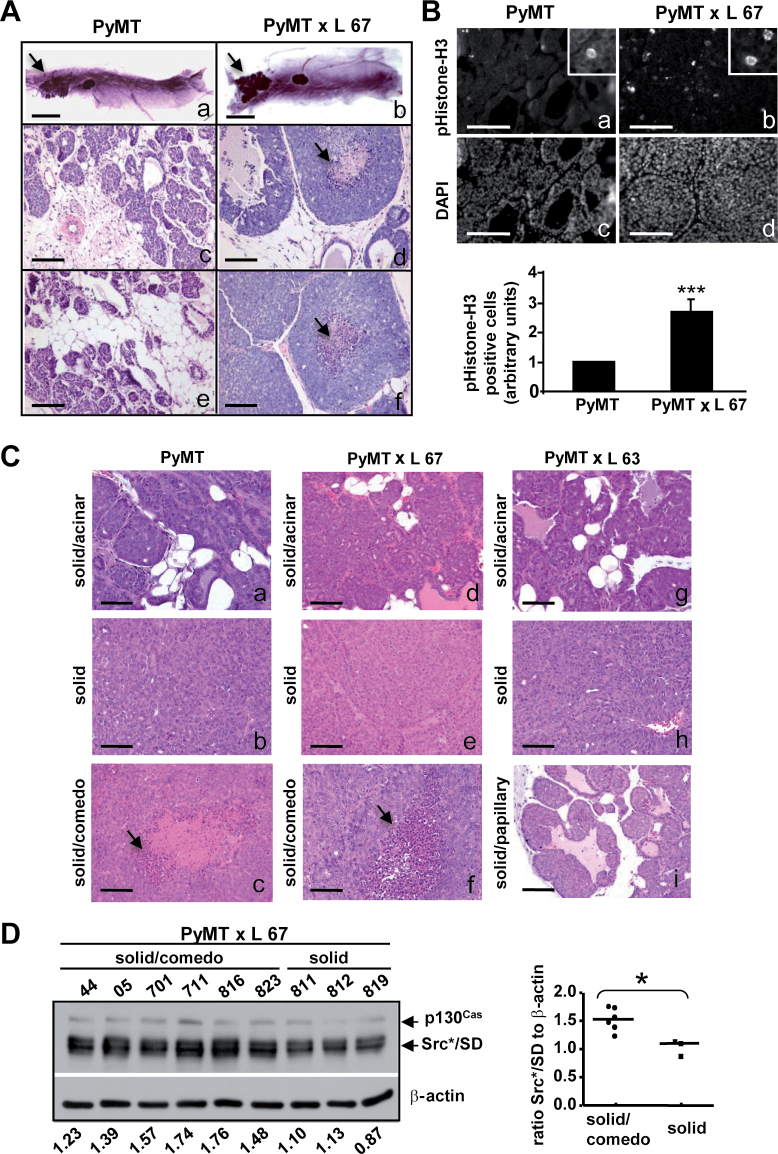
Due to the rapid tumor formation observed in the compound PyMT × Src*/SD (L 67) mice compared with PyMT mice, the effects of Src*/SD on earlier tumor formation were investigated. The inguinal mammary gland morphology and histologic structures in PyMT and PyMT × Src*/SD (L 67) mice at 4 weeks of age were examined (Figure 3A). In mammary gland whole-mount preparations from PyMT and L 67 mice, a single lesion developed close to the main collecting duct. Lesions of L 67 glands were denser but only slightly larger than the PyMT gland tumors potentially correlated with their earlier palpability (Figure 3A). Mammary tumor sections taken from the PyMT (n = 4 tumors) and PyMT × L 67 (n = 4 tumors) mice were analyzed for differences in morphology and mitotic index. Tumor histology was classified by a pathologist without knowledge of the sample genotype. PyMT × L 67 tumors were classified as ductal carcinoma in situ (DCIS) or solid/comedo type of tumors (Figure 3A, d and f) and all tumors contained pockets of necrosis. In contrast, PyMT mice examined developed less aggressive lesion types classified as hyperplasia or as carcinoma in situ (Figure 3A, c and e). The number of cells positive for pHistone-H3 in tumors formed by PyMT × L 67 animals were significantly higher (2.7-fold) compared with PyMT tumors (Figure 3B).
Fig. 3.
Comparison of tumor development in PyMT and PyMT × Src*/SD animals. (A) Whole-mount inguinal mammary glands isolated from 4-week-old PyMT and PyMT × Src*/SD (L 67) mice (a and b). Arrows indicate lesions. Hematoxylin and eosin-stained mammary gland tumor sections (c–f). Scale bars: 5mm (a and b) and 100 μm (c–f). (B) Nuclei of mammary gland tumors from 4-week-old PyMT and PyMT × Src*/SD (L 67) mice labeled by immunofluorescence for mitotic marker pHistone-H3 (a and b) and with DAPI (c and d). Insets show enlarged areas with stained cells. Scale bar: 100 μm (a–d). Quantification of tumor cells stained positive for pHistone-H3 (pH3; lower panel). ***P ≤ 0.001. (C and D) Mammary gland tumors were harvested at ~1cm in the largest dimension from PyMT and PyMT × Src*/SD (L 67 and L 63) mice. (C) Representative histological patterns of solid/acinar (panels a, d and g), solid (panels b, e and h), solid/comedo (panels c and f) and solid/papillary (panel i) identified for each line are shown. Arrows indicate necrotic core. Scale bar: 100 μm. (D) Src*/SD expression is higher in mammary gland tumors with comedo-type histology. Left panel, WCE (20 μg) were analyzed by IB with CasB and β-actin antibodies (loading control). Relative levels of Src*/SD for each tumor are listed on the bottom of the lanes. Right panel, comparison of Src*/SD levels in solid/comedo tumors versus solid tumors. *P ≤ 0.05.
To characterize late-stage carcinomas of PyMT × Src*/SD and PyMT animals, tumors were harvested when they each reached ~1cm in the largest dimension and analyzed by immunohistochemistry. Tumors from both PyMT × L 67 and PyMT mice were characterized as solid/acinar, solid and solid/comedo pathology types (Figure 3C). L 63 tumors were classified as solid/acinar, solid and solid/papillary types. When these late-stage comedo-type carcinoma sections were stained for pHistone-H3 and the Ki-67 proliferation markers, no substantial differences were observed between PyMT × L 67 and PyMT mammary tumors (Supplementary Figure 1, available at Carcinogenesis Online). To assess the relationship between the tumor pathology and the expression level of Src*/SD in L 67 animals, we examined the Src*/SD expression in solid/comedo and solid tumors by IB. Tumor samples from solid/comedo demonstrated higher levels of Src*/SD (P ≤ 0.05) compared with solid tumors (Figure 3D).
These results suggest that Src*/SD promotes the development of more aggressive tumors at relatively early stages of mammary tumor development when introduced into the PyMT mouse model particularly when high levels of the transgene are expressed.
Analysis of Src*/SD in PyMT cells in vitro
To better understand how the SD of p130Cas contributes to the malignant processes in PyMT × Src/SD mice, we studied these cells in vitro. Mammary carcinoma cells derived from PyMT mice (24) were stably transduced with the Dox-inducible HA-tagged Src*/SD and controls: Src* (attenuated Src kinase domain only), SrcKM/SD [SD fused to a kinase inactive (KM) Src kinase domain] and EV as described previously (11). At 48h of expression induction, cells were harvested and WCE were subjected to IB using HA, pTyr, PyMT and CasB antibodies. All the proteins were well expressed, and as expected, the Src*/SD was tyrosine phosphorylated, whereas the SrcKM/SD was not (Figure 4A and B). These cell populations were examined for their Erk1/2 activity and levels to test whether changes in this mitogen-activated protein kinase are in part responsible for the accelerated tumor development in PyMT × L 67 animals expressing high protein amounts of the transgene. Erk activity was investigated by IB as described previously. Src*/SD expression in PyMT cells significantly increased Erk1/2 phosphorylation (P ≤ 0.05), whereas no significant changes were noted with Src* and SrcKM/SD (Figure 4C). Furthermore, to examine the baseline of Erk1/2 phosphorylation and effects of serum stimulation on Erk1/2 activation, PyMT-EV and PyMT-Src*/SD cells were serum-starved for 24h and then stimulated with 20% FBS. A time course analysis of Erk1/2 activation in response to FBS treatment revealed a significant induction of Erk1/2 phosphorylation in PyMT-EV (P ≤ 0.05 at 15min), whereas no significant Erk1/2 activation was observed in PyMT-Src*/SD cells (Figure 4D). This can in part be explained by the finding that the baseline Erk1/2 activity in PyMT-Src*/SD cells was significantly increased to about 3-fold (P ≤ 0.05). An elevation of Erk1/2 phosphorylation was observed in Src*/SD cells compared with EV control cells at each time point.
To determine the effects of Src*/SD in the PyMT model on tumorigenicity in vitro, matrigel colony formation assays were performed using PyMT-Src*/SD, PyMT-Src*, PyMT-SrcKM/SD and PyMT-EV control cells (Figure 4E). Although all four cell populations formed colonies in matrigel after 10 days in culture, they exhibited different appearances ranging from rough to smooth. Quantification of three independent experiments indicated that PyMT-Src*/SD cells displayed 70% of rough and larger colonies, whereas EV, Src* and SrcKM/SD cells exhibited 60–70% of smooth and slightly smaller colonies. Because rough colonies are correlated with higher aggressiveness, these results further suggest that the phosphorylated SD may have an increased ability to promote the transformed phenotype in part by activation of Erk1/2 PyMT breast cancer cells.
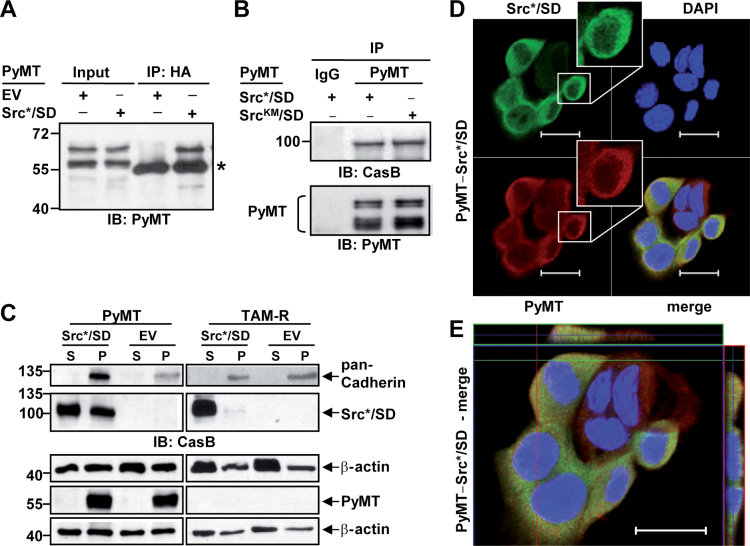
PyMT interacts with Src*/SD and targets it to membrane compartments
PyMT is a membrane-anchored protein, which has been found to interact with a diverse set of intracellular proteins (33–35). To identify a mechanism by which Src*/SD promotes the PyMT-mediated tumor development, we examined the possibility that Src*/SD and PyMT directly interact in cells. Extracts from PyMT cells expressing Src*/SD, SrcKM/SD or EV were immunoprecipitated with either HA, PyMT or mouse IgG antibodies. Subsequently, the immunoprecipitates were probed with PyMT antibodies. Using HA antibodies for IP of Src*/SD, PyMT was coimmunoprecipitated but not in EV control lysates (Figure 5A). These results revealed an association between PyMT and Src*/SD. PyMT antibodies immunoprecipitated both Src*/SD and SrcKM/SD (Figure 5B), indicating that Src*/SD interacts with PyMT in a manner that is independent of SD phosphorylation and the activity of the Src kinase domain. Thus, we hypothesized that the association of the Src*/SD with PyMT leads to the distribution of Src*/SD to membranes. Therefore, the cellular distribution of Src*/SD was investigated in WCE from PyMT-Src*/SD and PyMT-EV and control TAM-R-Src*/SD and TAM-R-EV cells by fractionation and IB analysis (Figure 5C). TAM-R cells were also included because Src*/SD alleviates the transformed phenotype of these breast cancer cells (11). The purity of the fractions was confirmed by the plasma membrane marker pan-Cadherin. PyMT was found only in the membrane fraction of PyMT cells. Supporting our hypothesis, in PyMT cells, a great portion of Src*/SD was found at membranes, whereas in TAM-R cells, only a small amount of Src*/SD was detected in this fraction (Figure 5C). This result was also confirmed by an alternative fractionation method as described previously in ref. 26 (Supplementary Figure 2, available at Carcinogenesis Online).
Fig. 5.
The phosphorylated Src*/SD interacts with PyMT in vitro and is targeted to membrane fractions. (A) WCE (500 μg) of PyMT-Src*/SD and PyMT-EV cells were subjected to IP with HA antibodies followed by IB with PyMT antibodies. *, heavy chain. (B) WCE (500 μg) of PyMT-Src*/SD and PyMT-SrcKM/SD cells were subjected to IP with PyMT antibodies followed by IB analysis with HA, CasB and PyMT antibodies. (C) Src*/SD is targeted to the membrane fraction in PyMT-expressing cells. WCE from PyMT and TAM-R cells expressing Src*/SD and EV were fractionated into cytoplasmic (S) and membrane/pellet (P) fractions as described previously in Materials and methods and analyzed by WB with pan-Cadherin, CasB, β-actin and PyMT antibodies. (D and E) Co-localization of PyMT and Src*/SD. Confocal analysis of immunofluorescence of DAPI, PyMT and Src*/SD in PyMT-Src*/SD cells was performed as described previously in Materials and methods. Bars 20 μM. (D) Insets show higher magnification of the indicated single cell. (E) Enlarged PyMT-Src*/SD cells with high-magnification z-axis images showing co-localization of Src*/SD and PyMT.
Moreover, the subcellular localization of PyMT and Src*/SD was determined using immunofluorescence staining and analyzed with a confocal laser scanning microscope (Figure 5D and E; Supplementary Figure 3, available at Carcinogenesis Online). The staining revealed a great overlap of PyMT and Src*/SD expression in 81.2% (±11.2%) of the double-positive cells particularly at intracellular punctuate areas and to some degree at the cell periphery, showing the co-localization of both proteins in these cells. Cell fractionation and immunofluorescence studies also showed that expression of Src*/SD does not alter the localization of PyMT (Figure 5D; Supplementary Figure 3A, available at Carcinogenesis Online).
Together these data indicate that PyMT binds to Src*/SD at the membrane most likely by interacting with the attenuated Src kinase domain of the Src*/SD chimera.
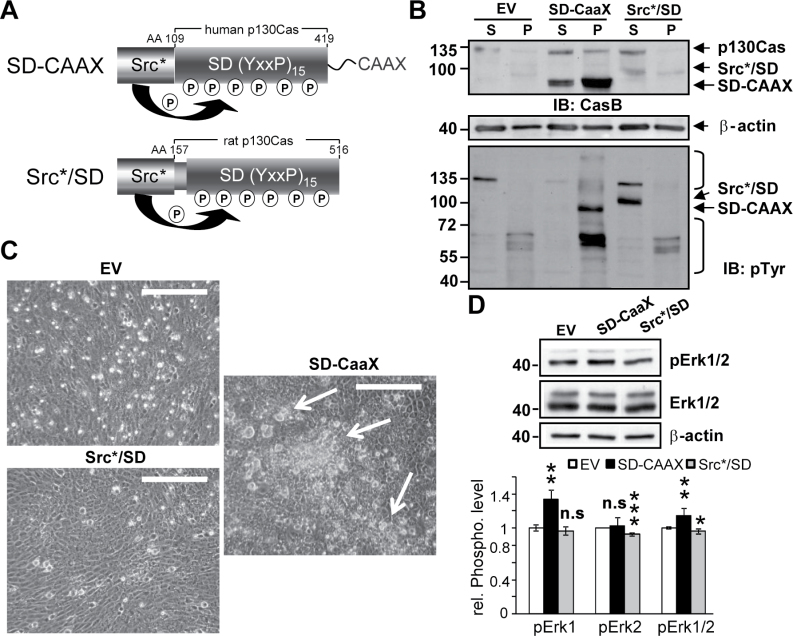
Targeting of the phosphorylated Src*/SD to membranes promotes a transformed phenotype in vitro
To further investigate the effects of Src*/SD membrane association on cellular transformation independent of PyMT, the H-Ras CAAX lipid anchor, essential for membrane localization of H-Ras (23), was fused to the C-terminus of Src*/SD (SD-CAAX) (Figure 6A). The Dox-inducible constructs for SD-CAAX, WT Src*/SD (SD-WT) and EV were transduced into 3Y1 rat fibroblast (Figure 6), which do not undergo spontaneous transformation. Cell fractionation studies confirmed that SD-CAAX is mostly found and phosphorylated in the membrane fraction, whereas SD-WT is predominantly expressed in the cytoplasmic fraction (Figure 6B). Moreover, SD-CAAX expression led to enhanced tyrosine phosphorylation of multiple proteins in the pellet fraction. To test for transforming properties of SD-CAAX, the transductants were assayed for focus formation (Figure 6C). Equal cell numbers of 3Y1-EV, 3Y1-SD-CAAX and 3Y1-SD-WT were seeded and analyzed by microscopy 5 days after seeding. Focus formation was only observed in 3Y1 cells expressing SD-CAAX. Erk activity was also investigated in 3Y1-EV, 3Y1-SD-CAAX and 3Y1-SD-WT cells by IB (Figure 6D). SD-CAAX expression in 3Y1 cells significantly increased total Erk1/2 phosphorylation (P ≤ 0.05) mediated mostly through enhanced Erk1 phosphorylation (P ≤ 0.01), whereas a slight reduction of Erk2 (P ≤ 0.001) was observed in SD-WT cells.
Fig. 6.
Targeting of the phosphorylated Src*/SD to membranes promotes a transformed phenotype in vitro. (A) Schematic of the Src*/SD constructs. AA, amino acids. CAAX, H-Ras lipid anchor. P, phosphorylation. (B–D) 3Y1 rat fibroblasts expressing the Dox-inducible constructs for EV, Src*/SD (SD-WT) and Src*/SD-CAAX (SD-CAAX) were analyzed after 2 days (B and D) and 5 days (C) of Dox induction (2 μg/ml). (B) Phosphorylated SD-CAAX is targeted to the membrane fraction in 3Y1 cells. WCE were fractionated into cytoplasmic (S) and membrane/pellet (P) fractions and analyzed by WB with CasB, β-actin and pTyr antibodies. Lower panel, brackets indicate proteins differentially tyrosine phosphorylated in EV, SD-CAAX and SD-WT cells. (C) Equal cell numbers (10 000) expressing the indicated constructs were seeded in 6-well plates and focus formation was analyzed by phase contrast microscopy after 5 days. Scale bars, 250 μm. (D) Upper panel, Erk1/2 phosphorylation was analyzed by IB in WCE (40 μg) of 3Y1 cells expressing the indicated constructs. Total ERK1/2 and β-actin antibodies were used as controls. Lower panel, the fold change in pErk1/2 was determined by densitometric analysis of two independent experiments setup in duplicates. n.s., not significant; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
These data suggest that recruitment of Src*/SD to membranes supports the loss of contact inhibition and thereby contributing to transformation.
Discussion
Our experiments examine the function of a constitutively phosphorylated SD (Src*/SD) of p130Cas in vivo. Here, we show that single MMTV-Src*/SD transgenic mice, which express endogenous p130Cas, are healthy, develop normal mammary glands and involution is normal as well. Tumor formation was not detected during an approximate 11 months observation period in single transgenic mice. Expression of the Src*/SD transgene in MMTV-PyMT mice accelerated tumor formation. Notably, all early developing tumors had histologic characteristics of aggressive comedo-type DCIS. We show evidence that this unexpected finding was potentially mediated by the recruitment of Src*/SD to membrane compartments in this model.
Src*/SD expression in vitro in v-crk transformed fibroblasts and in TAM-R attenuated the transformed phenotype (10,11). Importantly, cells that express lower levels of endogenous activated p130Cas (i.e. MCF-7 versus TAM-R cells) were less affected by Src*/SD (11). Accordingly, no adverse effects were noticed in single transgenic MMTV-Src*/SD mice expressing normal endogenous p130Cas levels. This is of special importance because the MMTV promoter is active also in some tissues other than the mammary gland (30,36). Therefore, these data indicate that the p130Cas SD decoy does not interfere with normal cellular functions in mice even when it is expressed at high levels.
Moreover, the effects of the decoy approach were investigated in vivo in the c-Src-dependent PyMT breast tumor model. PyMT is a membrane protein, with potent transforming activities due to activation of and interaction with numerous cellular proteins at membranes that when activated promote tumor formation (37). Unexpectedly, compared with PyMT single transgenic mice, PyMT × Src*/SD mice develop tumors more rapidly. High levels of Src*/SD gave rise to palpable tumors as early as just 4 weeks, whereas in single transgenic PyMT mice, palpable tumors were detected not before 7 weeks of age. Phosphorylated endogenous p130Cas exerts its activity mainly at cell membranes, specifically at focal adhesions, where it can interact with effectors such as c-Crk and activate the small guanosine triphosphatase Rac (1). Coupling of the phosphorylated SD to downstream effector molecules has been linked to cell migration and transformation (31). Thus, we hypothesize that Src*/SD increases the activation of signaling complexes at membranes in the PyMT × Src*/SD mice thereby promoting PyMT-induced tumor growth. Direct binding of c-Src to PyMT is mediated by the c-Src kinase domain (38), which is also present in Src*/SD. Importantly, c-Src mutants that lack the SH2 or SH3 domains associate better than WT c-Src with PyMT (38). The constitutive phosphorylation of the SD in Src*/SD was achieved by fusing an attenuated kinase domain (Src*) of c-Src, not containing the c-Src SH2 or SH3 domains, to the SD. Therefore, Src*/SD likely has a high affinity for the PyMT protein. Indeed, our results indicate that in the PyMT model, Src*/SD is recruited by PyMT to membranes, where it can activate signaling pathways. Moreover, we confirmed the transforming abilities of a membrane-targeted Src*/SD in 3Y1 rat fibroblasts by expressing a variant (SD-CAAX) containing a C-terminal H-Ras CAAX lipid anchor. We have shown that expression of Src*/SD in PyMT-transformed murine breast cancer cells and SD-CAAX in 3Y1 cells activates the mitogen-activated protein kinase/Erk signaling pathway. This is similar to the findings in p130Cas transgenic mice (7). Activated mitogen-activated protein kinase/Erk signaling promotes proliferation (39), correlating with the rapid tumor growth observed in PyMT × Src*/SD mice, in particular at early stages. Interestingly, these findings contrast our observations in TAM-R cells, where Src*/SD attenuates Erk1/2 phosphorylation and induces apoptosis (11). Potentially, the different Src*/SD functions are controlled by differing subcellular locations in the two systems. In the PyMT model, a substantial amount of Src*/SD translocates to membranes, whereas in the TAM-R cell system, it is primarily cytoplasmic, where it acts as decoy molecule. This is further supported by findings that MMTV-p130Cas transgenic mice develop extensive mammary epithelial hyperplasias (7) (activated full-length p130Cas translocates to focal adhesions), whereas mammary glands of our MMTV-Src*/SD mice are normal, even when Src*/SD is expressed at high levels. Together, these findings indicate that the activity of Src*/SD may depend strongly on its subcellular location. This also emphasizes the importance of the phosphorylated p130Cas SD specifically at cellular membranes in cancer progression and therefore underscore the importance of inhibiting p130Cas signaling in cancers with elevated/active p130Cas.
All of the early tumors of the PyMT × Src*/SD mice investigated at 4 weeks of age showed characteristics of comedo-type DCIS, whereas PyMT mice at that age had developed mainly hyperplasias or carcinomas in situ. Comedo-type DCIS are known to grow faster than the non-comedo types of DCIS (40,41) and confer the highest risk for progression into invasive carcinoma and/or recurrence (42). The enhanced proliferation in the PyMT × Src*/SD model measured by pHistone-H3 might account for the rapid development of the comedo-type tumors. Our results suggest that signaling pathways activated by the SD of p130Cas contribute to the more aggressive DCIS seen in PyMT × Src*/SD mice. Although unexpected, the PyMT × Src*/SD mice may represent an useful model for future studies of the development and progression of comedo-type DCIS, which represent about 40% of all DCIS cases (43).
Conclusion
We have elucidated the activity of a constitutively phosphorylated SD of p130Cas in vivo. We uncovered that when Src*/SD is targeted to membranes, it can contribute to PyMT-induced tumor growth. Moreover, because no other genetic mouse model for comedo DCIS exists, this double transgenic mouse model might represent a useful tool for studying comedo-type DCIS and for drug screening purposes targeting this aggressive premalignant lesion. Nevertheless, in single transgenic mice, this molecule does not interfere with normal functions and it also does not induce tumors by itself. Therefore, the development of inhibitors that block the interaction of the SD of endogenous p130Cas with SH2-containing binding partners might be feasible and open novel therapeutic approaches for breast cancer.
Supplementary material
Supplementary Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA106468 and CA143108 to K.H.K., CA096846 to A.H.B.); Department of Defense pre-doctoral fellowship (W81XWH-06-1-0379 to B-T.L.); Susan G. Komen for the Cure Breast Cancer Foundation postdoctoral fellowship (KG101208 to K.H.K. and J.K.).
Supplementary Material
Acknowledgements
The authors thank M.D.Layne for helpful comments and critical reading of the manuscript. We are grateful to A.de las Morenas for expert advice in the histological analysis of the tumor tissues and to V.Trinkaus-Randall for help with the confocal microscopy. We thank A.Belkina for generation of PyMT-stable cell lines. We gratefully acknowledge K.Ravid for providing the MMTV-Sv40-Bssk vector and R.Hasan for providing the PyMT tumor cell line, and S.Dilworth for providing the pAb762 PyMT antibody.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- cDNA
complementary DNA
- DAPI
4ʹ6-diamidino-2-phenylindole
- DCIS
ductal carcinoma in situ
- Dox
doxycycline
- Erk
extracellular signal-regulated kinase
- EV
empty vector
- FBS
fetal bovine serum
- HA
hemagglutinin epitope
- IB
immunoblotting
- IP
immunoprecipitation
- KM
kinase inactive
- MMTV
mammary tumor virus
- p130Cas
Crk-associated substrate
- pHistone-H3
phospho-Histone H3
- PyMT
polyoma middle T-antigen
- pTyr
phospho-tyrosine
- SD
substrate domain
- SD-CAAX
membrane-targeted variant of Src*/SD
- TAM-R
tamoxifen-resistant breast cancer cells
- WCE
whole-cell extracts
- WT
wild-type.
References
- 1. Cabodi S., et al. (2010). Integrin signalling adaptors: not only figurants in the cancer story. Nat. Rev. Cancer, 10, 858–870 [DOI] [PubMed] [Google Scholar]
- 2. Sakai R., et al. (1994). Characterization, partial purification, and peptide sequencing of p130,the main phosphoprotein associated with v-Crk oncoprotein. J. Biol. Chem., 269, 32740–32746 [PubMed] [Google Scholar]
- 3. Tikhmyanova N., et al. (2010). CAS proteins in normal and pathological cell growth control. Cell. Mol. Life Sci., 67, 1025–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouton A.H., et al. (2001). Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene, 20, 6448–6458 [DOI] [PubMed] [Google Scholar]
- 5. van der Flier S., et al. (2000). BCAR1/p130Cas expression in untreated and acquired tamoxifen-resistant human breast carcinomas. Int. J. Cancer, 89, 465–468 [DOI] [PubMed] [Google Scholar]
- 6. Konstantinovsky S, et al. (2010) Breast carcinoma cells in primary tumors and effusions have different gene array profiles. J. Oncol., 2010, 969084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabodi S., et al. (2006). p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res., 66, 4672–4680 [DOI] [PubMed] [Google Scholar]
- 8. Kumbrink J., et al. (2011). Targeting Cas family proteins as a novel treatment for breast cancer . In Gunduz E., Gunduz M. (eds) Breast Cancer - Current and Alternative Therapeutic Modalities. InTech, Rijeka, Croatia. ISBN: 978-953-307-776-5, pp. 37–62 [Google Scholar]
- 9. Klemke R.L., et al. (1998). CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol., 140, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirsch K., et al. (2002). A p130Cas tyrosine phosphorylated substrate domain decoy disrupts v-crk signaling. BMC Cell Biol., 3, 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soni S., et al. (2009). Expression of a phosphorylated p130(Cas) substrate domain attenuates the phosphatidylinositol 3-kinase/Akt survival pathway in tamoxifen resistant breast cancer cells. J. Cell. Biochem., 107, 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piwnica-Worms H., et al. (1987). Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c-src. Cell, 49, 75–82 [DOI] [PubMed] [Google Scholar]
- 13. Kmiecik T.E., et al. (1987). Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell, 49, 65–73 [DOI] [PubMed] [Google Scholar]
- 14. Knowlden J.M., et al. (2003). Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology, 144, 1032–1044 [DOI] [PubMed] [Google Scholar]
- 15. Kumbrink J., et al. (2012). Regulation of p130(Cas)/BCAR1 expression in tamoxifen-sensitive and tamoxifen-resistant breast cancer cells by EGR1 and NAB2. Neoplasia, 14, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hiscox S., et al. (2006). Elevated Src activity promotes cellular invasion and motility in tamoxifen resistant breast cancer cells. Breast Cancer Res. Treat, 97, 263–274 [DOI] [PubMed] [Google Scholar]
- 17. Ichaso N., et al. (2001). Cell transformation by the middle T-antigen of polyoma virus. Oncogene, 20, 7908–7916 [DOI] [PubMed] [Google Scholar]
- 18. Treisman R., et al. (1981). Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature, 292, 595–600 [DOI] [PubMed] [Google Scholar]
- 19. Lin E.Y., et al. (2003). Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol., 163, 2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guy C.T., et al. (1994). Activation of the c-Src tyrosine kinase is required for the induction of mammary tumors in transgenic mice. Genes Dev., 8, 23–32 [DOI] [PubMed] [Google Scholar]
- 21. Min C., et al. (2007). The tumor suppressor activity of the lysyl oxidase propeptide reverses the invasive phenotype of Her-2/neu-driven breast cancer. Cancer Res., 67, 1105–1112 [DOI] [PubMed] [Google Scholar]
- 22. Kumbrink J., et al. (2013). p130Cas acts as survival factor during PMA-induced apoptosis in HL-60 promyelocytic leukemia cells. Int. J. Biochem. Cell Biol., 45, 531–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hart K.C., et al. (1997). Derivatives of activated H-ras lacking C-terminal lipid modifications retain transforming ability if targeted to the correct subcellular location. Oncogene, 14, 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hulit J, et al. (2007) N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res., 67, 3106–3116 [DOI] [PubMed] [Google Scholar]
- 25. Bouton A.H, et al. (1997) Detection of distinct pools of the adapter protein p130CAS using a panel of monoclonal antibodies. Hybridoma, 16, 403–411 [DOI] [PubMed] [Google Scholar]
- 26. Zhao Y, et al. (2009) The lysyl oxidase pro-peptide attenuates fibronectin-mediated activation of focal adhesion kinase and p130Cas in breast cancer cells. J. Biol. Chem., 284, 1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirsch K.H, et al. (1998) Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G. J. Biol. Chem., 273, 25673–25679 [DOI] [PubMed] [Google Scholar]
- 28. Patwardhan P, et al. (2010) Myristoylation and membrane binding regulate c-Src stability and kinase activity. Mol. Cell. Biol., 30, 4094–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Min C, et al. (2009) A loss-of-function polymorphism in the propeptide domain of the LOX gene and breast cancer. Cancer Res. 69 6685–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ross S.R., et al. (1990). Negative regulation in correct tissue-specific expression of mouse mammary tumor virus in transgenic mice. Mol. Cell. Biol., 10, 5822–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cunningham-Edmondson A.C., et al. (2009). p130Cas substrate domain signaling promotes migration, invasion, and survival of estrogen receptor-negative breast cancer cells. Breast Cancer, 2009, 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riggins R.B., et al. (2006). Physical and functional interactions between Cas and c-Src induce tamoxifen resistance of breast cancer cells through pathways involving epidermal growth factor receptor and signal transducer and activator of transcription 5b. Cancer Res., 66, 7007–7015 [DOI] [PubMed] [Google Scholar]
- 33. Cheng J., et al. (2009). Cellular transformation by simian virus 40 and murine polyoma virus T antigens. Semin. Cancer Biol., 19, 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fluck M.M., et al. (2009). Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev., 73, 542–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winberry L.K., et al. (1985). Transformation by polyoma ts-a mutants. I. Characterization of the transformed phenotype. Virology, 144, 433–447 [DOI] [PubMed] [Google Scholar]
- 36. Stewart T.A., et al. (1988). Multiple regulatory domains in the mouse mammary tumor virus long terminal repeat revealed by analysis of fusion genes in transgenic mice. Mol. Cell. Biol., 8, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gottlieb K.A., et al. (2001). Natural biology of polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev., 65, 288–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dunant N.M, et al. (1996) Polyomavirus middle-T antigen associates with the kinase domain of Src-related tyrosine kinases. J. Virol., 70, 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 40. Jaffer S., et al. (2002). Histologic classification of ductal carcinoma in situ. Microsc. Res. Tech., 59, 92–101 [DOI] [PubMed] [Google Scholar]
- 41. Meyer J.S. (1986). Cell kinetics of histologic variants of in situ breast carcinoma. Breast Cancer Res. Treat., 7, 171–180 [DOI] [PubMed] [Google Scholar]
- 42. Fisher E.R., et al. (2007). Pathologic variables predictive of breast events in patients with ductal carcinoma in situ. Am. J. Clin. Pathol., 128, 86–91 [DOI] [PubMed] [Google Scholar]
- 43. Silverstein M.J., et al. (1996). A prognostic index for ductal carcinoma in situ of the breast. Cancer, 77, 2267–2274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.