Abstract
Distance determination from an echo intensity modulation obtained by pulsed double electron-electron resonance (DEER) experiment is a mathematically ill-posed problem. Tikhonov regularization yields distance distributions that can be difficult to interpret, especially in system with multiple discrete distance distributions. Here, we show that by using geometric fit constraints in symmetric homo-oligomeric protein systems, we were able to increase the accuracy of a model-based fit solution based on a sum of Gaussian distributions. Our approach was validated on two different ion channels of known oligomeric states, KcsA (a tetramer) and CorA (a pentamer). Statistical analysis of the resulting fits was integrated within our method to help the experimenter evaluate the significance of a symmetry-constrained vs. standard model distribution fit and to examine multi-distance confidence regions. This approach was used to quantitatively evaluate the role of the C-terminal domain (CTD) on the flexibility and conformation of the activation gate of the K+ channel KcsA. Our analysis reveals a significant increase in the dynamics of the inner bundle gate upon opening. Also, it explicitly demonstrates the degree to which the CTD restricts the motion of the lower gate at rest and during activation gating.
Keywords: pulsed EPR, spectroscopy, DEER, distance measurement, membrane protein, ion channel, KcsA, CorA
INTRODUCTION
Distance determination in macromolecules, and particularly in membrane proteins, is often at the core of arguments to decipher molecular mechanisms by which these nanomachines execute their biological function. 1–3 In transporters, distance determination has been extensively used to decipher not only the extent of molecular motion, but also the nature of the alternative access mechanism.4,5 Pulsed double electron-electron resonance (DEER) method is an unparalleled tool for measurement of long range distances in proteins (20–80 Å).6 EPR offers several advantages over other spectroscopic techniques such as fluorescence resonance energy transfer (FRET). First, the small size of the spin label improves its accessibility to protein target sites. Second, the very short linker significantly improves probe localization by limiting the probe’s diffusive region. Third, the same probe is used to label all target sites which greatly simplifies the labeling strategy. Fourth, in DEER experiments the signal is not polluted by possible under-labeling of the sample since the echo modulation arises exclusively from dipolar coupling. All these advantages translate into greater accuracy of probe position when applied to a macromolecule. The sensitivity and reliability of this technique depends on the optimization of the sample preparation, experimental conditions for measurement, and data analysis.4,5
Analysis of DEER measurements yields a distance distribution obtained by either a model-free (e.g., Tikhonov regularization) or model-based fit. In this work, we improved model-based distance distribution analysis by utilizing the symmetry of homomeric proteins. Experimentally, all subunits are spin-labeled at the same site. In such a multi-spin system, high precision distance determination has been obtained using a classical four pulse DEER protocol.7 However, broadening of the distance distribution may occur due to the signal contributions from the combinations of dipolar frequencies.8 Our geometric fit constraint method applies to a large class of proteins, including membrane proteins, which are often assembled into symmetrical multimeric entities. This is the case for Na+ 9, Ca2+ 10, K+ 11, Mg2+ 12, mechanosensitive13, and ligand-gated ion channels.14 The inherent symmetry of subunit organization enables the fit of a model-based distance distribution function using the known organization of the protein in question.
Generating a distance distribution P(r) of interprobe distance r from a DEER signal is a moderately mathematically ill-posed problem and as such, slight variations in the raw data’s signal-to-noise ratio (SNR) can generate a large difference in the distance distributions.15,16 Tikhonov regularization is an elegant and widely used method for model-free analysis of DEER signal.17 Briefly, this technique minimizes the squared error between simulated and measured dipolar evolution by balancing smoothness and resolution of the distance distribution using a dampening term. Since the width of the distance distribution is not known in advance, the optimal regularization parameter λ must be selected (typically by the L-curve criterion18). Conversely, the distance distribution P(r) can also be obtained by using a model-based approach, which has the advantage of enforcing smoothness and improving convergence, thereby improving the reliability of fitted distances provided that the chosen model is correct. Comparison and validation of any model-based fit to that of Tikhonov regularization is a critical component of such distance analysis. A recent study demonstrated that the interprobe distance distribution of two normally-distributed spin labels is described by a three-dimensional Rice (Rice3D) distribution rather than a Gaussian, which is especially important for accurate parameter estimation in broad distance distributions (e.g., μ/σ < 4 with mean interprobe distance μ and standard deviation σ of each spin label position).19
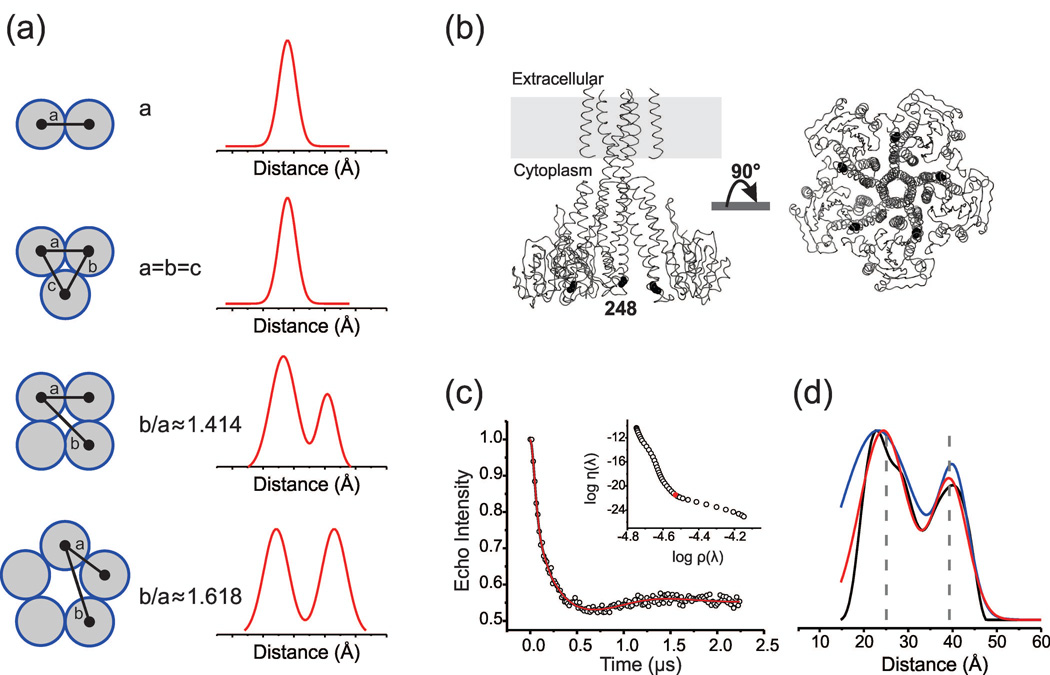
In this study, we sought to incorporate the intrinsic symmetry of ion channels into model-based distance distribution fits. These homomeric proteins are each arranged as a regular convex polygon with m vertices (subunits). In the biologically relevant range of m = 3–6, all geometries have vertex-to-vertex lengths characterized by 1 unique side (adjacent) and the following number of diagonals (dm): d3 = 0, d4,5 = 1, d6 = 2. By symmetry, the number of expected peaks in the distance distribution P(r) is equal to the sum of unique sides and diagonals (1+dm). Moreover, the diagonal and side lengths maintain a constant proportionality relation, independent of polygon size. In this paper we consider the geometries for m = 4–5, which both yield 2 unique mean distances between spin label paramagnetic centers, which we denote as <r1> for adjacent and <r2> for diagonal. In these cases, there is only one proportionality constant, given by the mean distance ratio <k> = <r2>/<r1>, and P(r) is appropriately modeled by a 2-component Rice3D distribution. In the case of polygon models with m>6, multiple distance ratios <ki> exist. If we consider a homopentameric (m=5) protein such as the Mg2+ channel CorA, where all subunits are spin-labeled at the same site, we expect <k> = (1+√5)/2 ≈1.618. Likewise, a homotetrameric (m=4) assembly such as the K+ channel KcsA is expected to have <k> = √2 ≈1.414. Expected distance distributions as a function of the oligomeric state are illustrated in Figure 1a. Given symmetric labeling, it is important to note that the mean distance ratio is equivalent for the mean position of the spin labels (e.g., nitroxide oxygen), as well as their corresponding labeling site (e.g., cysteine Cα).
Figure 1. Distance determination in multimeric proteins.
(a) Schematic representation of the expected distance distribution profile as a function of the oligomeric state of a homomeric protein. (b) Cysteine mutation in CorA is shown at position V248 as black spheres at Cβ. For clarity, only 3 subunits are shown. (c) The DEER refocused echo intensity is plotted (open circles) vs. the evolution time. Fits determined from Tikhonov regularization, 2 Rice3D and symmetry-constrained 2 Rice3D models are shown in black, blue, and red, respectively. In the inset, the optimal regularization parameter (λ=158) is shown at the corner of the L-curve. (d) Corresponding distance distributions are plotted using the same color code. The vertical dashed lines represent the average spin-spin distances calculated from rotameric libraries of the spin label conjugated to the V248C label site.31
We demonstrate that the distance ratio <k> can be utilized as a symmetry-based inequality constraint to: (1) improve the model fit of a distance distribution P(r) to DEER data, and (2) to aid interpretation of distances obtained from model-based fits. The method is especially suitable for poorly defined (flat) dipolar evolutions, as often obtained from flexible/dynamic systems. CorA V248C serves as an illustrative example of such a scenario (Figure 1b–c).
This new approach was implemented on two different ion channels of different oligomeric states and the result are compared with other fitting methods (2-Rice3D and Tikhonov regularization). We also performed a statistical analysis to compare the significance of different models and fitting procedures. The cross validation between our DEER data and the corresponding X-ray structures provides a solid framework to study conformational changes associated with the gating mechanism of KcsA and CorA.
Taking advantage of our method we sought to monitor the influence of the C-Terminal domain (CTD) helix bundle on the dynamics and the extent of motion of KcsA’s activation gate. Upon proton dependent activation, the K+ channel KcsA undergoes relatively large movements of its helical transmembrane segment TM2. These rearrangements have been extensively characterized by spectroscopic methods such as EPR20, fluorescence21, NMR22 and are in agreement with available open KcsA X-ray crystal structures.23,24 However, the influence of the CTD on the extent of the inner bundle gate opening has not been studied in absence of either crystal lattice contacts or crystallographic chaperones. Here we found that in the absence of the CTD, the lower gate opening is significantly increased with respect to the full length channel, with a concomitant increase in protein dynamics upon activation. Our results are discussed in the context of the C-type inactivation mechanism and existing crystal structures.
MATERIALS AND METHODS
Protein expression and purification
Single cysteine mutants of Thermotoga maritima CorA (TmCorA) channels and Streptomyces lividans KcsA channels were expressed and purified as previously described.25,26 Briefly, after expression the membrane fractions were solubilized with 10 mM DDM and loaded onto a pre-equilibrated cobalt affinity column (Clontech). During the purification, 5 mM β-Mercaptoethanol and 0.5 mM TCEP were used to prevent cysteine oxidation. Immediately after protein elution with 250 mM imidazole, cysteine mutants were labeled with two additions of a 10-fold molar excess of MTSL spin label probes (Toronto Research Chemicals) on ice for 30 min sequentially. Labeled proteins were then further purified by size exclusion chromatography on a Sephadex G-200 column (GE Healthsciences) previously equilibrated with buffer (Hepes 50 mM pH=7.0, NaCl 200 mM, MgCl2 40 mM, DDM 1 mM) for CorA mutants and (Hepes 50 mM pH=7.0 or McIlVaine buffer (phosphate/citrate) pH=3, KCl 200 mM, DDM 1 mM) for KcsA mutants. The fractions containing the main monodisperse peak were collected and pooled together, supplemented with 20% glycerol and concentrated to 100 µM final concentration of oligomer assembly using a Millipore Centricon 30 kDa centrifugal filter unit.
DEER experiment
Samples were loaded into a Quartz capillary (Vitrocom) and equilibrated at 80K under a flow of liquid N2 using an Oxford cryostat. A standard 4-pulse DEER sequence was conducted on a Bruker Elexsys 580 EPR Spectrometer equipped with a 3 mm split-ring resonator. The 4-pulse DEER sequence was set with pulses of 16 ns (90°) and 32 ns (180°), respectively, and evolution times were typically set to 1800–2500 ns depending on distance and signal quality. The pump pulse was placed at the center peak of the spectrum and the observation pulses were placed at a 75 MHz distance away on the low field side. Refocused echo intensity evolutions were recorded and these phase-corrected signals were background-corrected assuming a homogenous 3D distribution.
Distance distribution analysis
The distance distribution P(r) was recovered from DEER measurements by three different approaches: model-free fit by Tikhonov regularization, model fit of a 2-component Rice3D mixture, or model fit of a 2-component Rice3D mixture with distance ratio constraint. For all approaches, the DeerAnalysis 2011 program16 was used for analysis with software modifications described below. Each time domain dipolar evolution dataset was preprocessed using tools provided by DeerAnalysis to correct for experimental phase errors and to separate the intramolecular distances from the intermolecular background contribution. The background was subtracted from the dipolar evolution assuming a homogeneous distribution in 3 dimensions, appropriate for spin-labeled membrane proteins in detergent micelles. The origin of the background fit was determined by approximate Pake transformation in the frequency domain. The background-subtracted experimental dipolar evolution is referred to below as Vexp (t).
Model-free distance distributions by Tikhonov regularization
For pulsed EPR measurements, Tikhonov regularization minimizes the following functional:6
| (1) |
where K(r, t) is a kernel that represents the ensemble average of the dipolar coupling over all possible molecular orientations for a given spin label radial separation r (and is the shape of a Pake double dipolar signal in the time domain); P(r) represents the distance distribution of r. The first term on the right-hand side is the mean square deviation between the simulated and experimental dipolar evolution function; the second term on the right-hand side is the square norm of the second derivative of the distance distribution, which is multiplied by regularization parameter λ. Tikhonov regularizations were performed for a logarithmic sequence of regularization parameters on the range λ = [10−5, 103] with 4 points per decade. The optimal regularization parameter λ was calculated from this set of solutions by applying the L-curve criterion of maximum curvature.18 For all datasets, the optimal λ was confirmed to be located at the corner of the L-curve by visual inspection. Parameters from Tikhonov regularization solution at the optimal λ were analyzed and reported in Table 1. All dataset solutions had a distribution P(r) comprised of two primary distance peaks. The 1st peak (shorter distance) was interpreted as the mean adjacent distance 〈r1〉 and the 2nd peak (longer distance) was interpreted as the mean diagonal distance 〈r2〉.
Table 1.
Distance determination results for DEER data analyzed by various methods
| Cysteine Mutant |
Distribution Fit Method | 〈r1〉: Adjacent Distance (Å)* |
〈r2〉: Diagonal Distance (Å)* |
〈k〉 | Fit RMSD | Threshold RMSD (α=0.05) |
|---|---|---|---|---|---|---|
| CorA V248C | <X-ray structure> | 26.0 | 41.3 | 1.588 | N/A | 0.0090260 |
| Tikhonov (λ=158) | 22.8 | 40.35 | 1.770 | 0.0086750 | ||
| 2 Rice3D | 19.9 (±9) | 40.3 (±3.3) | 2.027 | 0.0086698 | ||
| 2 Rice3D [1.578 ≤ <k> ≤1.645] | 23.6 (±4.9) | 38.8 (±4.2) | 1.645 | 0.0088010 | ||
| CorA R252C | <X-ray structure> | 24.2 | 38.0 | 1.570 | N/A | 0.0080159 |
| Tikhonov (λ=31) | 24.2 | 39.1 | 1.616 | 0.0074304 | ||
| 2 Rice3D | 23.9 (±2.2) | 38.7 (±1.8) | 1.6185 | 0.0076829 | ||
| 2 Rice3D [1.587 ≤ <k> ≤1.629] | 23.9 (±2.3) | 38.7 (±1.8) | 1.6185 | 0.0076829 | ||
| 2 Rice3D [1.367 ≤ <k> ≤1.433] | 24.2 (±1.4) | 34.6 (±5.5) | 1.433 | 0.0125890 | ||
| KcsA R64C | <X-ray structure> | 22.0 | 32.0 | 1.454 | N/A | 0.0045779 |
| Tikhonov (λ=10) | 22.0 | 31.8 | 1.445 | 0.0056134 | ||
| 2 Rice3D | 22.1 (±0.7) | 31.4 (±3.6) | 1.4212 | 0.0043960 | ||
| 2 Rice3D [1.379 ≤ <k> ≤1.418] | 22.1 (±0.7) | 31.3 (±3.6) | 1.418 | 0.0043963 | ||
| 2 Rice3D [1.578 ≤ <k> ≤1.645] | 20.8 (±3.3) | 33.7 (±2.7) | 1.5780 | 0.0061192 | ||
Values reported in parentheses are fit values of σR from the 2 Rice3D model (i.e., not distance confidence intervals).
Model-based distance distributions by three-dimensional Rice mixture
A model fit minimizes the following functional:
| (2) |
where θ is a parameter vector that defines the probability density function of an analytical model distribution. Here, dampening is inherently introduced by the smooth distance distribution Pmodel(r; θ). The interprobe distance between two spin label centers, each normally distributed in space (x, y, z) with standard deviation σS about its center, is described by the Rice3D distribution,27,28 generalized to n-dimensions as:
| (3) |
where the mean interprobe distance μ > 0 and Rice standard deviation σR > 0 are real numbers; n is the dimension of the spin label normal random variables (n = 3 for labeled membrane proteins in detergent micelles); Iν denotes the modified Bessel function of the first kind of real order ν. Assuming all spin labels have equal spatial variance, the Rice standard deviation is related to the spin label spatial standard deviation by , which is a correction from that reported in 29. The Rice3D distribution can be simplified29 to a more computationally efficient form using the relation :
| (4) |
The probability density function of the 2-component mixture of Rice3D distributions is:
| (5) |
where w is the fraction of the 1st mixture component and θ = (μ1, σR,1 , μ2, σR,2 ,w) is the set of all parameters that defines the bimodal distance distribution. The probability density function P(r;θ) was fit to Vexp (t) by minimization of Equation 2 using nonlinear least-squares regression.
Symmetry-constrained model fit and software modification
DeerAnalysis 2011 software16 was modified and supplemented in MATLAB (The MathWorks, Inc) to introduce the use of nonlinear inequality constraints into the nonlinear minimization problem of fitting a model-based distance distribution to a measured dipolar evolution dataset using Equation 2. We primarily added the ability to perform constrained 2-component Rice3D and 2-component Gaussian mixture model fits where the mean distance ratio 〈k〉 = 〈r2〉/〈r1〉 = μ2/μ1 is constrained within user-defined limits [kmin, kmax] relevant to known symmetry of the protein under investigation. For model fits of a distance distribution, DeerAnalysis 2011 performs minimization in the least-squares sense using the built-in MATLAB function fminsearch with simplex search method. We implemented model fitting with nonlinear inequality constraints using the built-in MATLAB function fmincon with active-set algorithm. Both functions were verified to return the same solution when used under equivalent conditions. The ability to perform model fits with nonlinear inequality constraints will be implemented in a future release of DeerAnalysis software16, available at ETH Zurich http://www.epr.ethz.ch/software/.
RESULTS AND DISCUSSION
A symmetry-derived nonlinear constraint to fit DEER measurements
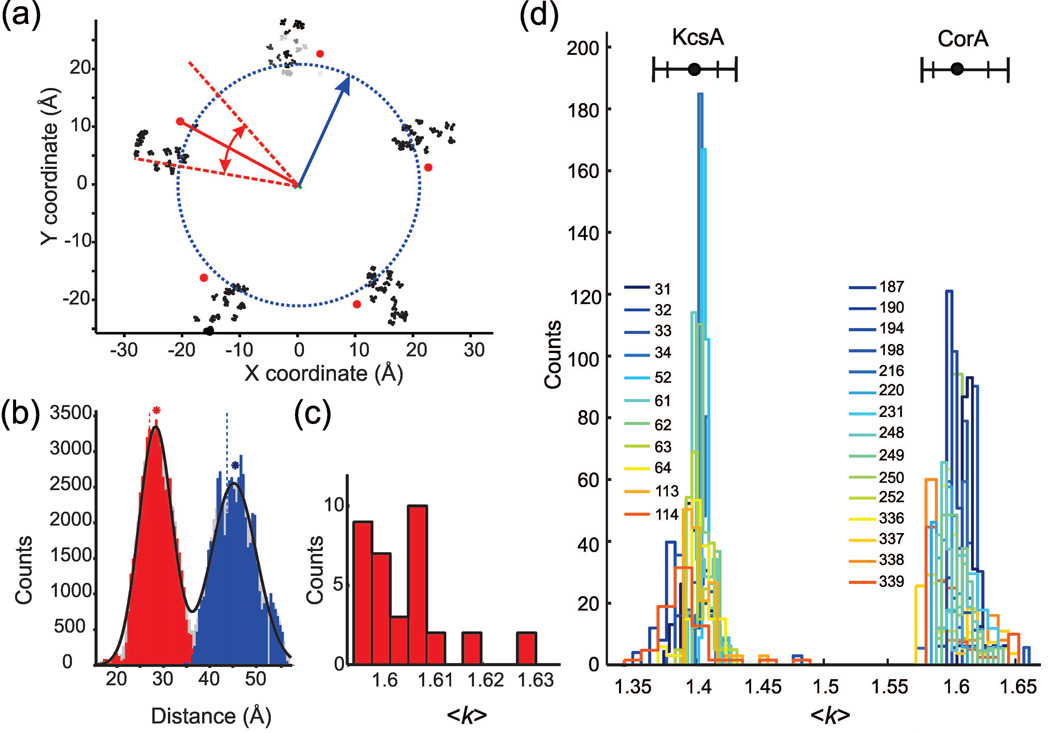
A poorly defined (flat) dipolar evolution is difficult to transform into a reliable distance distribution (Figure 1c). Here, we sought to incorporate intrinsic symmetry of homomeric ion channels into model-based distance distribution fits. Both homotetrameric and homopentameric assemblies have an adjacent <r1> and diagonal <r2> distances related by the ratio <k> = <r2>/<r1> with theoretical values of 1.414 and 1.618, respectively (Figure 1a). Of course in nature, symmetry is never perfect and membrane proteins are no exception. Simple measurement of Cβ-Cβ distances from the CorA crystal structure30 illustrates this deviation, resulting in a k interval of 1.57–1.68. Combined with thermal motion, this observation supports the idea that the distance ratio must be contained within two limits, rather than at a fixed value. Accordingly, we sought to evaluate in a rational and quantitative manner the distribution of <k>. Accordingly, we selected a total of 26 positions distributed along the sequence of KcsA and CorA and used an in-silico spin-labeling technique via the software MMM.31 For each mutant tested, the selected residue was mutated to cysteine and MTSL was attached with the 26 most probable rotameric conformations, thus producing a cloud of possible spin label conformations on each channel subunit. The nitroxide oxygen (O1) atom was used as an approximation of the paramagnetic center of the probe. To slightly smooth the MTSL O1 distance distribution and account for minimal conformational relaxation, each O1 point was replicated 5 times with a small amount of random noise added (sample points were randomly drawn from a uniformly distributed sphere with 0.5 Å radius, centered at the actual O1 position). To evaluate the effect on <k> from MTSL cloud distortion caused by conformational change or other factors, we implemented simulation tools to sequentially include or exclude rotameric conformations based on 3 criteria: (1) occupancy, (2) O1 radial distance to the symmetry axis, and (3) O1 cylindrical angle with respect to the labeled cysteine Cα (Figure 2a). The influence of these 3 parameters was homogeneously assayed by our sampling conditions such that only uniquely modified MTSL clouds were analyzed. For each modification condition, we calculated <k> and a histogram of all unique inter-subunit O1-O1 distances, which could be successfully fit by a 2 Rice3D model distribution (Figure 2b). From the set of all MTSL cloud modification conditions, we obtained a histogram for <k> (Figure 2c). The final result of this simulation analysis performed on all KcsA and CorA mutants is presented in Figure 2d. Remarkably, we found that the distance ratio remained extremely close to theoretical values: <k> = 1.402 for KcsA and <k> = 1.605 for CorA. Moreover, despite our efforts to distort the rotameric spatial distribution of spin labels at their binding site, the distribution of <k> remained tight. The 1–99% <k> interval for all mutants tested was 1.367– 1.433 for KcsA and 1.578–1.645 for CorA (Figure 2d). We assumed that the extensive number of labeling sites tested in addition to the comprehensive sampling strategy leads to a robust and reliable <k> distribution. Henceforth, we refer to this exercise as rotamer simulation.
Figure 2. Rotamer-based simulation of distance ratio distribution for KcsA and CorA.
(a) MTSL rotamers were attached to CorA at position 252 using the program MMM.31 Nitroxide oxygen coordinates were oversampled (see text) and are displayed as black dots. CorA 252C Cα are represented as red dots and the channel’s symmetry axis is marked as a green cross. MTSL rotamer clouds were independently modified by exclusion of points based on a linear scan of occupancy (step = 0.01), radial distance (step = 0.5 Å, and cylindrical angle (step = ±0.3°). For visualization of occupancy, rotamers in the uppermost subunit were grayscaled (black=high; white=low). The blue circle (dotted) and arrow represent the radial distance scan. The red lines (dashed) and arrows represent the cylindrical angle scan. (b) Distance histograms are shown to exemplify one MTSL cloud modification for one mutant; r1 is red, r2 is blue, and their combination is gray. Asterisks represent the mean of the distribution using the same color code; vertical dotted lines represent Cα distances. A 2 Rice3D fit of the combined distribution is displayed in black. (c) Histogram of <k> from the set of all MTSL cloud modifications for one mutant. (d) Histograms of <k> from the set of all MTSL cloud modifications for 11 KcsA (tetramer) and 15 CorA (pentamer) mutants. For both channels, the average <k> is plotted as a black circle and 1–99% and 5–95% intervals are plotted as tall and short ticks, respectively.
Symmetry-constrained analysis of a broad distance distribution
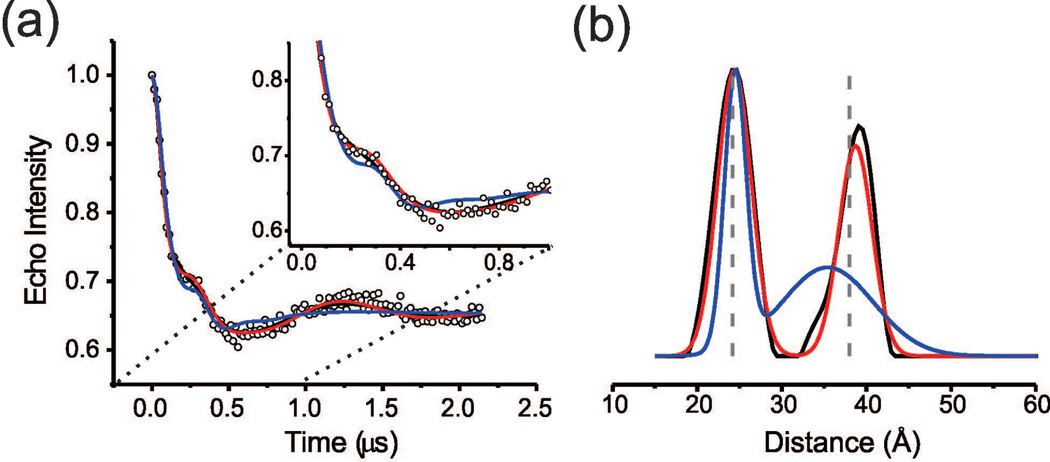
We sought to compare the experimentally derived P(r) from a poorly defined dipolar evolution obtained from CorA V248C analyzed by 3 different fit methods: Tikhonov regularization, 2 Rice3D, and symmetry-constrained 2 Rice3D (see Materials and Methods). Figure 1c shows that fits of the background-subtracted dipolar evolution by these 3 methods are almost indistinguishable visually, and have fairly similar root mean square deviation (RMSD) values (Table 1). However, the corresponding set of distance distributions are substantially different (Figure 1d). When we imposed rotamer simulation-based limits on the distance ratio (<k> = 1.578–1.645) to stabilize the correct solution, we obtained adjacent and diagonal distances that both agree remarkably well with the distances calculated from the CorA crystal structure12 using a rotameric library of the spin label.31 Notice that P(r) obtained by Tikhonov regularization is distorted from the idealized distribution, illustrated by emergence of a 3rd peak. We argue that this deviation from an idealized distance distribution is purely artificial and is not a true reflection of the structural dynamics of the system. This reasoning is based on the observation that when the dipolar evolution is better defined, i.e., when more periods are detected, P(r) calculated from a Tikhonov fit is virtually equivalent to a 2 Rice3D distribution (see Figure 3). Recently, an improved dipolar evolution was demonstrated by a groundbreaking paper by the Hubbell group where they used a bifunctional probe which reflects the motion of the protein with greater accuracy. When they compared P(r) obtained with the bifunctional vs. classical probe at an equivalent position, they obtained a sharper distribution and homogenous peak shape.32 This result supports the idea that our approach of using a symmetry-constrained 2 Rice3D model to derive the distance distribution is an idealization rather than an over-smoothing of the data. It is important to note that all mathematical tools used to extract P(r) require a certain smoothness of the distribution. 6 Several algorithms have shown that the introduction of constraints and smoothing in the distance domain can stabilize the solution. This has been shown for the approximate Pake pattern33 and is also at the core of Tikhonov regularization.16,34
Figure 3. Validation of the symmetry-constrained model fit method.
(a) The background-subtracted dipolar evolution (open circles) obtained for the CorA R252C mutant was fit using 3 different methods: Tikhonov regularization (λ=31) is shown in black, a 2 Rice3D fit with distance ratio <k> = 1.578–1.645 is shown in red, and a 2 Rice3D fit intentionally misconstrained such that <k> = 1.367–1.433 is shown in blue. The inset is a magnification to better illustrate fit disparities. (b) The corresponding distance distributions are shown using the same color code. The dashed vertical lines correspond to average distances calculated from the X-ray structure.30
Statistical analysis of symmetry-constrained distance distributions
Since over-smoothing the fit of the data is a legitimate concern, we sought to investigate the influence of our symmetry-constrained model on a more well-defined CorA mutant dataset. Here, we chose the CorA cysteine mutant R252C that yields data of much better quality as compared to V248C, i.e., a well-defined background-corrected dipolar evolution (Figure 3a). In this case, we observed no substantial difference between P(r) generated by Tikhonov regularization or by the 2 Rice3D model approach (Figure 3b). Interpretation of distances obtained by a symmetry-constrained model fit requires statistical analysis of the goodness of fit. A generalized model distribution function can be defined by parameter vector θ = (θ1, θ2,…, θp) with p fitted parameters. In absence of a symmetry constraint, the least squares estimate of θ, denoted by θ̂, minimizes the error sum of squares (S) between the experimental and simulated dipolar evolution. Note that application of a symmetry constraint does not alter the number of free parameters. In this case, the statistical significance of a constrained nonlinear regression can be assessed by the likelihood-ratio criterion, which defines the 100(1-α)% confidence region35,36 of the parameters as:
| (6) |
where the dipolar evolution has n observations and is the upper critical value of the Fp,n−p distribution at significance level α. Equation (6) defines a threshold at which variation of the parameters causes a fractional increase in S that is statistically significant at level α, typically set at α = 0.05 (2σ).
The likelihood-ratio allows rigorous estimation of parameter uncertainty by error surface analysis.37 The procedure is to first obtain the optimal parameter set θ̂ from a standard (unconstrained) model fit with minimum error, i.e., S(θ̂) or RMSD (θ̂). Next, the selected parameter is incremented away from its optimal value by a fixed step size and the regression is repeated (with all other parameters simultaneously optimized). This process continues until the fit error exceeds the threshold value. The parameter value that defines the exact confidence interval boundary is obtained by quadratic interpolation. This procedure is then repeated in the opposite direction to obtain the opposite boundary.
By inversion, the likelihood ratio also gives an F-test to determine if the model fit is significantly different in constrained vs. standard form. This F-test thus has the null hypothesis H0: θ = θ̂ and alternative hypothesis HA: θ ≠ θ̂. For example, if a symmetry-constrained fit yields S(θ) that is greater than the right-hand side of Equation (6), the parameter set is significantly different. We analyzed our data using the DeerAnalysis program16, which reports goodness of fit by RMSD. Therefore we used the equivalent form of Equation (6) to calculate all F-test threshold values reported in Table 1:
| (7) |
This F-test allows us to compare, without bias, the goodness of fit between a symmetry-constrained and standard form of the same model. As a proof of principle of this F-test, we intentionally misconstrained our fit of the R252C dataset using a 2 Rice3D distribution with distance ratio <k> = 1.367–1.433 (from rotamer simulation), as if CorA was a tetramer (Figure 3). As anticipated, the small variation in the fit of the dipolar evolution translates into a dramatic change in the distance distribution. The values reported in Table 1 show that the incorrect model has RMSD above the F-test threshold, letting the user know that the model fit is significantly different (worse) at α = 0.05. This demonstrates that the use of this F-test gives the user a valid and useful approach to compare different models and fitting procedures for analysis of DEER data.
Validation on a homotetrameric K+ channel
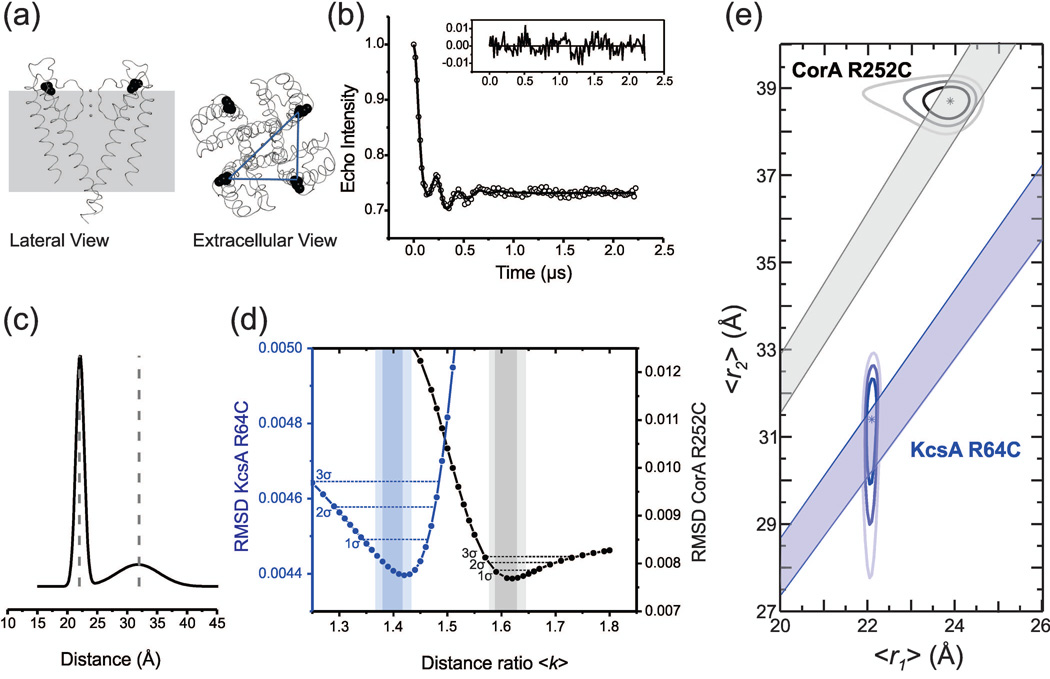
We further validated the method on another membrane protein harboring a different oligomeric state. We chose the well characterized K+ channel KcsA because we can easily compare the DEER-derived distance distribution against its well-accepted crystal structure.38 The residue R64, located in the outer vestibule of KcsA, has already been shown to be suitable for DEER measurement and was therefore our first choice.7 Remarkably, as illustrated in Figure 4, we were able to find excellent agreement between our R64C dataset fit by a 2 Rice3D distribution constrained by <k> = 1.367–1.433, the crystal structure39 and previously reported DEER measurements7 (Table 1). As another proof of principle for the F-test, we observed that when we intentionally misconstrained the fit with distance ratio <k> = 1.578–1.645, as if KcsA was a pentamer, the RMSD exceeds the F-test threshold value at α = 0.05. Interestingly, the distance ratio error surface (RMSD vs. <k>) shows that the error minimum is located close to the theoretical <k> value for both systems (CorA and KcsA). There are several important observations which apply to both systems: (1) the theoretical <k> values are contained within their respective 95% confidence intervals, and (2) there is no overlap between 95% confidence intervals of <k> (Figure 4d). This result indicates that the <k> from a 2 Rice3D fit can be used to distinguish between a homotetrameric and a homopentameric assembly. It also suggests that a 95% confidence interval (2σ) is likely to be the best compromise between statistical significance and stringency.
Figure 4. DEER measurements from the homotetrameric K+ channel KcsA R64C and approximate confidence regions.
(a) The R64 residue is shown in black on a ribbon representation of the X-ray structure. 39 (b) The background-corrected dipolar evolution is shown as open circles and the fitted dipolar evolution from a 2 Rice3D model is plotted as a solid line; fit residuals are shown in the inset. (c) The corresponding distance distribution is shown as a solid line together with the expected distances (dashed lines) from the crystal structure. (d) The error surface of the distance ratio <k> from 2 Rice3D model fits of KcsA R64C (blue) and CorA 252C (black). Distance ratio confidence intervals by F-test at α = 0.32 (1σ), α = 0.05 (2σ), and α = 0.003 (3σ) significance levels are shown as dashed horizontal lines. Shadowed areas represent the 5–95% and 1–99% <k> intervals determined by rotamer simulation. (e) Plots of the 68%, 95%, and 99.7% approximate confidence regions of mean distances (〈r1〉, 〈r2〉) obtained by 2 Rice3D model analysis of DEER measurement from CorA R252C and KcsA R64C. The asterisk is positioned at the optimal parameter set θ̂. Shadowed areas represent the 1–99% <k> intervals.
Distance confidence regions as a measure of uncertainty for DEER measurements
The ability to perform an F-test on the RMSD from a DEER fit allowed us to determine distance confidence regions for model-based fits of our DEER data. Distance confidence region analysis conveys the most complete representation of the confidence of recovered distances. Figure 4e shows various confidence regions for mean distances (〈r1〉, 〈r2〉) obtained by 2 Rice3D analysis of CorA R252C and KcsA R64C datasets. Each dataset was fit by the model at fixed combinations of mean distance (〈 r1〉, 〈 r2〉) within a uniformly-spaced 2D grid of test points that encompassed the solution basin. At each test point (〈r1〉, 〈r2〉), all other parameters were simultaneously optimized. The RMSD from the set of grid solutions was then interpolated (cubic) at 10× higher resolution to obtain a smooth expectation surface, followed by isocontour evaluation at selected levels of significance. The 100(1 −α)% confidence interval of a selected parameter (e.g., 〈r1〉), can be obtained by projection of the 100(1 − α)% confidence region onto the selected parameter axis. For these well-defined datasets, several observations can be made about the Rice3D model and its parameter estimation. First, the optimal solution (and the majority of the 68% confidence region) lies within the respective <k> intervals obtained by rotamer simulation. Second, the confidence regions are approximately elliptical, with slight distortion for the CorA R252C dataset, indicating low degree of nonlinearity (a linear model has elliptical confidence regions). Third, the relative roundness of the confidence regions indicates stable parameters, which means that the estimation precision of both distance parameters is balanced and there is low parameter-effects curvature (i.e., well-conditioned parameterization). Fourth, the small off-axis tilt of the confidence regions indicates low parameter correlation. Combined, these observations validate our proposed use of statistical inference for the 2 Rice3D model.
Influence of the C-terminal domain on activation gating of KcsA
The activity of ion channels is often regulated by interaction with ligand or other subunits via their cytoplasmic domain (N- or C-terminus). For example, Ca2+ activation of BK channels is mediated through its binding at the cytoplasmic domain RCK.40,41 In some prokaryotic K+ channels, the cytoplasmic domain plays an important role in protein folding and thermal stability.42–44 The KcsA CTD directly connects the gating transmembrane helices TM2 and fold as a 4 helix-bundle that projects ~70 Å into the cytoplasm.44,45 Using a chaperone assisted crystallography approach, the structure of the full length (FL) KcsA was recently solved in an open/inactivated conformation.24 These studies have revealed that the CTD remains a four helix bundle during the gating process and by exerting a physical strain on the lower gate, restricts the motion of the gating helices.23,24 This observation correlates well with the increased rate and extent of C-type inactivation when the CTD is truncated.46 Moreover, binding of crystallographic chaperones (Fab fragments) has functional consequences: slowing down the rate of C-type inactivation.24 Since C-type inactivation is mechanically coupled to the opening of the lower gate,23,46 it is reasonable to propose that the Fab fragment used for crystallography provides additional strain on the lower gate that would further stabilize the closed conformation.
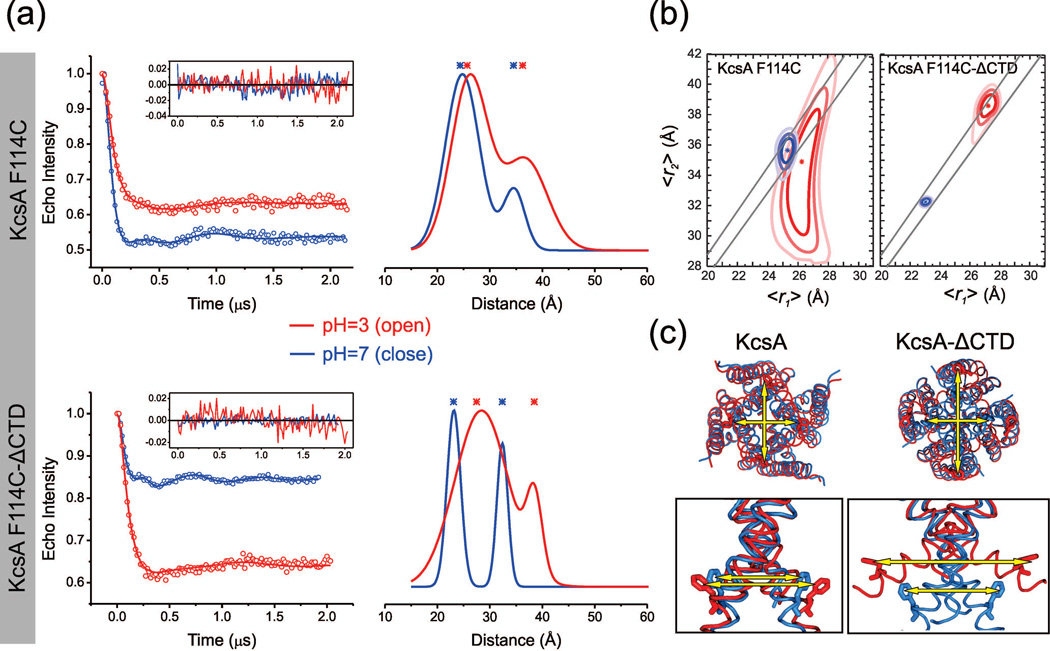
Taking advantage of our ability to reliably determine distance in homomeric proteins, we sought to monitor the influence of the KcsA CTD on the extent of lower gate opening in the absence of any antibody or artificial crystal lattice contact. The outward facing position 114 located at the lower gate was an ideal reporter candidate because interprobe separation is compatible with the distance range of the DEER technique (20–80 Å). The experiments were conducted for both constructs (FL and ΔCTD) at pH=7 which stabilizes the closed state and at pH=3 which favors the open conformation. In the closed state (pH=7), the truncation of the CTD is known to produce a substantial structural rearrangement of the lower gate as detected semi-quantitatively by FRET46 and revealed in further detail by X-ray crystallography.45 Our DEER experiments for F114C clearly show that the interprobe distances change at pH=7 when KcsA is truncated (Figure 5). The mean distances (<r1>,<r2>) decrease from (25.3, 35.6 Å) for the FL channel to (23.1, 35.6 Å) for the ΔCTD channel. Although small, this modest rearrangement is reliably reported by our DEER experiment as there is no overlap of the distance confidence regions at the 99.7% (3σ) confidence level (Figure 5b). This distance difference seems fully compatible with the 15° outward tilting of TM2 after residue 110 observed by crystallography when truncated structures are compared to the full length channel.45
Figure 5. Influence of the C-Terminal Domain on activation gating of KcsA.
(a) Background subtracted dipolar evolution of KcsA-F114C FL and ΔCTD are shown in upper and lower panels, respectively. Data recorded at pH=3 and pH=7 are shown in red and blue respectively. For each construct, the inset displays the fit residuals and the right panel shows the corresponding distance distribution obtained by a symmetry-constrained 2 Rice3D model fit constrained with <k> = 1.367–1.433. For each dataset <r1> and <r2> are shown by asterisks. (b) Plots of the 68%, 95%, and 99.7% approximate confidence regions of mean distances (〈r1〉, 〈r2〉), obtained by symmetry-constrained 2 Rice3D model analysis of DEER measurements from KcsA-F114C FL and ΔCTD. Measurements obtained at pH=7 (closed) and pH=3 (open) are colored blue and red, respectively. An asterisk is positioned at each optimal parameter set θ̂. The two diagonal lines represent the 1–99% <k> interval (1.367–1.433). (c) KcsA closed (3EFF)45 and open (3PJS)24 structures are aligned and ribbon-represented using the same color code. The inset represents a lateral magnification of the activation gate where F114 is stick-represented. An analogous representation for the truncated channel is shown in the lower panels using 1K4C38 and 3F7V23 PDB accession codes.
In the full length channel, we observe a very modest change in interprobe distances when the channel is triggered to open (Figure 5a). In the open state, the dipolar evolution is flatter and the distance ratio constraint <k> is necessary to find a satisfactory solution (Figure 5). The confidence region analysis revealed no overlap at the 68% (1σ) level in the allowed distance region between the pH=7 and pH=3 datasets. However, there is a significant overlap at the 95% (2σ) level, indicating that uncertainties of the DEER measurements at this confidence level are greater than the inferred motion when KcsA-FL opens.
One major conclusion of the experiment is that the CTD truncation allows wider opening of the activation gate at position 114. This is evidenced by the large change in the distance distribution of TM2. During opening, the mean interprobe distance change is more than 6 Å for the diagonal distance: (<r1>, <r2>) increases from (23.1, 32.3 Å) at pH=7 to (27.5, 38.6 Å) at pH=3 (Figure 5 and Table 2). Several KcsA-ΔCTD open structures have been crystallized, with lower gate openings (using T112 Cα-Cα diagonal distance as a reporter) ranging from 15, 17, 23, 32 Å open (the closed state being 12 Å).23 The average distance change observed by DEER seems in excellent agreement with the intermediate opening states (structures 15 Å and 17 Å open) captured by crystallography23 with distance changes in Cα-Cα at residue T112 to be 4–6 Å from the closed state. The broad range of opening captured by crystallography is likely correlated to the wider distance distribution observed by DEER spectroscopy at pH=3. Interestingly, the widest opening state captured by crystallography23 seems at odds with the average conformation observed in our ensemble DEER measurement, suggesting that this very wide open state is either transient or only occurs in the presence of stabilizing crystal lattice forces.
Table 2.
Distance determination at KcsA’s activation gate from constrained 2 Rice3D analysis
| Construct | Experimental Conditions |
〈r1〉: Adjacent Distance (Å)* |
〈r2〉: Diagonal Distance (Å)* |
Fit RMSD | Threshold RMSD (α=0.05) |
|---|---|---|---|---|---|
| KcsA F114C | pH=7 (closed) | 25.3 (± 3) | 35.6 (± 1.8) | 0.0088333 | 0.0092203 |
| pH=3 (open) | 26.5 (± 2.9) | 36.2 (± 4.5) | 0.0087260 | 0.0090727 | |
| KcsA F114C-ΔCTD | pH=7 (closed) | 23.1 (± 1.3) | 32.3 (± 1) | 0.0029793 | 0.0031230 |
| pH=3 (open) | 27.5 (± 5.3) | 38.6 (± 1.2) | 0.0062729 | 0.0065581 | |
Values reported in parentheses are fit values of σR from the 2 Rice3D model (i.e., not distance confidence intervals).
Interestingly, the dipolar evolution from F114C obtained in the open state (pH=3) is systematically flatter than its closed state (pH=7) counterpart, leading to a much broader distance distribution. This observation can be interpreted as an increase in protein dynamics at the lower gate suggesting that upon activation, the lower gate is sampling a larger conformational space than when the channel is closed. This observation is not likely to arise from an increase in probe dynamics, because the probe mobility does not increase upon activation as measured by CW-EPR.20 Since the opening of lower gate is physically coupled to the inactivation of the selectivity filter via allosteric coupling,46 determining the correlation between the dynamics of the lower gate with the conformational fluctuations of the selectivity filter represents an important step towards the detailed understanding of ion conduction and C-type inactivation in K+ channels.
CONCLUSIONS
In summary, we have implemented a simple but efficient strategy to stabilize the fitting solution of DEER data using the implicit organization of homomeric proteins. The methods described here will be available in a future release of DeerAnalysis software,16 available at http://www.epr.ethz.ch/software/. The symmetry-constrained model fit allows quantitative and statistical evaluation of theoretical vs. experimentally derived distance distributions. We have experimentally shown that our approach is valid for a homotetramer as well as for a homopentamer. We hope that the method presented here will be particularly advantageous for challenging targets and highly dynamic systems where the distance distributions are broader, and thus more difficult to accurately fit due to a flatter dipolar evolution.
We have used the potential of this method to analyze the influence of the KcsA C-terminal domain on the extent of lower gate motion. We confirmed that in absence of antibody and crystal lattice contact, the CTD restricts lower gate movement, a finding in excellent agreement with the increased rate and extent of C-type inactivation accompanied by CTD truncation.46 We also found that in absence of any crystallographic lattice contact or Fab fragment attached to the protein, KcsA’s lower gate opens approximately 6– 8 Å, suggesting that the widest opening (32 Å) captured by crystallography might be a rare or transient event possibly captured by stabilizing crystal lattice forces. Finally, the broadening of our distance distribution when KcsA opens has revealed the dynamic nature of the lower gate suggesting that upon activation, the lower gate populates multiple conformationally open states.
ACKNOWLEDGMENTS
The authors thank Dr. Gunnar Jeschke for feedback and helpful advice with DEER Analysis. We also thank Dr. Luis G. Cuello and members of the Perozo lab for technical advice and manuscript review. O.D. thanks the organizers of the EPR Workshop 2010: Cutting-Edge Biomedical EPR Methods, for a stimulating experience on the pulsed EPR technique. This work was supported by NIH grant GM088406.
REFERENCES
- 1.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Nature. 2002;418:942. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 2.Perozo E, Cuello LG, Cortes DM, Liu YS, Sompornpisut P. Novartis Found Symp. 2002;245:146. doi: 10.1002/0470868759.ch10. [DOI] [PubMed] [Google Scholar]
- 3.Claxton DP, Quick M, Shi L, de Carvalho FD, Weinstein H, Javitch JA, Mchaourab HS. Nat Struct Mol Biol. 2010;17:822. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smirnova I, Kasho V, Choe JY, Altenbach C, Hubbell WL, Kaback HR. Proc Natl Acad Sci U S A. 2007;104:16504. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borbat PP, Surendhran K, Bortolus M, Zou P, Freed JH, Mchaourab HS. PLoS Biol. 2007;5:e271. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeschke G, Polyhach Y. Phys Chem Chem Phys. 2007;9:1895. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 7.Endeward B, Butterwick JA, MacKinnon R, Prisner TF. J Am Chem Soc. 2009;131:15246. doi: 10.1021/ja904808n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeschke G, Sajid M, Schulte M, Godt A. Phys Chem Chem Phys. 2009;11:6580. doi: 10.1039/b905724b. [DOI] [PubMed] [Google Scholar]
- 9.Payandeh J, Scheuer T, Zheng N, Catterall WA. Nature. 2011;475:353. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catterall WA. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Science. 1998;280:69. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 12.Lunin VV, Dobrovetsky E, Khutoreskaya G, Zhang R, Joachimiak A, Doyle DA, Bochkarev A, Maguire ME, Edwards AM, Koth CM. Nature. 2006;440:833. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Science. 1998;282:2220. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 14.Hilf RJ, Dutzler R. Nature. 2008;452:375. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 15.Schlick S. Wiley InterScience (Online service) Hoboken, N.J.: Wiley-Interscience; 2006. p. xiv. [Google Scholar]
- 16.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl Magn Reson. 2006;30:473. [Google Scholar]
- 17.Tikhonov AN, Arsenin VIA. Solutions of ill-posed problems. Winston: 1977. [Google Scholar]
- 18.Hansen PC, O'Leary DP. SIAM Journal on Scientific Computing. 1993;14:1487. [Google Scholar]
- 19.Domingo Kohler S, Spitzbarth M, Diederichs K, Exner TE, Drescher M. J Magn Reson. 2011;208:167. doi: 10.1016/j.jmr.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Perozo E, Cortes DM, Cuello LG. Science. 1999;285:73. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 21.Blunck R, Cordero-Morales JF, Cuello LG, Perozo E, Bezanilla F. J Gen Physiol. 2006;128:569. doi: 10.1085/jgp.200609638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Nat Struct Mol Biol. 2007;14:1089. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuello LG, Jogini V, Cortes DM, Perozo E. Nature. 2010;466:203. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uysal S, Cuello LG, Cortes DM, Koide S, Kossiakoff AA, Perozo E. Proc Natl Acad Sci U S A. 2011;108:11896. doi: 10.1073/pnas.1105112108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalmas O, Cuello LG, Jogini V, Cortes DM, Roux B, Perozo E. Structure. 2010;18:868. doi: 10.1016/j.str.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perozo E, Cortes DM, Cuello LG. Nat Struct Biol. 1998;5:459. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 27.Miller KS. Multidimensional Gaussian distributions. New York: Wiley; 1964. [Google Scholar]
- 28.Jakeman E, Tough RJA. J Opt Soc Am A. 1987;4:1764. [Google Scholar]
- 29.Kohler DS, Spitzbarth M, Diederichs K, Exner TE, Drescher M. J Magn Reson. 2011;208:167. doi: 10.1016/j.jmr.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Eshaghi S, Niegowski D, Kohl A, Martinez Molina D, Lesley SA, Nordlund P. Science. 2006;313:354. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 31.Polyhach Y, Bordignon E, Jeschke G. Phys Chem Chem Phys. 2011;13:2356. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 32.Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kalai T, Hideg K, Hubbell WL. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeschke G, Koch A, Jonas U, Godt A. J Magn Reson. 2002;155:72. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 34.Jeschke G, Bender A, Paulsen H, Zimmermann H, Godt A. J Magn Reson. 2004;169:1. doi: 10.1016/j.jmr.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Beale EML. J Roy Stat Soc B. 1960;22:41. [Google Scholar]
- 36.Seber GAF, Wild CJ. Nonlinear regression. New York: Wiley; 1989. [Google Scholar]
- 37.Straume M, Frasier-Cadoret S, Johnson M. In: Topics in fluorescence spectroscopy. Lakowicz J, editor. New York: Plenum Press; 1991. p. 177. [Google Scholar]
- 38.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Nature. 2001;414:43. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 39.Morais-Cabral JH, Zhou Y, MacKinnon R. Nature. 2001;414:37. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 40.Yuan P, Leonetti MD, Hsiung Y, MacKinnon R. Nature. 2012;481:94. doi: 10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Yang Y, Ye S, Jiang Y. Nature. 2010;466:393. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iscla I, Wray R, Blount P. Protein science : a publication of the Protein Society. 2011;20:1638. doi: 10.1002/pro.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pau V, Zhu Y, Yuchi Z, Hoang Q, Yang D. The Journal of biological chemistry. 2007;282:29163. doi: 10.1074/jbc.M703277200. [DOI] [PubMed] [Google Scholar]
- 44.Cortes DM, Cuello LG, Perozo E. J Gen Physiol. 2001;117:165. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uysal S, Vasquez V, Tereshko V, Esaki K, Fellouse FA, Sidhu SS, Koide S, Perozo E, Kossiakoff A. Proc Natl Acad Sci U S A. 2009;106:6644. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuello LG, Jogini V, Cortes DM, Pan AC, Gagnon DG, Dalmas O, Cordero-Morales JF, Chakrapani S, Roux B, Perozo E. Nature. 2010;466:272. doi: 10.1038/nature09136. [DOI] [PMC free article] [PubMed] [Google Scholar]