Abstract
Skin cancer is the most common form of cancer in the USA, with an estimated two million cases diagnosed annually. Tumor progression locus 2 (Tpl2), also known as MAP3K8, is a serine/threonine protein kinase in the mitogen-activated protein kinase signal transduction cascade. Tpl2 was identified by our laboratory as having a tumor suppressor function in skin carcinogenesis, with the absence of this gene contributing to heightened inflammation and increased skin carcinogenesis. In this study, we used gene expression profiling to compare expression levels between Tpl2 +/+ and Tpl2 − /− keratinocytes. We identified over 2000 genes as being differentially expressed between genotypes. Functional annotation analysis identified cancer, cell growth/proliferation, cell death, cell development, cell movement and cell signaling as the top biological processes to be differentially regulated between genotypes. Further microarray analysis identified several candidate genes, including Mmp1b, Mmp2, Mmp9 and Mmp13, involved in migration and invasion to be upregulated in Tpl2 − /− keratinocytes. Moreover, Tpl2 −/− keratinocytes had a significant downregulation in the matrix metalloproteinase (MMP) inhibitor Timp3. Real-time PCR validated the upregulation of the MMPs in Tpl2 −/− keratinocytes and zymography confirmed that MMP2 and MMP9 activity was higher in conditioned media from Tpl2 −/− keratinocytes. Immunohistochemistry confirmed higher MMP9 staining in 12-O-tetradecanoylphorbol-13-acetate-treated skin from Tpl2 −/− mice and grafted tumors formed from v-rasHa retrovirus-infected Tpl2 −/− keratinocytes. Additionally, Tpl2 −/− keratinocytes had significantly higher invasion, malignant conversion rates and increased endothelial cell tube formation when compared with Tpl2 +/+ keratinocytes. In summary, our studies reveal that keratinocytes from Tpl2 −/− mice demonstrate a higher potential to be invasive and metastatic.
Summary
Tpl2has a tumor suppressor function in skin carcinogenesis. Moreover, using microarray complemented with cellular assays we identified a role for Tpl2 in skin cancer progression and demonstrated the absence of the gene contributes to increased invasive and metastatic potential.
Introduction
Tumor progression locus 2 (Tpl2), also known as MAP3K8, is a serine/threonine kinase in the mitogen-activated protein kinase signal transduction cascade (1). As a MAPKKK family member, activation of Tpl2 by inflammatory stimuli allows Tpl2 to dissociate from its ternary complex with nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 [nuclear factor κB (NFκB1)] and A20-binding inhibitor of NFκB2 (ABIN-2) (2). This renders Tpl2 free to activate its downstream target MAPK/ERK kinase and stimulate the extracellular signal-regulated MAP kinase (ERK) signal transduction pathway. Additionally, Tpl2 activation can stimulate other downstream pathways including p38, JNK, ERK1/2 and ERK5 (3–5).
The Tpl2/MAP3K8 gene was initially isolated from a human thyroid carcinoma cell line and was first characterized by its ability to cause morphological transformation of NIH3T3 and SHOK cells (6). An oncogenic role forTpl2/MAP3K8 has been well established, as numerous reports have found elevated MAP3K8 activity in a number of human cancers including breast, endometrial, thymomas, lymphomas, lung, Hodgkin’s disease and nasopharyngeal carcinoma (3,7–10). However, Tpl2 can act as a tumor suppressor in several cancer models (11–15). We have previously reported that Tpl2 −/− mice develop significantly more chemically induced skin tumors than wild-type (WT) mice, with the increased tumorigenesis associated with an upregulation in NFκB, inflammatory cytokines and prostanoid signaling (11,12). However, it is unknown whether the skin tumors arising in Tpl2 −/− mice are more likely to be invasive and metastatic than skin tumors that develop in WT mice.
The extracellular matrix (ECM) is a network of protein and polysaccharide macromolecules that provide structural support for cells (16). Degradation of the ECM is an important event for development, remodeling, morphogenesis and repair (17). Under normal physiological conditions, the ECM is degraded in a highly regulated fashion, but when dysregulated it can facilitate invasion of cancer cells to surrounding tissues (16). Several types of proteinases are implicated in ECM degradation, but the key enzymes are considered to be a family of 23 zinc-dependent endopeptidases known as matrix metalloproteinases (MMPs) (18).
Historically, MMPs are classified into six categories based on their preferred ECM substrate: gelatinases, collagenases, matrilysins, stromelysins, membrane type and other MMPs (19). Expression of MMPs is a highly controlled process including regulation at the level of gene transcription, enzyme activation and through inhibition by soluble factors known as tissue inhibitors of metalloproteinases (TIMPs) (20). Basal expression of MMPs is typically low or absent in normal tissues and can be upregulated by various factors including inflammatory cytokines, growth factors, hormones or contact with components of the ECM (19). MMPs are secreted as inactive precursors and become activated upon exogenous or autocatalytic cleavage of the N-terminal propeptide domain (19). Additionally, the proteolytic activity of the MMPs is carefully regulated by the inhibitory effects of endogenous TIMPs.
Disruption of the MMP/TIMP balance through an upregulation in MMPs or downregulation in TIMPs can lead to all aspects of cancer cell progression, playing a role in invasion, metastasis and angiogenesis (21). Among the many MMPs, the gelatinase MMP9 and the collagenases MMP1 and MMP13 have received considerable attention for their role in cancer progression (18,22–25). Elevated levels of MMP9 are documented in melanoma, breast, brain, ovarian, pancreatic, colorectal, bladder, prostate and lung cancers, as well as in several leukemias and lymphomas (21). In early stages of cancer, MMP9 can activate growth factors, help evade apoptosis and release angiogenic factors (17). In later stages of cancer, MMP9 is associated with tumor aggressiveness and poor patient prognosis (21). Similar to MMP9, overexpression of the collagenases MMP1 and MMP13 is associated with many cancer types, including lung, breast and melanoma, and often correlates with a poor clinical prognosis (26–32). These two collagenases are also associated with early metastasis (27,29).
In the present study, we used gene expression profiling to compare expression levels between Tpl2 +/+ and Tpl2 −/− keratinocytes, specifically focusing on markers of invasion and metastasis. We found upregulation of numerous MMPs in Tpl2 −/− keratinocytes, both basally and upon stimulation with 12-O-tetradecanoylphorbol-13-acetate (TPA). This upregulation was confirmed through quantitative PCR (qPCR) and zymography. Additionally, we found downregulation of the MMP endogenous inhibitor Timp3. Moreover, Tpl2 −/− keratinocytes had increased invasion, endothelial cell tube formation and malignant conversion as well as altered migration rates. Taken together, we postulate a role for Tpl2 in mouse skin carcinogenesis with the absence of this gene contributing to an increased invasive and metastatic potential.
Materials and methods
WT and transgenic mice
Male and female WT (Tpl2 +/+) and knockout (Tpl2 −/−) C57BL/6 mice were engineered as described previously (7). Tpl2 +/+ WT control mice were generated from the same colony as the Tpl2 −/− mice. All mice were bred and maintained at an National Institutes of Health animal facility (Bethesda, MD). Tpl2 −/− status was regularly confirmed by PCR. All animal work was performed following National Institutes of Health guidelines under an approved animal protocol.
Microarray analysis
Primary keratinocytes were isolated from Tpl2 −/− and Tpl2 +/+ mice pups at 1–2 days of age according to standard protocols (33). Primary keratinocytes were treated with 10 ng/ml TPA for 1 or 4 h or received dimethyl sulfoxide as a vehicle control. RNA was extracted, isolated and purified from keratinocytes using Qiagen RNeasy kit per manufacturer’s instructions (Qiagen, Valencia, CA). Samples were DNase-treated during purification to remove any genomic DNA contamination. Pure RNA was resuspended in RNase-free ultrapure water and RNA concentrations were determined using a GE NanoVue Spectrophotometer (GE Healthcare Biosciences, Pittsburgh, PA). RNA quality and concentration were confirmed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) prior to microarray analysis.
Twenty-four Affymetrix microarray chips were used for microarray analysis corresponding to two genotypes (Tpl2 +/+ and Tpl2 −/−), three treatments (no TPA, 1 h TPA and 4 h TPA) and four replicates. Each microarray chip contained probesets to measure over 34 000 transcripts on a single array (GeneChip Mouse Genome 430 2.0; Affymetrix, Santa Clara, CA). Microarray experiments using the Affymetrix chips were performed by the Laboratory of Molecular Technology (National Cancer Institute, Frederick, MD) and microarray data analysis was conducted by the Center for Cancer Research Bioinformatics Core (CCRIFX, National Cancer Institute, Bethesda, MD). Principal components analysis was performed to confirm that experimental replicates clustered together. The intensity values were then transformed using the binary logarithm (log2-transformation) to normalize intensity values and replicates for each experimental group were averaged.
Analysis of the microarray gene expression data was performed through Partek® Genomic Suite™ (PGS, v. 6.12.0712; Partek, St Louis, MO). PGS Gene Expression workflow was used to identify differentially expressed genes (DEGs) in all comparisons. Fold change (FC) values were calculated as a ratio of average gene expression in each condition. Based on previous work, a list of Mmps and Timps related to skin cancer was extracted and further selected for Mmps and Timps with absolute FC ≥1.8 in at least one comparison. Heatmaps of genes were produced with R heatmap.2 script (v. 2.15.0; R Foundation for Statistical Computing, Vienna, Austria) and clustered on genes.
Quantitative PCR
qPCR was used to determine Mmp1b, Mmp2, Mmp9, Mmp13, Timp1, Timp2, Timp3 and Timp4 expression levels. For qPCR analysis, the SuperScript® III Platinum® SYBR® Green One-Step qPCR Kit w/ROX was used per manufacturer’s instructions (Invitrogen, Carlsbad, CA) using 200 ng of purified total RNA as template for amplifications. qPCR was performed on an Mx3005P QPCR System (Agilent Technologies). Threshold cycle values were calculated with the MxPro QPCR software (v. 4.10), using the Pfaffl method and dissociate curves for each reaction were checked to confirm that only a single PCR product was obtained (34). Threshold cycle values were normalized with β-actin, and relative expression was calculated comparing differences between Tpl2 +/+ and Tpl2 −/− samples. An unpaired t-test between triplicates was used to determine level of significance. All experiments were repeated a minimum of three times. The specific oligonucleotide primers were purchased from Invitrogen for the sequences provided below:
| Primer | Forward sequence | Reverse sequence |
|---|---|---|
| Mmp1b | TGCTATAATTACATATCGGGGG | CATCGATCAAAGGTTCTGGC |
| Mmp2 | CCTTAAAAGTATGGAGCGACGTCA | AGCGTTCCCATACTTTACGCG |
| Mmp9 | ACCCTGTGTGTTCCCGTTCAT | GATACTGGATGCCGTCTATGTCGT |
| Mmp13 | TGATGAAACCTGGACAAGCA | GGTCCTTGGAGTGATCCAGA |
| Timp1 | CCAGAACCGCAGTGAAGAGT | TCTCCAAGTGCACAAGCCTA |
| Timp2 | GAGCGAGAAGGAGGTGGATT | GGTTCAGGCTCTTCTTCTGG |
| Timp3 | TTGGCACTCTGGTCTACACT | GTCCTGTGTACATCTTGCCT |
| Timp4 | GCCAGATTCTCAGTGATGGA | TGGTACATGGCACTGCATAG |
| Actb | GGCTACAGCTTCACCACCAC | GGCTACAGCTTCACCACCAC |
Grafting cells on athymic nude mice
Tpl2 −/− or WT primary keratinocytes isolated from newborn mice were infected with the v-rasHa retrovirus, then trypsinized and used for grafting on day 8 as described previously (11). Briefly, a graft site was prepared on the backs of 24 eight-week-old athymic nude mice. Mice received six million keratinocytes (either WT or Tpl2 −/−) mixed with six million fibroblasts (either WT or Tpl2 −/−). Tumor measurements began 10 days after grafting and continued once a week for 28 days. Tumors were collected, fixed in 10% normal-buffered formalin overnight and processed into paraffin blocks from which 4 μm sections were cut and stained with hematoxylin and eosin.
Immunohistochemistry
Immunohistochemistry was performed on grafted tumors (n = 6/group) and skin sections from mice receiving TPA treatment for 24 h or acetone vehicle controls (n = 4/group). Slides were rehydrated followed by antigen retrieval and endogenous peroxidase activity was quenched using methanol and hydrogen peroxide. Following washing and protein block (DAKO, Carpinteria, CA), the slides were incubated overnight at 4°C with rabbit anti-mouse MMP9 (1:100; Abcam, Cambridge, MA). Following washing, the slides were incubated for 40 min at room temperature with anti-rabbilt secondary antibody (Cell Signaling, Danvers, MA). ABC reagent (Vector Labs, Burlingame, CA) was then applied for amplification of primary antibody binding and 3,3′-diaminobenzidine was applied for visualization. Sections were melanin-bleached, and counterstained with Gill’s hematoxylin. The sections were dehydrated through graded alcohols, immersed in xylene and mounted with coverslips. Representative areas were photographed using an Eclipse E800 digital camera (Nikon, Melville, NY) at ×20 magnifications.
In vitro scratch/wound healing assay
Primary keratinocytes were isolated from Tpl2 −/− and Tpl2 +/+ mice pups at 1–2 days of age and infected with the v-rasHa retrovirus for 48 h as described previously (33). Keratinocytes were grown until fully confluent in 24-well ImageLock plates (Essen BioScience, Ann Arbor, MI). Mitomycin C (Sigma–Aldrich, St Louis, MO) was added at a final concentration of 10 µg/ml 2 h prior to scratching. Scratches were made using an Essen 24-well wound maker. Dishes were incubated in an Essen IncuCyte Live Cell Imaging System contained within a 36°C incubator with 7% CO2. The IncuCyte system photographed each scratched well under phase-contrast microscopy every 4 h for 48 h, and measurements of the percent of wound closure were calculated using IncuCyte software. All treatments were performed in quadruplicate and replicated a minimum of three times.
Zymography
To measure Mmp2 and Mmp9 enzymatic activity, we performed zymography using 10% Tris-Glycine gels containing 0.1% gelatin (Invitrogen). Total protein in conditioned media from Tpl2 +/+ and Tpl2 −/− keratinocytes infected with a replication-defective v-rasHa retrovirus, treated with 10 ng/ml TPA, or infected and treated with TPA was measured using a BCA Protein Assay Kit (Thermo Scientific, Rockland, IL). Fifteen micrograms of protein sample was loaded into each well, along with 2X loading buffer free of β-mercaptoethanol, and gels electrophoresed at 125 V for 90 min in Tris-Glycine/sodium dodecyl sulfate running buffer. After renaturation to quench sodium dodecyl sulfate and allow the MMPs to regain their tertiary structure and enzymatic activities, developing solution was added, gels were washed and stained using Invitrogen SimplyBlue™ SafeStain (Invitrogen) followed by destaining in deionized water. Invitogen Novex® Sharp Protein Standard and MMP2- and MMP9-positive controls (EMD Millipore, Billerica, MA) were included on all gels. Bands were quantified using National Institutes of Health ImageJ (v.1.44) (35). Zymography experiments were repeated a minimum of three times.
Invasion assay
Invasion was measured using the BD BioCoat Tumor Invasion System (BD Biosciences, San Jose, CA) as specified by the manufacturer. Briefly, 2.5×104 Tpl2 +/+ and Tpl2 −/− keratinocytes infected with a replication-defective v-rasHa retrovirus or receiving dimethyl sulfoxide as a vehicle control were seeded in 500 μl of serum-free Invitrogen low calcium (0.05 mM) Dulbecco’s modified Eagle’s medium (DMEM) media in triplicate. Cells were seeded into the apical chambers of Matrigel-coated BD Falcon Fluoroblok 24-Multiwell plates and 750 μl of DMEM + 10% was added to the basal chambers as a chemoattractant. Invasive cells passed through an 8.0 μm pore size polyethylene terephthalate membrane that was coated with BD Matrigel matrix. Twenty-two hours later, inserts were removed from plates, transferred to a new 24-well plate containing 500 μl/well of 4 μg/ml calcein AM, incubated for 1 h at 37°C and 5% CO2 and fluorescence measured at 494/517nm in a filtermax F5 microplate reader (Molecular Devices, Sunnyvale, CA).
In vitro conversion assay
An in vitro conversion assay was performed as described by Morgan et al. (36). In brief, primary keratinocytes were plated in eighteen 60 mm plates for each genotype, infected with v-rasHa retrovirus for 48 h and grown in Invitrogen low calcium (0.05 mM) DMEM media. After 1 week, the media was changed to high calcium (0.5 mM) DMEM media. Media was changed twice per week for 12 weeks until the experiment was terminated. After 12 weeks, dishes were fixed, and foci were stained with rhodamine in 10% formalin, microscopically photographed and quantified.
In vitro endothelial cell tube formation assay
The tube formation assay was performed as described previously (37). Briefly, 3B11 murine endothelial cells were serum-starved in DMEM with 0.2% fetal bovine serum for 24 h. Growth factor reduced Matrigel™ (250 μl; BD Biosciences) was added to a 24-well plate and allowed to set. One hundred thousand 3B11 endothelial cells were mixed with conditioned media from either Tpl2 −/− or Tpl2 +/+ keratinocytes (amount of conditioned media was normalized to keratinocyte cell number), layered on the solidified Matrigel™, then incubated in a 37°C incubator with 5% CO2 overnight. Wells were examined for tube formation 18 h later with a Zeiss Axiovert microscope (Thornwood, NY) and the number of total branch sites (control- and v-rasHa retrovirus-infected keratinocytes) or the number of branch sites in five fields of view (in vitro conversion samples) were quantified. Samples were plated in triplicate and experiments were repeated a minimum of three times.
Statistical analysis
Microarray, qPCR and invasion data were analyzed through t-tests. Migration data were analyzed through t-tests and non-parametric Wilcoxon rank-sum tests. A simple Bonferroni correction using an adjusted threshold P value of 0.002 was used to correct for multiple comparisons.
Results
Microarray analysis of primary keratinocytes from Tpl2 −/− mice identifies upregulated genes involved in cancer, cell movement, cell death and inflammation
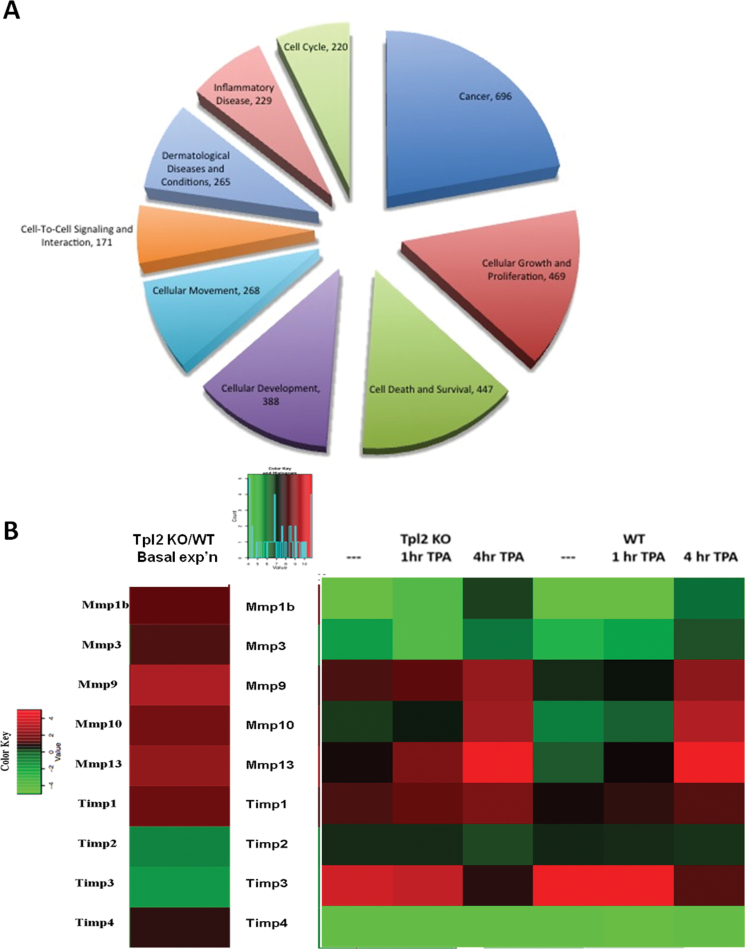
We performed microarray analysis on untreated or TPA-treated keratinocytes from Tpl2 −/− or Tpl2 +/+ mice to determine differentially regulated genes between genotypes. After normalizing data and eliminating weakly expressed genes, the expression ratio of each gene was calculated and gene expression profiles were compared between genotypes. Only the DEGs with a false discovery rate ≤0.05 and FC ≥1.8 were further analyzed to ensure accuracy of results. This filtering criteria identified 2191 genes of the 34 000 on the chip as being either significantly up- or downregulated in at least one of the samples compared with baseline expression in Tpl2 +/+ cells at 0 h. Functional annotation analysis using Database for Annotation, Visualization and Integrated Discovery identified cell death, inflammatory response, transcription regulation, protease activity and phosphorylation among the top biological processes in the list of genes (38,39). IPA® (Ingenuity® Systems, Redwood City, CA) was used for further pathway analysis of top functional processes, including cancer, cell growth/proliferation, cell death, cell development, cell movement, cell signaling, dermatological diseases and inflammation for the globally DEGs (Figure 1A).
Fig. 1.
Global microarray analysis of primary keratinocytes from Tpl2 −/− keratinocytes identifies upregulated genes involved in cancer, cell movement, cell death and inflammation. Log-transformed and median-centered gene expression from basal- or TPA-stimulated Tpl2 +/+ and Tpl2 −/− keratinocytes are represented. With a stringent filtering criteria of false discovery rate ≤0.05 and FC ≥1.8, 2191 genes of the 34 000 on the chip were identified as being either significantly up- or downregulated in at least one sample compared with a 0h, WT baseline. Of the globally DEGs, ingenuity pathway analysis used an input set of 1856 genes for analysis. The top functional pathways enriched are depicted as a pie chart (A). Select Mmps and Timps involved in skin carcinogenesis were analyzed for basal- (B, left) or TPA-stimulated (B, right) differences. Genes in green are downregulated, genes in red are upregulated and genes in black are neither down- nor upregulated.
Microarray analysis identifies upregulation in genes involved in migration and invasion in primary keratinocytes from Tpl2 −/− mice
Further microarray analysis identified candidate genes involved in migration and invasion. Multiple MMPs were upregulated basally in Tpl2 −/− keratinocytes compared with Tpl2 +/+ keratinocytes (Figure 1B). The most pronounced expression differences occurred in Mmp1b (1.5-fold), Mmp9 (3-fold) and Mmp13 (2.5-fold). TPA treatment increased MMP expression in both genotypes but remained significantly higher in Tpl2 −/− keratinocytes compared with Tpl2 +/+ keratinocytes (Figure 1B, right). Moreover, the expression of Timp3 was 1.8-fold lower in Tpl2 −/− keratinocytes.
Tpl2 −/− primary keratinocytes have elevated gene expression of MMPs and reduced expression of TIMP inhibitors
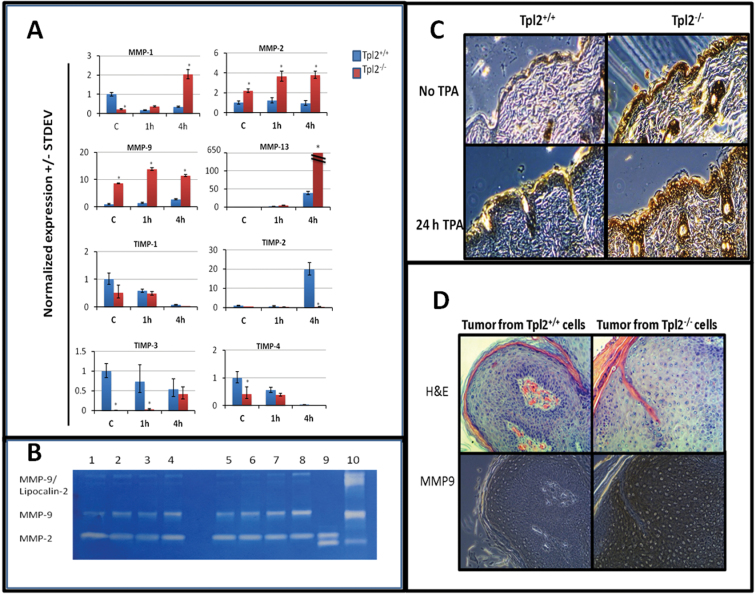
The microarray analysis was validated with qPCR for Mmp1b, Mmp9 and Mmp13 messenger RNA (mRNA) and their corresponding TIMP inhibitors (Figure 2A). Additionally, we analyzed gene expression of Mmp2 as this gelatinase is often upregulated in cancer and was determined to be higher in Tpl2 −/− keratinocytes in microarray samples, but was below the 1.8-fold cutoff. The qPCR results showed significant upregulation of basal levels of Mmp2 (2.1-fold) and Mmp9 (9-fold) in Tpl2 −/− keratinocytes. Mmp13 (3-fold), Mmp9 (9.8-fold), Mmp2 (3-fold) and Mmp1b (2-fold) mRNA remained significantly higher in Tpl2 −/− keratinocytes when TPA was administered for 1 h. Mmp13 (17-fold), Mmp9 (4.1-fold), Mmp2 (4-fold) and Mmp1b (5.8-fold) expression remained significantly higher in Tpl2 −/− keratinocytes after 4 h of TPA treatment. All results are significant at the P = 0.01 level.
Fig. 2.
Tpl2 −/− primary keratinocytes have elevated gene expression of MMPs, reduced expression of Timp inhibitors and increased levels of active Mmp9 in vivo and in vitro. (A) qPCR results for Mmp1, 2, 9 and 13 and Timp1, 2, 3 and 4. Total RNA from basal- or TPA-stimulated Tpl2 +/+ and Tpl2 −/− keratinocytes was used for qPCR analysis. Note the broken graph for Mmp13 due to the extremely high gene expression in Tpl2 −/− keratinocytes. Asterisks denote significance at the P ≤ 0.001 level. (B) Gelatin zymography for Mmp2/Mmp9. Protein from conditioned media was electrophoresed and analyzed for expression of active Mmp2 and Mmp9. Lane 1: Tpl2 +/+ control; Lane 2: Tpl2 +/+ v-rasHa; Lane 3: Tpl2 +/+ TPA; Lane 4: Tpl2 +/+ v-rasHa + TPA; Lane 5: Tpl2 −/− control; Lane 6: Tpl2 −/− v-rasHa; Lane 7: Tpl2 −/− TPA; Lane 8: Tpl2 −/− v-rasHa + TPA; Lane 9: Mmp2-positive control; Lane 10: Mmp9-positive control. (C) Immunohistochemistry of Mmp9 in TPA-treated skin. Shaved WT and Tpl2 −/− mice were treated with TPA (10 μg) for 0 or 24h and analyzed for expression of Mmp9. (D) Immunohistochemistry of Mmp9 in grafted tumors. Tumors developed on nude mice with v-rasHa-transduced Tpl2 +/+ (left) or Tpl2 −/− (right) keratinocytes mixed with fibroblasts from the same genotype were analyzed for Mmp9 expression. Magnification = ×20.
Additionally, qPCR confirmed reduced expression of the MMP inhibitors Timp1, Timp2, Timp3 and Timp4 in Tpl2 −/− keratinocytes (Figure 2A). Timp1 (2-fold) and Timp4 (2.5-fold) mRNA were reduced under basal conditions. Timp3 expression was reduced the most in untreated cells and was undetectable in Tpl2 −/− cells. Moreover, Timp3 mRNA was significantly higher (20-fold) in Tpl2 +/+ keratinocytes after 1 h of TPA treatment. Timp2 was significantly higher in Tpl2 +/+ keratinocytes following 4 h of TPA treatment (Figure 2A). Similar trends were observed in the microarray data.
Tpl2 −/− primary keratinocytes have higher levels of active Mmp9
We recapitulated the in vivo tumor responses from our previous studies by transducing keratinocytes with a replication-defective v-rasHa retrovirus (11,12). Gelatin zymography was then used to detect the enzymatic activity of Mmp2 and Mmp9. Tpl2 −/− primary keratinocytes infected with v-rasHa retrovirus and/or treated with TPA displayed higher Mmp2 and Mmp9 enzymatic activity when compared with Tpl2 +/+ keratinocytes. Basal expression of Mmp9 was 1.8-fold greater in Tpl2 −/− primary keratinocytes when compared with control cells (Figure 2B). v-rasHa, TPA or v-rasHa plus TPA all induced expression of Mmp9 in both genotypes, but to a larger extent in Tpl2 −/− primary keratinocytes, reaching levels of 1.9-, 3.6- and 2.9-fold higher, respectively as determined by densitometry using ImageJ. Mmp9 can also form a high molecular weight complex with lipocalin 2 (Lcn2) (40). Expression of the Mmp9/Lcn2 complex was similar among all treatment groups, except the v-rasHa plus TPA treatment group, and was 3.5-fold greater in Tpl2 −/− primary keratinocytes. Neither Tpl2 −/− nor Tpl2 +/+ primary keratinocytes had differences in MMP2 activity at basal levels, but upon treatment with TPA, the activity in Tpl2 −/− primary keratinocytes was 2-fold higher (Figure 2B).
Tumors and TPA-treated skin from Tpl2 −/− mice have higher expression of MMP9
We performed immunohistochemistry to correlate our in vitro results with the expression levels of Mmp9 in TPA-treated mouse skin and grafted tumors. Basal expression of Mmp9 in the epithelium of Tpl2 −/− mouse skin is notably higher than in WT mouse skin (Figure 2C). Upon treatment with TPA, the differences become even more pronounced. In a similar fashion, tumors developed on nude mice with v-rasHa-transduced Tpl2 −/− cells have markedly higher expression of Mmp9 when compared with tumors developed on nude mice with v-rasHa-transduced Tpl2 +/+ cells (Figure 2D).
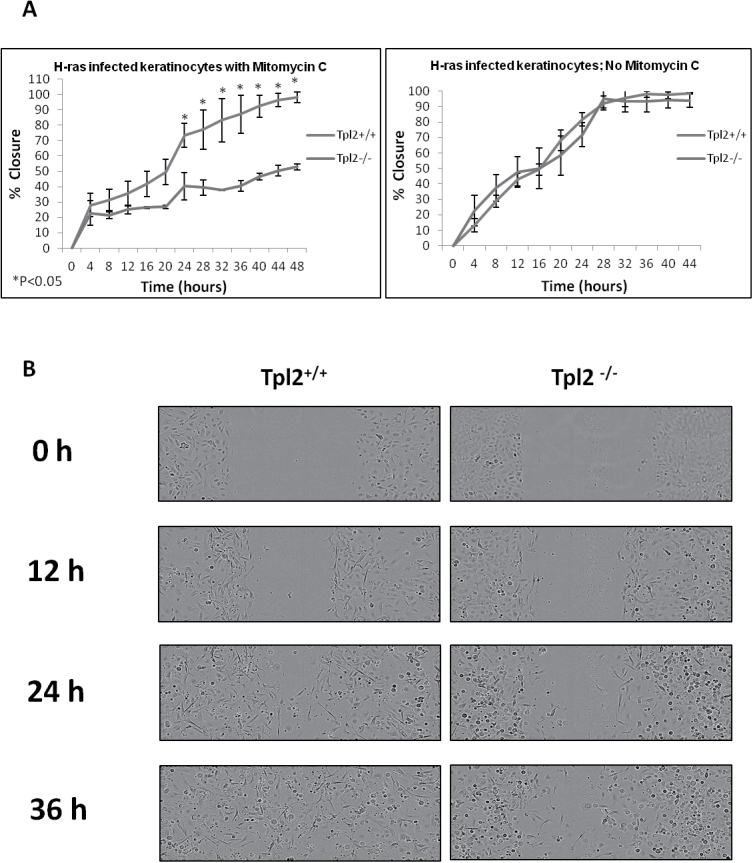
Tpl2 +/+ primary keratinocytes migrate faster than Tpl2 −/− primary cells in the presence of mitomycin C
A scratch assay was performed to assess migration differences between Tpl2 +/+ and Tpl2 −/− primary keratinocytes. Surprisingly, when treated with mitomycin C, an inhibitor of proliferation, Tpl2 +/+ cells migrated significantly faster than Tpl2 −/− cells, closing the wound within 48 h (Figure 3A, left panel; Figure 3B). In contrast, v-rasHa retrovirus-transduced Tpl2 −/− keratinocytes reached only 50% closure within 48 h (Figure 3A, left panel; Figure 3B). However, v-rasHa retrovirus-transduced Tpl2 +/+ and Tpl2 −/− keratinocytes did not have significantly different migratory rates when mitomycin C was not added (Figure 3A, right panel), suggesting when proliferation is not inhibited, wound healing occurs at a comparable rate due to the faster cell cycle of Tpl2 −/− keratinocytes (12).
Fig. 3.
Tpl2 +/+ primary keratinocytes migrate faster than Tpl2 −/− primary keratinocytes in the presence of mitomycin C. (A) Scratch test assay in v-rasHa retrovirus-infected Tpl2 +/+ and Tpl2 −/− primary keratinocytes treated with (left) or without (right) 10 μg/ml mitomycin C. Cells were photographed every 4h and closure calculated using IncuCyte software. Error bars were calculated using standard deviation from quadruplicate samples. Significance at P < 0.05 was determined through Student’s t-tests. (B) Scratch test assay in v-rasHa retrovirus-infected keratinocytes. Tpl2 +/+ keratinocytes (left panel) and Tpl2 −/− keratinocytes (right panel) were scratched, treated with 10 μg/ml mitomycin C and visualized using IncuCyte Live Cell Imaging System at ×10 magnification.
Tpl2 −/− primary keratinocytes infected with v-rasHa are more invasive than Tpl2 +/+ primary keratinocytes
v-rasHa retrovirus-infected or vehicle-treated Tpl2 −/− or Tpl2 +/+ keratinocytes were assessed for ability to invade Matrigel. Control Tpl2 −/− or Tpl2 +/+ keratinocytes had a low degree of invasion with invasive ability not significantly different between genotypes (Figure 4A). v-rasHa retrovirus-infected Tpl2 +/+ cells also had little invasive potential, being statistically similar to control cells. However, invasion was significantly (P < 0.02) higher in v-rasHa retrovirus-infected Tpl2 −/− keratinocytes, achieving levels three times higher than Tpl2 +/+ keratinocytes.
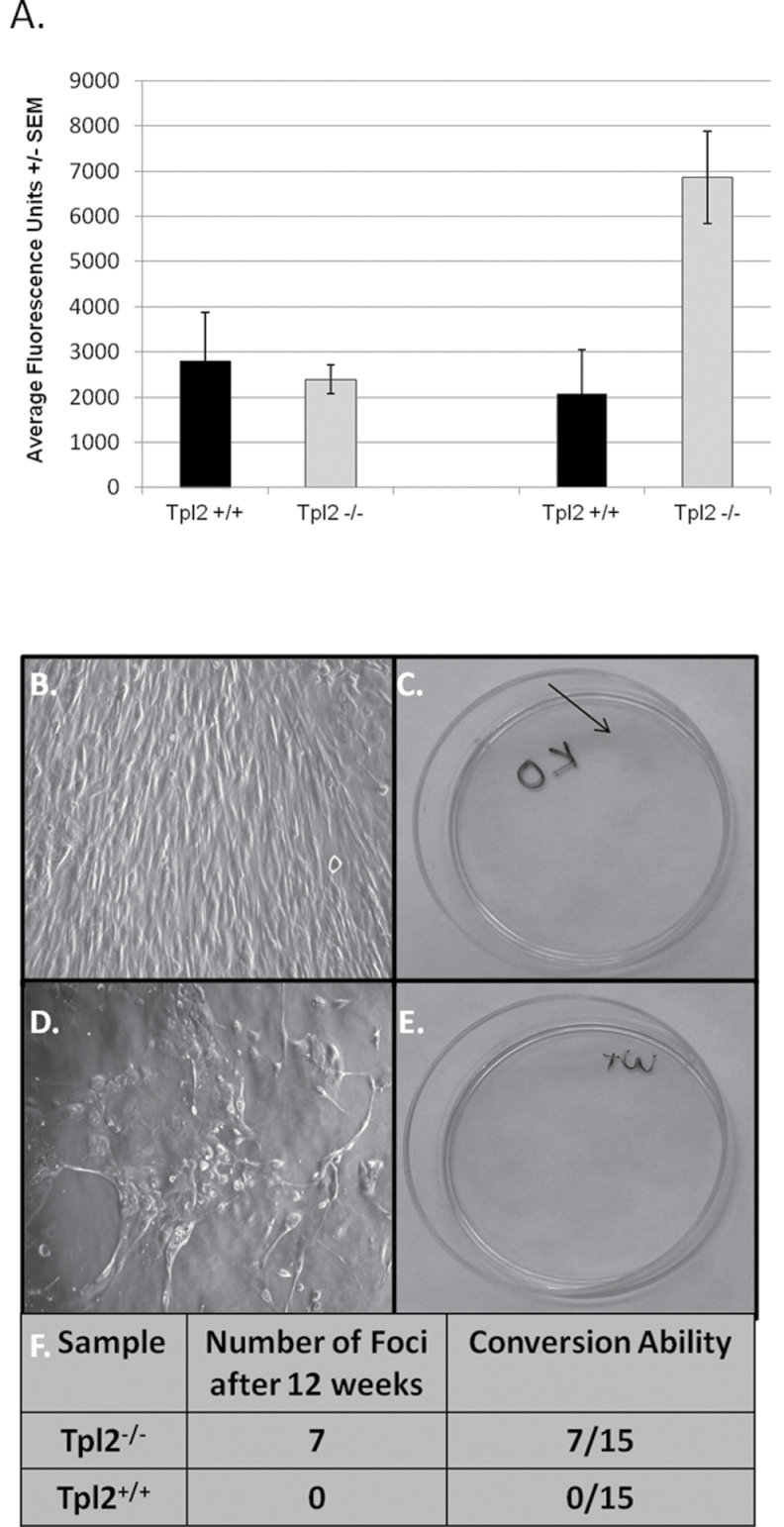
Fig. 4.
Tpl2 −/− primary keratinocytes infected with v-rasHa are more invasive than Tpl2 +/+ primary keratinocytes and undergo malignant conversion to a larger degree. (A) Invasion assay of v-rasHa-infected or vehicle-treated Tpl2 +/+ or Tpl2 −/− keratinocytes. Keratinocytes were allowed to invade Matrigel-coated inserts. Invasive cells passing through membrane were stained with calcein and fluorescence measured. (B–F) Conversion assay of v-rasHa-infected keratinocytes. The Tpl2 −/− dishes containing foci display a morphology resembling a transformed state (B) and stain pink with rhodamine (C). In contrast, the v-rasHa-infected keratinocytes in the Tpl2 +/+ dishes do not form foci (D) and therefore did not stain with rhodamine (E). The conversion rate in Tpl2 −/− and Tpl2 +/+ dishes (F).
Tpl2 −/− primary keratinocytes infected with v-rasHa undergo in vitro malignant conversion to a greater degree than Tpl2 +/+ primary keratinocytes
An in vitro conversion assay was used to compare the ability of Tpl2 +/+ and Tpl2 −/− keratinocytes to convert to a malignant phenotype. Primary keratinocytes from both genotypes were infected with v-rasHa retrovirus. Cells uninfected with v-rasHa virus undergo terminal differentiation, whereas the remaining v-rasHa-infected cells formed a monolayer with potential for converting into malignant cells (36). Foci formed in the Tpl2 −/− cultures, whereas none formed in the Tpl2 +/+ cultures (Figure 4B, D and F). Rhodamine dye was used to differentiate the foci in the Tpl2 −/− cultures from unstained keratinocytes in the Tpl2 +/+ cultures (Figure 4C and E).
Angiogenic factors secreted from Tpl2 −/− cells promote blood vessel formation
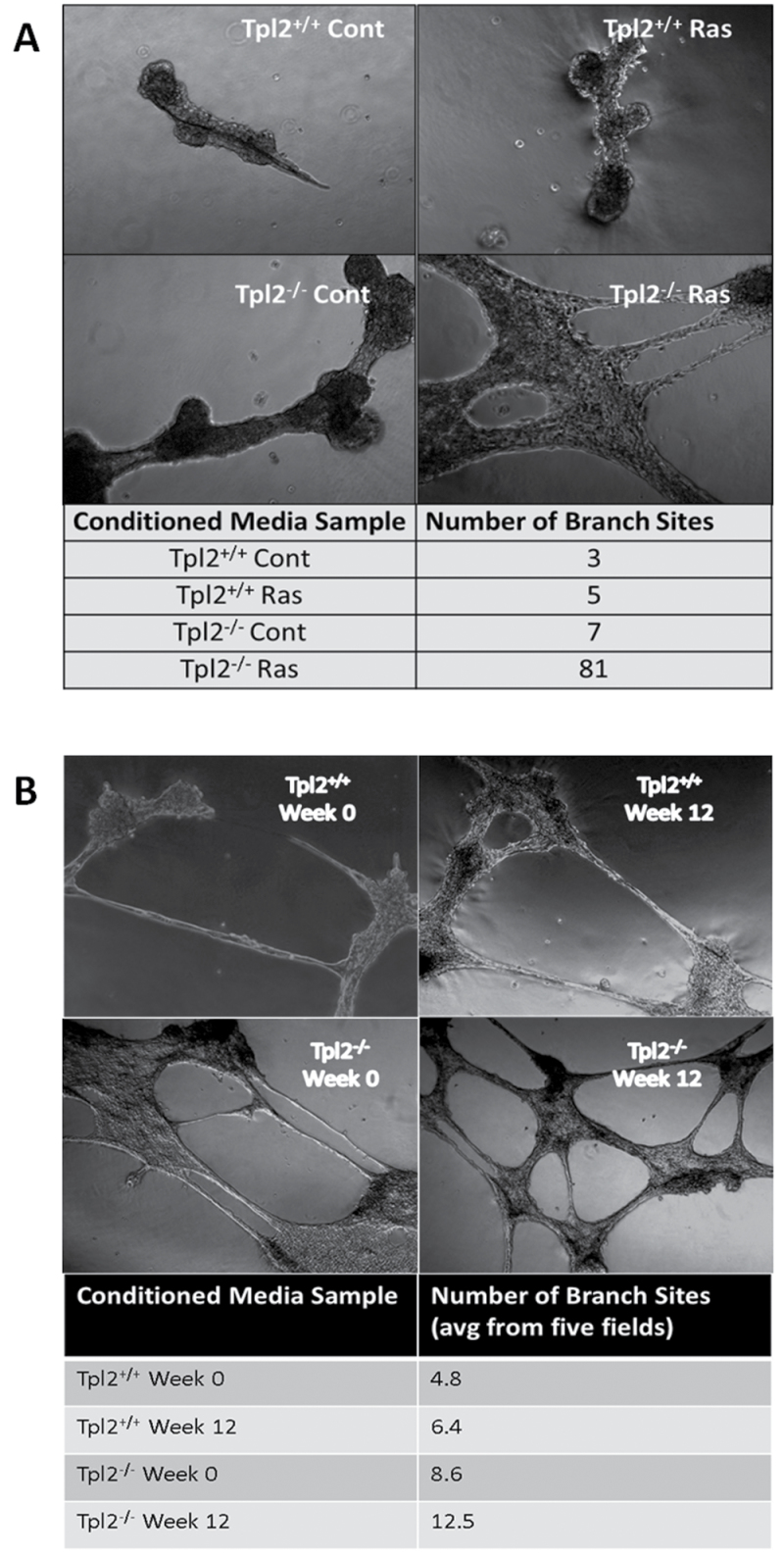
An endothelial cell tube formation assay was performed to determine if conditioned media from Tpl2 −/− cells had a greater potential to induce in vitro blood vessel formation than Tpl2 +/+ cells. Conditioned media was obtained from both genotypes and from both genotypes following transduction with v-rasHa. Endothelial cells cultured in conditioned media from Tpl2 −/− cells formed a much more extensive blood vessel network after 18 h than those cultured in conditioned media from Tpl2 +/+ cells (Figure 5). Cells in conditioned media from uninfected Tpl2 +/+ cells formed a total of three branch sites, whereas those in media from v-rasHa retrovirus-infected Tpl2 +/+ cells formed five branch sites. In contrast, cells cultured in untreated Tpl2 −/− cell media formed seven branch sites, but produced 81 branch sites when cultured with media from v-rasHa-infected Tpl2 −/− cells (Figure 5A). Moreover, conditioned media from Tpl2 −/− cells caused the formation of twice as many branch sites as media from Tpl2 +/+ cells (8.6 branch sites/five fields of view versus 4.8), both initially and after being subjected to an in vitro conversion assay for 12 weeks (12.5 branch sites/five fields of view versus 6.4) (Figure 5B).
Fig. 5.
Angiogenic factors secreted from Tpl2 −/− cells promote blood vessel formation to a larger extent than factors secreted by Tpl2 +/+ cells. Conditioned media from control- or v-rasHa retrovirus-infected Tpl2 +/+ or Tpl2 −/− keratinocytes (A) or from conversion assay samples (B) were combined with serum-starved 3B11 endothelial cells and overlaid on Matrigel™. Eighteen hours later, images were taken with a Zeiss Axioplan II microscope at ×10 magnification and number of branch sites was quantified.
Discussion
Tpl2 is a serine/threonine protein kinase that acts as a tumor suppressor in mouse skin carcinogenesis (12). The absence of Tpl2 leads to increases in proinflammatory cytokines and constitutively active NFκB, both of which contribute to an increase in chemically induced papillomas and squamous cell carcinomas (SCCs) through their downstream signaling pathways (11,12). A global microarray analysis was performed on keratinocytes from Tpl2 +/+ or Tpl2 −/− mice. Over 2000 genes were found to be differentially expressed between genotypes with the largest differences occurring in pathways related to cancer, inflammation, cellular growth, death and movement. Expression differences in factors involved in cancer cell invasion and metastasis were among the most DEGs. Thus, we focused on several candidate genes involved in migration and invasion to see which genes were differentially regulated between genotypes. We identified several MMPs whose expression by complementary DNA microarray and qPCR was significantly higher in Tpl2 −/− keratinocytes both basally and upon stimulation with TPA. Moreover, we found a significant downregulation in an endogenous MMP inhibitor, Timp3, in Tpl2 −/− keratinocytes.
The MMP family consists of 23 zinc-dependent endopeptidases integral to SCC invasion and metastasis (17,18,41). Members of this family degrade the ECM and are critical to multiple processes involved in cancer cell progression, invasion, migration, metastasis and angiogenesis (18,23). Numerous MMPs are secreted by human keratinocytes, with highest activity for MMP1, MMP2, MMP3, MMP9, MMP10 and MMP13 (24,42). Moreover, each of these MMPs has been found to be overexpressed in SCCs (24). Likewise, mouse keratinocytes have high expression of these same MMPs, although in rodents there is a duplication of Mmp1, resulting in Mmp1a and Mmp1b genes (43). However, the significance and exact role of the duplication of Mmp1a and Mmp1b in tumor cell invasion has not been fully elucidated.
In this report, we identify several MMPs, including Mmp1b, Mmp9 and Mmp13, with significantly higher expression in Tpl2 −/− in TPA-treated or untreated keratinocytes. qPCR confirmed upregulation of these MMPs with genotypic differences magnified even further presumably due to the increased sensitivity of qPCR over microarray hybridization (44). Moreover, the expression of Mmp2 was 4-fold higher when analyzed by qPCR.
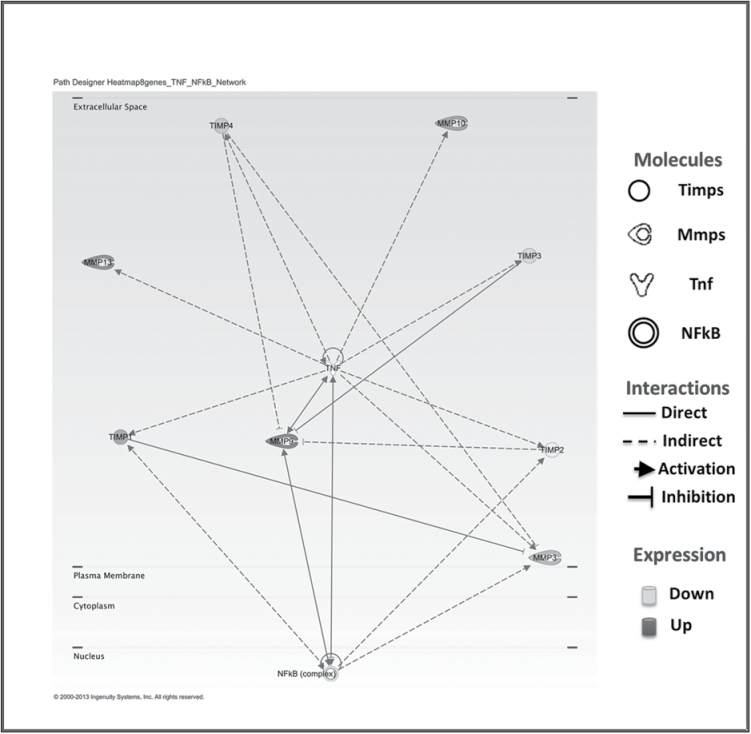
Regulation of MMPs occurs through gene transcription, mRNA stability, enzyme activation and through inhibition by soluble factors known as TIMPs (20). Many MMPs are coexpressed in response to proinflammatory stimuli through common cis-acting elements in their promoter regions (45,46). It has been reported that MMP1, MMP9 and MMP13 are all induced by proinflammatory cytokines working either directly or indirectly through NFκB (45–47). We have previously reported that Tpl2 −/− keratinocytes have constitutively active NFκB and heightened production of proinflammatory cytokines (12). Mmp1 and Mmp13 gene expressions are significantly higher in Tpl2 −/− keratinocytes. MMP13 is a MMP strongly associated with SCC and tumor invasion (48,49). Both MMP1 and MMP13 activation can lead to an increase in Mmp9 gene expression, which then can indirectly cause the induction of MMP1 and MMP13 in a positive feedback manner (Figure 6) (41,45,50).
Fig. 6.
Proposed Ingenuity model describing the relationship between MMPs and TIMPs. Mmp1 and Mmp13 can induce Mmp9 transcription, and Timp3 can inhibit Mmp9. An increase in Mmp9 can activate major signaling pathways like NFκB and mitogen-activated protein kinase (17). However, Mmp9 can also be stimulated by these same pathways in a positive feedback manner resulting in increased invasion, metastasis and angiogenesis.
MMPs enzymatic activities are also regulated through mRNA stability or association with their specific TIMP inhibitors (17). Since Mmp1 and Mmp13 can each affect the transcription of Mmp9, we used zymography to measure the enzymatic activity of Mmp9. Our previous in vivo tumor studies used a two-stage chemical carcinogenesis protocol and to recapitulate this design in vitro keratinocytes were transfected with a replication-defective v-rasHa retrovirus and treated with TPA. We found increased Mmp9 enzymatic activity in Tpl2 −/− keratinocytes before and after TPA treatment in agreement with the microarray data. This induction in Mmp9 in vitro also corresponded with higher expression of Mmp9 in vivo, as Mmp9 expression was induced in both TPA-treated skin from Tpl2 −/− mice and in tumors grafted with v-rasHa retrovirus-infected Tpl2 −/− keratinocytes. Mmp9 forms a high molecular weight complex with Lcn2 (51). The activity of this complex was highest in v-rasHa retrovirus/TPA-stimulated Tpl2 −/− keratinocytes. Lcn2 can stabilize MMP9 by preventing its autodegradation (40). Further, this protein is increased in multiple cancer types and is linked to cancer cell invasion and metastasis (40,51–58).
MMPs are inhibited through the formation of complexes with one of four TIMP proteins: TIMP1, TIMP2, TIMP3 and TIMP4. We found downregulation of Timp3 in Tpl2 −/− keratinocytes in our complementary DNA microarray and qPCR analyses. In addition to inhibiting several MMPs, TIMP3 is the only physiological antagonist for Kdr, one of two tyrosine kinase receptors for Vegfa (59). By binding directly to Kdr, TIMP3 inhibits vascular endothelial growth factor A-mediated downstream signaling pathways. Consequently, the loss or hypermethylation of Timp3 has been identified in several cancer types and can contribute to cancer cell progression (60–62).
In addition to their role in invasion and metastasis, MMPs also are involved in cell migration (21). Despite the fact that Tpl2 −/− keratinocytes display higher expression of Mmp9, Mmp1b and Mmp13, when cell proliferation was inhibited by mitomycin C, migration of v-rasHa-infected keratinocytes was more pronounced in keratinocytes from Tpl2 +/+ mice than in Tpl2 −/− keratinocytes. This data agrees with previous literature in thrombin-stimulated fibroblasts in which Tpl2 increased the rate of migration primarily through ERK activation (63). Interestingly, wound closure was similar in Tpl2 −/− keratinocytes that were not treated with mitomycin C. We have previously reported that Tpl2 −/− keratinocytes have a faster cell cycle than Tpl2 +/+ keratinocytes (12). Therefore, we postulate that the increased cell cycle in Tpl2 −/− keratinocytes compensates for a slower migration rate to produce an overall ability to migrate at a level comparable with Tpl2 +/+ keratinocytes.
An in vitro conversion assay compared conversion differences between Tpl2 +/+ and Tpl2 −/− keratinocytes by observing the spontaneous conversion of v-rasHa retrovirus-infected primary keratinocytes into foci (36). Cells uninfected with v-rasHa retrovirus underwent terminal differentiation in high calcium media, whereas the remaining v-rasHa-infected cells had the potential to form proliferative foci. Therefore, this experiment uses an in vitro assay to mimic the cellular and phenotypic changes seen in malignant conversion in vivo (36,64). We found nearly half of the Tpl2 −/− cultures produced foci, whereas no foci were formed in any of the Tpl2 +/+ cultures. As others have reported, C57BL/6 mice are much more resistant to malignant conversion of keratinocytes than other strains of mice (65). Our previous in vivo data support this notion as a low percentage of Tpl2 +/+ mice develop papillomas and SCCs in a two-stage skin carcinogenesis model, as well as when grafted on nude mice (11,12). Thus, the inherent resistance of keratinocytes from Tpl2 +/+ mice to form foci was maintained during the in vitro malignant conversion assay. In contrast, keratinocytes from Tpl2 −/− mice had significantly increased ability for malignant conversion. Although the precise factors responsible for this heightened susceptibility are unknown, other studies have linked malignant progression of keratinocytes to changes in proinflammatory cytokines, adhesion molecules, Jun transcription factors and transforming growth factor-beta signaling (65). We previously published increased inflammation markers in Tpl2 −/− mice (11,12). The degree to which these markers contribute to malignant conversion in vitro should be further investigated.
Tpl2 −/− keratinocytes have higher expression of multiple invasive genes, including several MMPs, and thus we performed an invasion assay using vehicle-treated Tpl2 +/+ and Tpl2 −/− keratinocytes or keratinocytes infected with a replication-defective v-rasHa retrovirus. As these are primary non-transformed cells, invasion levels were low in vehicle-treated cells and were not statistically different between genotypes. v-rasHa retrovirus-infected Tpl2 −/− cells achieved over three times higher invasion than v-rasHa retrovirus-infected Tpl2 +/+ cells, which had levels similar to control cells. This data agrees with the conversion data showing an inherent resistance to invasion and conversion in Tpl2 +/+ keratinocytes but not in Tpl2 −/− keratinocytes.
MMPs also play a pivotal role in angiogenesis in both normal and cancerous conditions (17). Angiogenesis is a complex process that requires numerous stages, including degradation of the ECM, proliferation and migration of endothelial cells and synthesis of new matrix components (66). Many of these stages are facilitated by MMPs (18). It has been documented that MMP9 degradation of ECM can release several proangiogenic factors, including vascular endothelial growth factor A, fibroblast growth factor 2 and transforming growth factor B1, which increase migration and proliferation of endothelial cells (21). Since we found the levels of Mmp9 and other proangiogenic MMPs were significantly higher in Tpl2 −/− keratinocytes, we measured angiogenesis through an in vitro tube formation assay. We found that conditioned media from either untreated or v-rasHa retrovirus-infected Tpl2 −/− keratinocytes could stimulate endothelial development of blood vessels to a significantly larger extent than conditioned media from v-rasHa retrovirus-infected Tpl2 +/+ cells. It is currently unknown which angiogenic factors play the largest role in inducing angiogenesis by Tpl2 −/− keratinocytes and whether these factors stimulate angiogenesis by degrading ECM, modulating endothelial cells or degrading an angiogenesis inhibitor.
In summary, our microarray analysis identified nearly 2200 genes differentially expressed between Tpl2 +/+ and Tpl2 −/− keratinocytes. The majority of differences occurred in pathways related to cancer growth or invasion. Further analysis of genes involved in invasion and metastasis identified several factors including Mmp1b, Mmp2, Mmp9 and Mmp13 upregulated in Tpl2 −/− keratinocytes. Moreover, Tpl2 −/− keratinocytes had increased invasion, endothelial cell tube formation and malignant conversion as well as altered migration rates. Thus, we propose a role for Tpl2 in skin cancer progression, with the absence of this gene contributing to an increased invasive and metastatic potential.
Funding
Intramural Research Program of the National Cancer Institute at the National Institutes of Health ; National Cancer Institute grant (UA5CA152907).
Acknowledgements
We would like to thank Dr Elizabeth Malloy of American University, Washington, DC, for her assistance with statistical analyses.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- DEG
differentially expressed gene
- DMEM
Dulbecco’s modified Eagle’s medium;
- ECM
extracellular matrix
- ERK
extracellular signal-regulated MAP kinase
- FC
fold change
- Lcn2
lipocalin 2
- MMP
matrix metalloproteinase
- mRNA
messenger RNA
- NFκB
nuclear factor κB
- qPCR
quantitative PCR
- SCC
squamous cell carcinoma
- TIMP
tissue inhibitors of metalloproteinase
- TPA
12-O-tetradecanoylphorbol-13-acetate
- Tpl2
tumor progression locus 2
- WT
wild-type.
References
- 1. Aoki M., et al. (1991). Identification and characterization of protein products of the cot oncogene with serine kinase activity. Oncogene, 6, 1515–1519 [PubMed] [Google Scholar]
- 2. Lang V., et al. (2004). ABIN-2 forms a ternary complex with TPL-2 and NF-kappa B1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol., 24, 5235–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salmeron A., et al. (1996). Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J., 15, 817–826 [PMC free article] [PubMed] [Google Scholar]
- 4. Chiariello M., et al. (2000). Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol. Cell. Biol., 20, 1747–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patriotis C., et al. (1993). Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl Acad. Sci. USA, 90, 2251–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyoshi J., et al. (1991). Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol. Cell. Biol., 11, 4088–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceci J.D., et al. (1997). Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev., 11, 688–700 [DOI] [PubMed] [Google Scholar]
- 8. Tsatsanis C., et al. (2000). The role of oncogenic kinases in human cancer (Review). Int. J. Mol. Med., 5, 583–590 [DOI] [PubMed] [Google Scholar]
- 9. Sourvinos G., et al. (1999). Overexpression of the Tpl-2/Cot oncogene in human breast cancer. Oncogene, 18, 4968–4973 [DOI] [PubMed] [Google Scholar]
- 10. Clark A.M., et al. (2004). Mutational activation of the MAP3K8 protooncogene in lung cancer. Genes. Chromosomes Cancer, 41, 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeCicco-Skinner K.L., et al. (2013). Altered prostanoid signaling contributes to increased skin tumorigenesis in Tpl2 knockout mice. PLoS One, 8, e56212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Decicco-Skinner K.L., et al. (2011). Loss of tumor progression locus 2 (tpl2) enhances tumorigenesis and inflammation in two-stage skin carcinogenesis. Oncogene, 30, 389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gkirtzimanaki K., et al. (2013). TPL2 kinase is a suppressor of lung carcinogenesis. Proc. Natl Acad. Sci. USA, 110, E1470–E1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsatsanis C., et al. (2008). Tpl2 and ERK transduce antiproliferative T cell receptor signals and inhibit transformation of chronically stimulated T cells. Proc. Natl Acad. Sci. USA, 105, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koliaraki V., et al. (2012). Tpl2 regulates intestinal myofibroblast HGF release to suppress colitis-associated tumorigenesis. J. Clin. Invest., 122, 4231–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S.H., et al. (2011). Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol., 209, 139–151 [DOI] [PubMed] [Google Scholar]
- 17. Folgueras A.R., et al. (2004). Matrix metalloproteinases in cancer: from new functions to improved inhibition strategies. Int. J. Dev. Biol., 48, 411–424 [DOI] [PubMed] [Google Scholar]
- 18. Hua H., et al. (2011). Matrix metalloproteinases in tumorigenesis: an evolving paradigm. Cell. Mol. Life Sci., 68, 3853–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagase H., et al. (2006). Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res., 69, 562–573 [DOI] [PubMed] [Google Scholar]
- 20. Sternlicht M.D., et al. (2001). How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol., 17, 463–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bauvois B. (2012). New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim. Biophys. Acta, 1825, 29–36 [DOI] [PubMed] [Google Scholar]
- 22. Balduyck M., et al. (2000). Specific expression of matrix metalloproteinases 1, 3, 9 and 13 associated with invasiveness of breast cancer cells in vitro . Clin. Exp. Metastasis, 18, 171–178 [DOI] [PubMed] [Google Scholar]
- 23. Deryugina E.I., et al. (2006). Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev., 25, 9–34 [DOI] [PubMed] [Google Scholar]
- 24. Kerkelä E., et al. (2003). Matrix metalloproteinases in tumor progression: focus on basal and squamous cell skin cancer. Exp. Dermatol., 12, 109–125 [DOI] [PubMed] [Google Scholar]
- 25. Kudo Y., et al. (2012). Matrix metalloproteinase-13 (MMP-13) directly and indirectly promotes tumor angiogenesis. J. Biol. Chem., 287, 38716–38728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murray G.I., et al. (1996). Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat. Med., 2, 461–462 [DOI] [PubMed] [Google Scholar]
- 27. Airola K., et al. (1999). Expression of collagenases-1 and -3 and their inhibitors TIMP-1 and -3 correlates with the level of invasion in malignant melanomas. Br. J. Cancer, 80, 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakopoulou L., et al. (1999). Matrix metalloproteinase-1 and -3 in breast cancer: correlation with progesterone receptors and other clinicopathologic features. Hum. Pathol., 30, 436–442 [DOI] [PubMed] [Google Scholar]
- 29. Nikkola J., et al. (2002). High expression levels of collagenase-1 and stromelysin-1 correlate with shorter disease-free survival in human metastatic melanoma. Int. J. Cancer, 97, 432–438 [DOI] [PubMed] [Google Scholar]
- 30. Poola I., et al. (2005). Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med., 11, 481–483 [DOI] [PubMed] [Google Scholar]
- 31. Shah S.A., et al. (2010). Differential matrix metalloproteinase levels in adenocarcinoma and squamous cell carcinoma of the lung. J. Thorac. Cardiovasc. Surg., 139, 984–990; discussion 990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giricz O., et al. (2010). Variability in melanoma metalloproteinase expression profiling. J. Biomol. Tech., 21, 194–204 [PMC free article] [PubMed] [Google Scholar]
- 33. Lichti U., et al. (2008). Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc., 3, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res., 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider C.A., et al. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgan D., et al. (1992). Development of an in vitro model to study carcinogen-induced neoplastic progression of initiated mouse epidermal cells. Cancer Res., 52, 3145–3156 [PubMed] [Google Scholar]
- 37. Arnaoutova I., et al. (2010). In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat. Protoc., 5, 628–635 [DOI] [PubMed] [Google Scholar]
- 38. Huang da W., et al. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 39. Huang da W., et al. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res., 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chakraborty S., et al. (2012). The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochim. Biophys. Acta, 1826, 129–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westermarck J., et al. (1999). Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J., 13, 781–792 [PubMed] [Google Scholar]
- 42. Tandara A.A., et al. (2011). MMP- and TIMP-secretion by human cutaneous keratinocytes and fibroblasts–impact of coculture and hydration. J. Plast. Reconstr. Aesthet. Surg., 64, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Balbín M., et al. (2001). Identification and enzymatic characterization of two diverging murine counterparts of human interstitial collagenase (MMP-1) expressed at sites of embryo implantation. J. Biol. Chem., 276, 10253–10262 [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., et al. (2006). Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics, 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vincenti M.P., et al. (2007). Signal transduction and cell-type specific regulation of matrix metalloproteinase gene expression: can MMPs be good for you? J. Cell. Physiol., 213, 355–364 [DOI] [PubMed] [Google Scholar]
- 46. Clark I.M., et al. (2008). The regulation of matrix metalloproteinases and their inhibitors. Int. J. Biochem. Cell Biol., 40, 1362–1378 [DOI] [PubMed] [Google Scholar]
- 47. Lin T.H., et al. (2011) 15-deoxy-Δ(12,14) -prostaglandin-J2 and ciglitazone inhibit TNF-α-induced matrix metalloproteinase 13 production via the antagonism of NF-κB activation in human synovial fibroblasts. J. Cell. Physiol., 226, 3242–3250 [DOI] [PubMed] [Google Scholar]
- 48. Lee S.Y., et al. (2011). Expression of matrix metalloproteinases and their inhibitors in squamous cell carcinoma of the tonsil and their clinical significance. Clin. Exp. Otorhinolaryngol., 4, 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johansson N., et al. (2000). Expression of collagenase-3 (MMP-13) and collagenase-1 (MMP-1) by transformed keratinocytes is dependent on the activity of p38 mitogen-activated protein kinase. J. Cell Sci., 113(Pt 2), 227–235 [DOI] [PubMed] [Google Scholar]
- 50. Steenport M., et al. (2009). Matrix metalloproteinase (MMP)-1 and MMP-3 induce macrophage MMP-9: evidence for the role of TNF-alpha and cyclooxygenase-2. J. Immunol., 183, 8119–8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leung L., et al. (2012). Lipocalin2 promotes invasion, tumorigenicity and gemcitabine resistance in pancreatic ductal adenocarcinoma. PLoS One, 7, e46677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fernández C.A., et al. (2005). The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin. Cancer Res., 11, 5390–5395 [DOI] [PubMed] [Google Scholar]
- 53. Iannetti A., et al. (2008). The neutrophil gelatinase-associated lipocalin (NGAL), a NF-kappaB-regulated gene, is a survival factor for thyroid neoplastic cells. Proc. Natl Acad. Sci. USA, 105, 14058–14063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kubben F.J., et al. (2007). Clinical evidence for a protective role of lipocalin-2 against MMP-9 autodegradation and the impact for gastric cancer. Eur. J. Cancer, 43, 1869–1876 [DOI] [PubMed] [Google Scholar]
- 55. Laurell H., et al. (2006). Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J. Gastroenterol., 12, 3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leng X., et al. (2009). Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res., 69, 8579–8584 [DOI] [PubMed] [Google Scholar]
- 57. Lim R., et al. (2007). Neutrophil gelatinase-associated lipocalin (NGAL) an early-screening biomarker for ovarian cancer: NGAL is associated with epidermal growth factor-induced epithelio-mesenchymal transition. Int. J. Cancer, 120, 2426–2434 [DOI] [PubMed] [Google Scholar]
- 58. Zhang H., et al. (2007). Upregulation of neutrophil gelatinase-associated lipocalin in oesophageal squamous cell carcinoma: significant correlation with cell differentiation and tumour invasion. J. Clin. Pathol., 60, 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qi J.H., et al. (2003). A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat. Med., 9, 407–415 [DOI] [PubMed] [Google Scholar]
- 60. Fendrich V., et al. (2005). Alterations of the tissue inhibitor of metalloproteinase-3 (TIMP3) gene in pancreatic adenocarcinomas. Pancreas, 30, e40–e45 [DOI] [PubMed] [Google Scholar]
- 61. Lui E.L., et al. (2005). DNA hypermethylation of TIMP3 gene in invasive breast ductal carcinoma. Biomed. Pharmacother., 59 (s uppl. 2), S363–S365 [DOI] [PubMed] [Google Scholar]
- 62. Masson D., et al. (2010). Loss of expression of TIMP3 in clear cell renal cell carcinoma. Eur. J. Cancer, 46, 1430–1437 [DOI] [PubMed] [Google Scholar]
- 63. Hatziapostolou M., et al. (2011). Tumor progression locus 2 mediates signal-induced increases in cytoplasmic calcium and cell migration. Sci. Signal., 4, ra55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Greenhalgh D.A., et al. (1989). Spontaneous Ha-ras gene activation in cultured primary murine keratinocytes: consequences of Ha-ras gene activation in malignant conversion and malignant progression. Mol. Carcinog., 2, 199–207 [DOI] [PubMed] [Google Scholar]
- 65. Woodworth C.D., et al. (2004). Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis, 25, 1771–1778 [DOI] [PubMed] [Google Scholar]
- 66. Bergers G., et al. (2000). Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol., 2, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]