SUMMARY
The generation of human induced Pluripotent Stem (iPS) cells holds great promise for development of regenerative medicine therapies to treat a wide range of human diseases. However, the generation of iPS cells in the absence of integrative DNA vectors remains problematic. Here we report a simple, highly reproducible RNA-based iPS generation approach that utilizes a single, synthetic self-replicating VEE-RF RNA replicon that expresses four reprogramming factors, OCT4, KLF4, SOX2 with c-MYC or GLIS1 at consistent high levels prior to regulated RNA degradation. A single VEE-RF RNA transfection into newborn or adult human fibroblasts resulted in efficient generation of iPS cells with all the hallmarks of stem cells, including cell surface markers, global gene expression profiles and in vivo pluripotency to differentiate into all three germ layers. The VEE-RF RNA-based approach has broad applicability for the generation of iPS cells for ultimate use in human stem cell therapies in regenerative medicine.
INTRODUCTION
The generation of human induced Pluripotent Stem (iPS) cells by retroviral expression of four reprogramming factors opened the potential for regenerative medicine therapies based on patient-specific, personalized stem cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). However, the insertional mutagenic potential of retroviruses combined with the potential for latent reprogramming factor gene activation, especially c-MYC, all but eliminates integrative DNA-based approaches for use in regenerative medicine therapies (Okita and Yamanaka, 2011; González et al., 2011; Hussein et al., 2011; Ben-David and Benvenisty, 2011). Several methods based on DNA, RNA, miRNAs and proteins have been developed to generate integration free iPS cells, and the advantages and disadvantages have been discussed elsewhere (González et al., 2011; Hussein et al., 2011; Mochiduki and Okita, 2012; Okita and Yamanaka, 2011;). Of all these methods, RNA-based iPS cell approaches using Sendai virus (Fusaki et al., 2009), miRNAs and mRNA transfection (Warren et al., 2010) avoid potential integration problems associated with DNA-based approaches and at this point in time, appear inherently safer methods for future clinical applications.
Expression of pluripotent factors by infection with Sendai virus, a negative-sense, single stranded RNA virus that does not go through a DNA intermediate, offers a highly efficient iPS approach in the absence of concerns for integration into the genome (Fusaki et al., 2009; Ban et al., 2011). However, due to persistent Sendai virus replication in iPS cell clones, this approach requires a negative selection step followed by one or more recloning steps from the single cell level to isolate virus-free iPS cells. A temperature-sensitive mutant of Sendai virus is a successful alternative method to remove the virus (Ban et al., 2011), though it requires a higher biosafety due to production of infectious virus particles. One of the more promising non-DNA based iPS approaches involves transfection of four individual RF mRNAs generated by in vitro transcription (Warren et al., 2010; Angel et al., 2010; Warren et al., 2012). Recent work has shown that activation of the innate immune system enhances the overall efficiency of iPS cell generation by repeated mRNA transfection (Lee et al., 2012). However, due to the rapid degradation of reprogramming factor mRNAs, this approach requires repetitive daily transfection of four individual mRNAs into the same target cells over the 14 day reprogramming period. Although both Sendai virus and mRNA transfection approaches have been shown to generate iPS cells, there remains a significant need for a simple, highly reproducible, non-DNA based approach to generate human iPS cells.
To develop an RNA-based iPS generation strategy, we focused our efforts on an approach that utilizes: 1) a single RNA species capable of self-replicating for a limited number of cell divisions, thereby reducing the number of transfections; 2) was capable of encoding at least four reprogramming factor open reading frames (ORFs); 3) consistently expressed all reprogramming factor genes at high threshold levels over multiple cellular divisions; and 4) could be selectively retained and degraded in a controlled fashion. To ectopically express all four reprogramming factors, we modified a non-infectious (non-packaging), self-replicating Venezuelan Equine Encephalitis (VEE) virus RNA replicon (Kinney et al., 1989; Petrakova et al., 2005) that is currently being investigated as an expression platform for vaccine development (Davis et al., 2002; Durbin and Whitehead, 2010). The VEE replicon is a positive-sense, single stranded RNA that mimics cellular mRNA with a 5′-Cap and poly(A) tail and does not utilize a DNA intermediate, so there is no potential for genomic integration (Kinney et al., 1989). Here we report on the generation of human iPS cells by a single transfection of a self-replicating VEE RNA species that expresses four reprogramming factor ORFs (OCT4, SOX2, KLF4 with either c-MYC or GLIS1). VEE reprogramming factor RNA generated human iPS cells that were free of VEE RNA and had all the hallmarks of human stem cells (expression of ES cell markers, global gene expression, differentiation in vivo into all three germ lineages). The VEE-RF RNA can also be selectively retained or removed from cells. The non-DNA and non-integrating, self-replicating VEE RNA approach has the potential to simplify the generation of human iPS cells for use in disease cell modeling studies and eventual cell therapy applications.
RESULTS
Development of self-replicative VEE RNA to express reprogramming factors
To develop a single RNA iPS generation approach, we focused on a polycistronic, self-replicative RNA system that would consistently express the reprogramming factors over multiple cellular divisions. We modified a non-infectious, self-replicating VEE RNA (Petrakova et al., 2005). VEE RNA is a positive stranded RNA that encodes four non-structural replication complex proteins (nsP) as a single ORF in the 5′ end of the RNA that is separated from the viral structural protein ORFs in the 3′ end (Figure 1A). Petrakova et al. showed the ability to express exogenous proteins by replacing the 3′ structural protein ORFs with GFP (Petrakova et al., 2005). To evaluate the VEE RNA replicon in primary human fibroblasts, we replaced the 3′ORF with GFP, followed by an internal ribosomal entry site (IRES) (Pelletier and Sonenberg, 1988) and a Puromycin resistance gene (Puror) (Figure 1A). Current SP6 or T7 RNA in vitro transcription kits can transcribe RNAs in excess of 25 kb in length (Schelle and Thiel, 2007). VEE-GFP RNA was produced using either SP6 or T7 RNA polymerases from a standard in vitro transcription kit followed by 5′-capping, and poly(A) tail addition resulting in a high yield, full length 11,500 nt RNA transcript. In our hands, both SP6 and T7 RNA polymerases readily produced high yield in vitro transcripts in excess of 14,000 nt (Figure S1A).
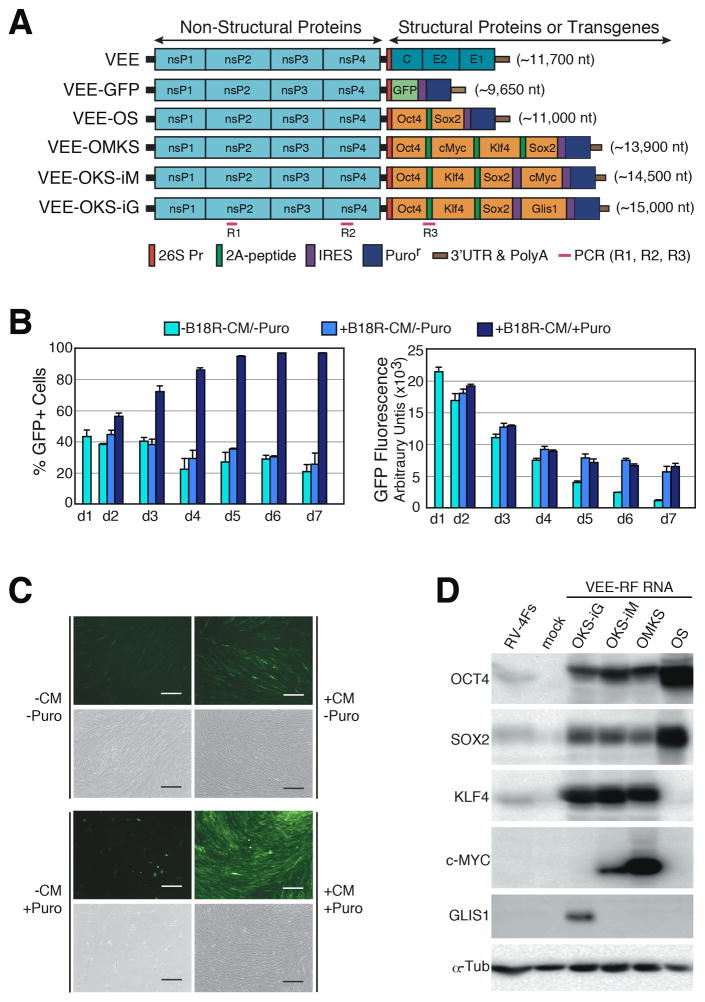
Figure 1. Construction and Persistence of Synthetic VEE-RF RNA Replicons in Primary Human Fibroblasts.
(A) Schematic of VEE-RF RNA replicons. 5′ end nsP1-4: non-structural proteins1–4; 3′ end C, E2, E1: Structural proteins. Location of 26S internal promoter, ribosome shifting 2A peptide, IRES sequence, Puromycin (Puro) resistance gene and PCR detection of replicon as indicated.
(B) B18R-CM Conditioned Media and puromycin selection are required for persistence of VEE-GFP RNA over 7 days. HFF cells were transfected on day 0 with VEE-GFP RNA and treated as indicated. GFP fluorescence of GFP positive cell population was measured by flow cytometry.
(C) B18R-CM and puromycin are required for retention of VEE-GFP RNA. Photographs of GFP expression on day 7 as indicated. Bar, 200 μm.
(D) Immunoblot analysis of VEE RNA expressed reprogramming factors expressed in HFFs cells on day 1 versus retrovirus (RV-4Fs: Oct, Sox2, Klf4, cMyc) expression.
See also Figure S1.
Exposure of cells to single stranded VEE RNA induces a strong IFN-α/β innate immune response. To mitigate the innate immune response to VEE-GFP RNA, we utilized B18R protein from Western Vaccinia virus that binds to and neutralizes type I IFNs (Alcamí et al., 2000). We compared GFP expression in primary human foreskin fibroblasts (HFFs) transfected with VEE-GFP RNA alone or co-transfected with B18R mRNA. Consistent with induction of a strong innate immune response to cells exposed to single stranded RNA, in the absence of B18R, we observed little to no GFP expression one day after transfection (Figure S1B). In contrast, co-transfection of VEE-GFP RNA replicon with B18R mRNA resulted in high levels of GFP expression in HFFs (Figure S1B), showing that B18R is required for efficient expression of proteins from the VEE RNA replicon.
The generation of iPS cells requires consistent, high level expression of reprogramming factors for >7 days; therefore, we examined the persistence of the VEE-GFP RNA replicon in human primary fibroblasts over 7 days. To continuously suppress the innate immune response over several weeks while avoiding daily transfection of B18R mRNA, we prepared conditioned media harvested from human fibroblasts expressing B18R protein (B18R-CM) (Figure S1C and S1D). HFFs were co-transfected with VEE-GFP RNA replicon and B18R mRNA (3:1 ratio) on day 0, then cultured in the presence or absence of 20% B18R-CM plus/minus puromycin on day 1 (Figures 1B). Puromycin selection in the presence of B18R-CM resulted in a >90% GFP positive population, while puromycin selection in the absence of B18R-CM resulted in <1% viable GFP cells (Figure 1B and 1C). We also observed that the level of GFP expression in the presence of B18R-CM gradually decreased from day 1 to day 4, but then remained steady out to day 7. In contrast, the level of GFP expression in the absence of B18R-CM continuously dropped to <10% intensity (Figures 1B). VEE GFP replicon persistence was dose dependent on B18R-CM (Figure S1E and S1F). We note the persistence of high levels of GFP expression from VEE-GFP RNA treated fibroblasts for over a month when continuously cultured in the presence of B18R-CM and puromycin (data not shown). Taken together, these results showed both the necessity of B18R protein to overcome the VEE RNA-induced innate immune response and also demonstrated the ability to selectively retain or degrade the VEE RNA replicon from cells by exposure to or withdrawal from B18R-CM.
Generation of iPS cells by VEE RNA replicon
We next engineered the VEE RNA replicon 3′ ORF to encode four reprogramming factors OCT4, KLF4, SOX2, with c-MYC or GLIS1, which avoids the potential genomic instability induced by c-MYC (Nakagawa et al., 2008; Maekawa et al., 2011). We generated and compared several VEE-RNA construct configurations (Figure 1A) using the following nomenclature: VEE-OMKS = OCT4, c-MYC, KLF4, SOX2 separated by internal ribosomal skipping 2A peptides (Szymczak et al., 2004) followed by an internal ribosomal entry site (IRES) and Puror ORF; VEE-OKS-iM = OCT4, KLF4, SOX2 separated by 2A peptides followed by an IRES then c-MYC and a second IRES and Puror ORF; and VEE-OKS-iG = OCT4, KLF4, SOX2 separated by 2A peptides followed by IRES then GLIS1 and a second IRES and Puror ORF (Figure 1A). Similar to the VEE-GFP RNA protocol, VEE reprogramming factor (VEE-RF) RNAs were produced by SP6 or T7 in vitro transcription, 5′-capping, and poly(A) tail addition resulting in full length VEE-OKS-iM RNA, VEE-OMKS RNA and VEE-OKS-iG RNA (Figure S1A). Transfection of various VEE-RF RNA replicons with B18R mRNA into human fibroblasts resulted in high levels of expression of all four reprogramming factors that exceeded reprogramming factor expression levels from retroviruses on day 1 and day 10 (Figure 1D and S2B). Together, these observations demonstrated the ability to express four reprogramming factors and a Puror gene from a single, synthetic VEE-RF RNA replicon in primary human cells, while utilizing B18R to block the innate immune response.
To develop a VEE-RF RNA replicon based iPS cell protocol, we evaluated several parameters, including number and timing of VEE-RF RNA transfections, selection for VEE-RF RNA replicon retention by puromycin, and the genetic organization of the VEE-RF RNA replicon (Figures 1A, 2A and S2). Human HFF or BJ fibroblasts were co-transfected with VEE-RF RNA replicons and B18R mRNA on day 1, then cultured in the presence of 20% B18R-CM plus puromycin.
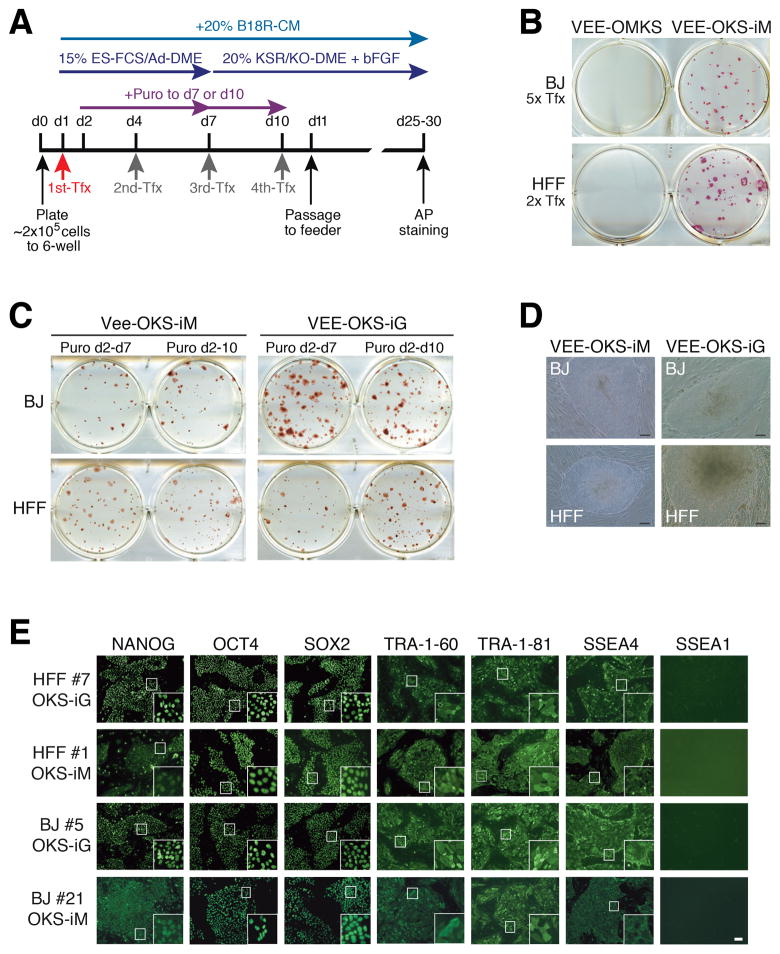
Figure 2. Generation of iPS cells by VEE-RF RNA.
(A) Schematic of epigenetic VEE-RF RNA iPS cell generation protocol. Human fibroblasts were plated on day 0 (d0) and co-transfected (Tfx) with VEE-RF RNA replicon plus B18R mRNA on day 1 (confluent, ~4×105 cells) and treated with puromycin until day 7 (or 10) as indicated. Cells were cultured in B18R-CM until iPS cell colonies were isolated on day 25 (to 30).
(B) iPS cell colonies stained with Alkaline Phosphatase were generated with VEE-OKS-iM RNA, but not VEE-OMKS RNA. Transfection was performed on day 1 and 3 (2x Tfx), or 1, 3, 5, 7 and 9 (5x Tfx).
(C) Alkaline Phosphatase staining of iPS cell colonies generated from BJ or HFFs from d1, 4, 7, 10 transfection protocol as indicated.
(D) Typical images of iPS cell colonies on day 26 by VEE-OKS-iM RNA and day 22 for VEE-OKS-iG RNA from BJ or HFFs fibroblasts as indicated. Bar, 100 μm.
(E) Immunohistochemistry staining of pluripotent ES marker genes in isolated iPS cell clones generated as indicated. Similar results obtained for 26 additional iPS cell clones (30 clones total). Bar, 100 μm; insert, 10× amplification.
See also Figures S2, S5 and S6.
Based on expression results of VEE-RF RNAs (Figures 1D and S2), we initially compared iPS cell generation from either twice (day 1 and 3) or 5-times (day 1, 3, 5, 7 and 9) transfections of VEE-OMKS RNA or VEE-OSK-iM RNA. Strikingly, Alkaline Phosphatase (AP) positive staining iPS colonies were only generated from VEE-OKS-iM RNA in both BJ and HFFs, and no iPS cell colonies were observed from VEE-OMKS RNA (Figure 2B). One significant difference between the two RNA vectors is the relative level of c-MYC expression to the other reprogramming factors with lower c-MYC levels expressed from the VEE-OKS-iM RNA, where c-Myc is the last ORF with a 5′ IRES, and higher levels of c-Myc expressed from the VEE-OMKS RNA non-iPS generating replicon, where c-Myc is the second ORF and utilizes a 2A ribosome skipping peptide (Figures 1A, 1D and S2B). Moreover, we noted an inverse c-MYC expression sensitivity to generating iPS cells using retroviral vectors, where high c-Myc levels were correlated with a decreased number of iPS cell colonies. Using VEE-OKS-iM RNA, we further optimized conditions for iPS cell generation and found that B18R-CM was required until the appearance of iPS cell colonies on feeding culture, whereas puromycin could be removed at the point of plating onto feeder cultures (Figure S2C).
To avoid the potential for genomic instability induced by c-MYC, we also generated a related VEE-OKS-iG RNA construct that substituted GLIS1 for c-MYC (Figure 1A). Initially, we found that several transfections of either VEE-OKS-iM RNA or VEE-OKS-iG RNA in the presence of B18R-CM and puromycin selection over the first seven days resulted in the highest generation of AP positive colonies (Figures 2C and 2D; Table S1). Starting with 1 well of a 6-well format (4 ×105 cells/well), we generally observed >100 iPS colonies per starting well from both HFF and BJ fibroblasts when using three (or more) VEE-RF RNA transfections (Table S1). Although iPS cells were generated in the absence of puromycin selection, we observed a substantially reduced efficiency. To further refine the approach, we changed the type of media used during the first 7 days after transfection to either Advanced-DME or Pluriton, both of which resulted in a large number of AP positive colonies from a single transfection of either VEE-OKS-iM RNA or VEE-OKS-iG RNA in BJ or HFF fibroblasts (Table 1). Moreover, a single transfection of VEE-OKS-iM RNA or VEE-OKS-iG RNA produced from either SP6 or T7 RNA polymerases into human adult NHDFc (aged 50) and HDF (aged 58) fibroblasts generated AP positive colonies with a characteristic iPS cell morphology (Table 1). Thus, we refined the methodology to generate iPS cells from a single transfection of the VEE-RF RNA replicon into both newborn and adult human fibroblasts.
Table 1.
iPS Generation by Single Transfection of VEE-RF RNA Replicon
| RNA Replicon | Cell | Age | Tfx Day | Transfection Media | AP+ Colonies per starting well |
|---|---|---|---|---|---|
| SP6-OKS-iM T7-OKS-iM |
BJ BJ |
Newborn Newborn |
Day 1 Day 1 |
Pluriton Pluriton |
89 181 |
| SP6-OKS-iM | BJ | Newborn | Day 1 | Ad-DME | 6 |
| SP6-OKS-iG T7-OKS-iG |
BJ BJ |
Newborn Newborn |
Day 1 Day 1 |
Pluriton Pluriton |
173 167 |
| SP6-OKS-iM T7-OKS-iM |
HFF HFF |
Newborn Newborn |
Day 1 Day 1 |
Pluriton Pluriton |
245 169 |
| SP6-OKS-iM T7-OKS-iM |
HFF HFF |
Newborn Newborn |
Day 1 Day 1 |
Ad-DME Ad-DME |
757 422 |
| T7-OKS-iG | HFF | Newborn | Day 1 | Ad-DME | 51 |
| SP6-OKS-iM T7-OKS-iM |
NHDFc NHDFc |
Adult Adult |
Day 1 Day 1 |
Ad-DME Ad-DME |
59 5 |
| SP6-OKS-iG T7-OKS-iG |
NHDFc NHDFc |
Adult Adult |
Day 1 Day 1 |
Ad-DME Ad-DME |
31 44 |
| SP6-OKS-iM T7-OKS-iM |
HDF HDF |
Adult Adult |
Day 1 Day 1 |
Ad-DME Ad-DME |
8 4 |
| Retroviruses O,K,S,M |
BJ | Newborn | - | Ad-DME | 117 |
| Retroviruses O,K,S,M |
HFF | Newborn | - | Ad-DME | 294 |
| Retroviruses O,K,S,M |
NHDFc | Adult | - | Ad-DME | 22 |
VEE-RF RNAs were generated with SP6 or T7 RNA polymerase as indicated. HFF or BJ newborn fibroblasts or NHDFc (age 50) and HDF (age 58) adult fibroblasts were co-transfected once with VEE-RF RNA replicon and B18R mRNA on day 1. Cells were selected with puromycin from day 2–5. Pluriton medium was present day 1–10, and Advanced-DME (Ad-DME) was present day 1–7, then changed to ES medium. All cultures were passaged to STO feeder cells and cultured in ES medium on day 10. Cells received daily 20% B18R-CM conditioned medium. iPS colonies were stained with AP on day 24 and number of colonies was indicated based on starting well (4 × 105 cells).
Characterization of iPS cell clones
We mechanically isolated >100 iPS cell colonies from multiple independent VEE-OKS-iM RNA and VEE-OKS-iG RNA protocols and had a >95% success rate for the ability of isolated iPS clones to continuously divide and retain a human embryonic stem cell (hESC) morphology. Of the >100 iPS morphology-like clones isolated, we analyzed 30 clones for expression of stem cell markers by immunofluorescence. All 30 VEE-RF RNA iPS clones analyzed (6x HFF VEE-OKS-iM clones, 12x BJ VEE-OKS-iM clones, 6x HFF VEE-OKS-iG clones, 6x BJ VEE-OKS-iG clones) showed strong nuclear staining of endogenous OCT4, SOX2 and NANOG, and strong cell surface staining of SSEA4, TRA-1-60 and TRA-1-81, with negative staining of SSEA1 (Figure 2E). In addition, we examined six clones from human adult HDF fibroblasts and three clones from adult NHDFc fibroblasts by immunofluorescence and found that all clones expressed Tra-1-60, Tra-1-81, SSEA4 and Nanog, but did not express SSEA1 (Figures S5 and S6).
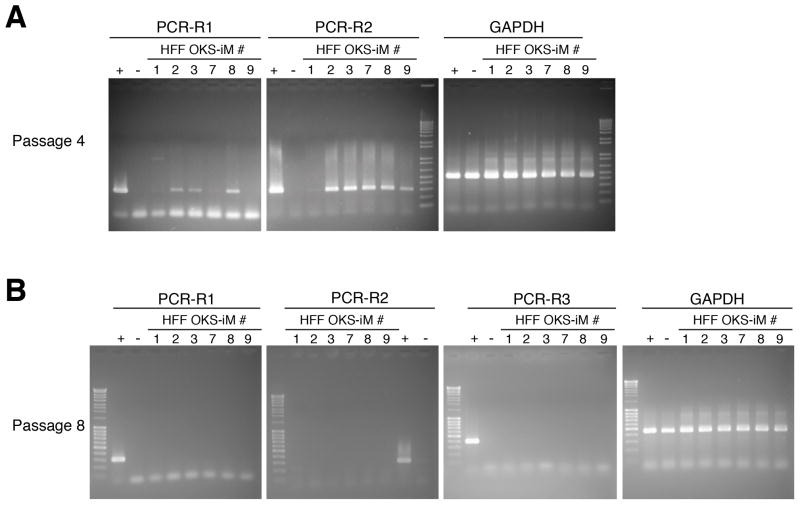
Continuous exposure to B18R-CM was essential for both retention of the VEE-RF RNA replicon and iPS cell generation (Figure 1B and S2C). Withdrawal of B18R-CM from iPS culture medium resulted in the elimination of the VEE-RF RNA replicon. To confirm the complete loss of VEE RF-RNA replicons, we developed a highly sensitive and specific PCR protocol capable of detecting <10 femtogram of the VEE RF-RNA replicon (Figure S3A). RT-PCR analysis of isolated RNAs showed that all iPS cell clones had lost the VEE RF-RNA replicon by passage 8, while most clones lost the RNA replicon in passage 5 or 6 (Figure 3; Table S2). Geuking et al. (2009) reported that non-retroviral RNA viruses under extreme conditions can recombine with endogenous retrotransposon genetic elements and result in a reverse transcription into DNA followed by genomic integration. Therefore, we examined VEE-RF induced iPS clones for the presence of DNA copies. However, consistent with a RNA-only vector that does not go through a DNA intermediate, we did not detect any genomic integrations of VEE-RF by genomic PCR analysis or by Southern blot analysis (Figure S3B–E). A consistent concern for iPS generation protocols is the generation of aneuploid or tetraploid iPS cell clones (Yu et al., 2007). By flow cytometry DNA analysis, we observed several tetraploid iPS cell clones generated from VEE-RF OKS-iM RNA, but no tetraploid colonies were detected from VEE-RF OKS-iG RNA (data not shown). However, karyotype analysis of 4 independent iPS cell clones generated from both OKS-iM and OKS-iG VEE-RF RNA replicons that showed normal DNA content by flow cytometry contained normal diploid karyotypes (Figure S4).
Figure 3. RT-PCR Analysis for Persistent VEE-RF RNA Replicon in iPS Cell Clones.
(A, B) RT-PCR of HFF-OKS-iM iPS cell clones from total RNA prepared from passage 4 (A) and passage 8 (B), as indicated. +, positive control, total RNA was prepared from one day after transfection of OKS-iM-RNA replicon. −, negative control, total RNA was prepared from mock transfected HFFs.
See also Table S2.
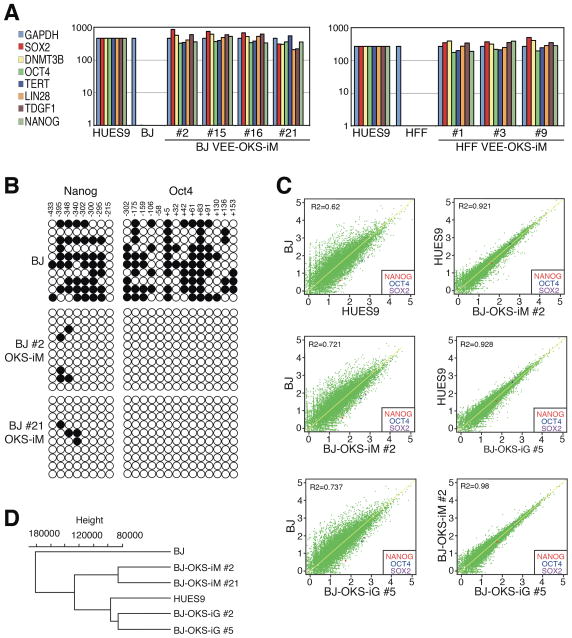
To further characterize the established iPS cell clones, we examined expression of human ES marker genes by qRT-PCR. Consistent with expression levels in human HUES9 ES cells, iPS clones generated from both parental BJ and HFF fibroblasts with either the VEE-OKS-iM RNA or VEE-OKS-iG RNA protocol expressed robust levels of endogenous OCT4, SOX2, NANOG, LIN28, TDGF1, DNMT3B and TERT, in contrast to low or no expression levels in starting parental BJ and HFF fibroblasts (Figure 4A). Likewise, VEE-OKS-iM RNA or VEE-OKS-iG RNA generated iPS clones from human adult HDF and NHDFc fibroblasts also expressed OCT4, SOX2, NANOG, LIN28, TDGF1, DNMT3B and TERT by qRT-PCR (Figures S5 and S6). A hallmark of induced pluripotency is reduced DNA methylation of CpG dinucleotides in the OCT4 and NANOG promoter regions (González et al., 2011). Bisulfite genomic sequencing of both the OCT4 and NANOG promoter regions showed extensive demethylation in iPS cell clones compared to parental fibroblasts (Figure 4B). To investigate mRNA expression profiles in iPS cell clones, we performed whole genome RNA sequencing (RNA-seq). All four iPS cell clones analyzed by RNA-seq showed unsupervised hierarchical clustering and expression signatures characteristic of human HUES9 ES cells that were highly divergent from parental human fibroblasts (Figures 4C and 4D). Together, these results demonstrate that VEE-RF RNA replicon generated iPS cells have all of the expression profile hallmarks of human ES cells.
Figure 4. Characterization of VEE-RF RNA iPS Cell Clones.
(A) Expression of ES maker genes by qRT-RCR analysis from indicated BJ and HFF VEE-RF RNA iPS cell clones.
(B) DNA methylation analysis of NANOG and OCT4 promoter regions. Solid circle, methylated; Open circle, demethylated. Numbers indicate CpG position relative to transcription start site.
(C) Genome-wide mRNA sequence profile scatter plot analysis of BJ-OKS-iM #2 and BJ-OKS-iG #5 compared to parental human BJ fibroblasts and human HUES9 embryonic stem cells with pluripotency NANOG, OCT4, SOX2 indicated.
(D) Unsupervised hierarchical dendrogram of genome-wide RNA sequences analysis showing clustering of four independent iPS cell clones with HUES9 compared to BJ fibroblasts. See also Figures S4, S5, S6 and Table S1.
Differentiation of VEE-RNA Replicon Generated iPS cell clones
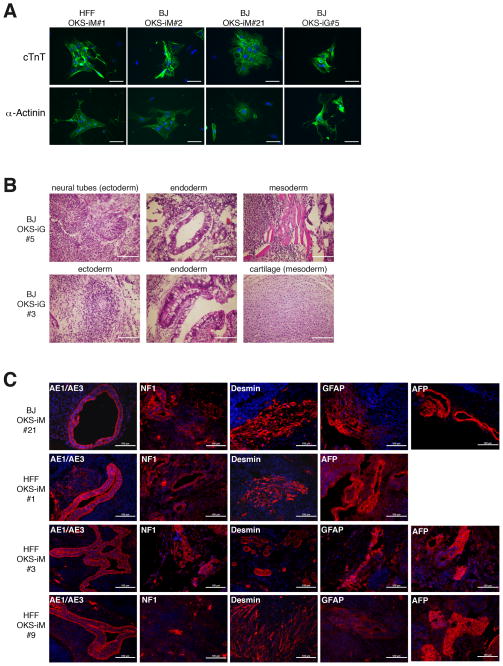
Lastly, we tested the pluripotency of VEE-RF RNA replicon generated human iPS cell clones to differentiate in vitro and in vivo. First, we differentiated iPS cells in vitro into cardiomyocytes per the method of Yang et al. (2008). Four independent iPS cell clones derived from either the OKS-iM or OKS-iG VEE-RF replicons (HFF OKSiM#1, BJ OKS-iM#2 & #21, BJ OKS-iG #5) were treated with activin A, bone morphogenetic protein 4 (BMP4), basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF) and dickkopf homolog 1 (DKK1) in serum-free media. Differentiated embryoid bodies (EB) from all four iPS clones began spontaneously contracting at day 7 and were stably contracting on day 15 (Movie S1). Using antibodies against cTNT and α-Actinin (Figure 5A), immunohistochemistry revealed that all four differentiated iPS EBs were positive for cardiomyocyte markers. These data combined with the spontaneously contracting EBs, confirm the ability of VEE-RNA replicon generated iPS cell clones to differentiate into cardiomyocytes.
Figure 5. Differentiation assays of VEE-RF RNA iPS Cell Clones.
(A) VEE-RF RNA iPS clones were differentiated into cardiomyocytes as described in Experimental Procedures. Contractile embryoid bodies (EBs) were recorded (see supplemental movie S1), and then dissociated and replated onto slides for immunofluorescence staining with mouse anti-Cardiac Troponin or mouse anti-alpha-Actinin, Anti-Mouse IgG Alexa Fluor 488, and DAPI. Bar, 50 μm.
(B) Teratoma formation of VEE-RF RNA iPS clones in nude mice. H&E staining; Bar, 100 μm.
(C) Immunohistochemistry staining of VEE-RF RNA iPS cell clone teratomas in nude mice. AE1/AE3 (cytokeratin), NF-1 (neuronal cells) and GFAP (neuronal cells) used for markers of ectoderm; Desmin (muscle cells) used for marker of mesoderm; and AFP (primitive and definitive endoderm) used for marker of endoderm. Bar, 100 μm.
To test for in vivo pluripotency to differentiate into cells of all three germ layers, human VEE-RF RNA iPS cell clones were injected into immunodeficient mice to generate differentiated teratomas. H&E staining of sections from two independent iPS cell clones contained representative cell types of all three germ layers, ectoderm, endoderm and mesoderm, that spread throughout the sections (Figure 5B). Immunohistochemistry staining of four additional, independent iPS cell derived teratomas were positive for ectoderm markers AE1/AE3 (cytokeratin), NF-1 (neuronal cells) and GFAP (neuronal cells); mesoderm marker Desmin (muscle cells); and endoderm marker AFP (primitive and definitive endoderm) (Figures 5C). Collectively, these observations confirm the ability of both VEE-OKS-iM and VEE-OKS-iG RNA replicons to efficiently generate pluripotent human iPS cells.
DISCUSSION
The generation of iPS cells has great potential for the development of personalized stem cell therapies (Okita and Yamanaka, 2011; González et al., 2011; Hussein et al., 2011). Here we devised a simple, straightforward and highly reproducible RNA-based approach to generate human iPS cells by a single transfection of a synthetic, polycistronic self-replicating VEE-RF RNA replicon that expresses four independent reprogramming factors plus a selectable marker (Puror) for positive selection of the RNA replicon. We found that both VEE-OKS-iM RNA and VEE-OKS-iG RNA replicons efficiently and consistently generated iPS cells from newborn and adult human fibroblasts that acquired full pluripotency by rigorous in vivo biological and molecular criterion that paralleled human ES cells. By expressing four reprogramming factors at consistent levels and ratios over time in the same cell combined with replication of the VEE-RF RNA for a limited number of multiple cell generations, the VEE-RF RNA approach solves both of the major inefficiency problems associated with generating iPS cells by repetitive daily transfections of four individual RF mRNAs.
Consistently maintaining the ratio of reprogramming factors to each other appears to be critically important for efficiently generating iPS cells. As an example, we note that expression of too much c-Myc relative to the other factors by the VEE-OMKS RNA replicon dramatically inhibited iPS cell generation, whereas the lower c-Myc ratio expressing VEE-OKS-iM RNA replicon was highly efficient at generating iPS cells. In contrast to simultaneously transfecting multiple mRNAs for each factor into cells, having the fixed factor ratio built into the single VEE-RF replicon RNA consistently results in the appropriate reprogramming factor expression levels and ratios. The generation of the VEE RF-RNA transcript utilizes a standard SP6 or T7 RNA polymerase in vitro transcription kit that does not require special conditions and thereby, further simplifies the approach for broad use. Although previously restricted to generating limited transcript lengths, current in vitro RNA transcription kits now exceed 25 kb in length (Schelle and Thiel, 2007), significantly longer than the ~15 kb length of the VEE-RNA replicons generated here. The ability to generate very long in vitro transcripts allowed us to generate VEE-RNA replicons with four pluripotent factor ORFs plus a puromycin selection gene (five ORFs).
Activation of the innate immune system has recently been reported to enhance iPS cell generation (Lee et al., 2012). However, in our hands, inhibiting the innate immune response by B18R exposure was critical for increased efficiency of iPS cell generation. We speculate that exposure of VEE-RF RNA harboring cells to B18R-CM may sufficiently decreased the innate immune response to allow for VEE-RF RNA persistence, while retaining a low level innate immune response to stimulate iPS cell generation. Although we generated the data presented here by a fixed protocol of B18R mRNA co-transfection on day 1 followed by addition of B18R conditioned media, we note in subsequent studies that addition of B18R condition media on day 1, in lieu of B18R mRNA co-transfection, followed by daily addition of B18R-CM also sufficed to suppress the innate immune response and generated iPS cells.
The VEE-RF RNA approach is an ectopic hit-and-run approach that does not utilize a DNA intermediate and therefore, there is no opportunity for integrative mutation that can occur with DNA vector-based types of iPS cell approaches. Moreover, the timing of VEE-RF RNA replicon loss by degradation can be regulated by B18R-CM withdrawal from the media. Here we used a combination of four reprogramming factors (OCT4, KLF4, SOX2 with c-MYC or GLIS1 ) to reproduce the retrovirus iPS cell generation. However, many factors to enhance reprogramming or generate safer iPS cells have now been reported to generate iPS cells (González et al., 2011; Hussein et al., 2011; Ben-David and Benvenisty, 2011; Mochiduki and Okita, 2012). Therefore, we note that the VEE-RF RNA approach has the versatility to be engineered to express alternative reprogramming factor combinations and/or insertion of additional reprogramming factor ORFs into the VEE RNA backbone for refining iPS cell generation from specific cell types or for use in driving transdifferentiation. In summary, the non-DNA based, self-replicating VEE-RF RNA approach has broad applicability for the efficient generation of human iPS cells for use in disease cell modeling studies and eventual human cell therapy regenerative medicine applications.
EXPERIMENTAL PROCEDURES
Cells
BJ foreskin fibroblasts and STO cell line were obtained from ATCC, primary human foreskin fibroblasts (HFF) were kindly obtained from M. Haas (UCSD) and HUES-9 human ES cell line D. Melton (HMS). Adult donor cells of normal human dermal fibroblasts (NHDF-c) and human dermal fibroblasts (HDF) were obtained from PromoCell (c-12302) and Cell Applications (106-05a), respectively. BJ, HFFs, NHDF-c, HDFs and STO were cultured in DMEM containing 10% FBS, MEM Non-Essential Amino Acids (NEAA), Pyruvate, penicillin, and streptomycin. HUES-9 and iPS cells were cultured with ES culture medium in Knockout D-MEM containing 20% Knockout SR, GlutaMAX, NEAA, 2-Mercaptoethanol, penicillin, streptomycin, and bFGF (10 ng/ml). STO feeder cells were prepared by mitomycin C treatment (10 μg/ml, Sigma). Matrigel (BD Bioscience) coated wells and conditioned medium of STO feeder cells were used for feeder free culture.
Plasmid construction
GFP/Pac genes and partial 3′UTR in p5′VEE/S/GFP/Pac (VEE backbone plasmid, a kind gift from I. Frolov) were deleted with XbaI/MfeI digestion, and then introduced the multiple cloning sites (Table S3), IRES and Puromycin resistance gene. This vector was renamed as pVEE-IRES-Puro. Multicistronic expression of RFs with viral 2A peptide sequences (Table S3) and IRES were constructed in pBluescript SK+ vector, and then cloned into pVEE-IRES-Puro to generate VEE-OS, VEE-OMKS, VEE-OKS-iM, VEE-OKS-iG plasmids. To generate RNA with T7 RNA polymerase, SP6 promoter (ATTTAGGTGACACTATAG) was replaced to T7 promoter (TAATACGACTCACTATAG) by PCR (Table S1) using the SacI/BstZ17I fragment of VEE vector as a template (SP6 promoter is located on next to the SacI site). B18R (D01019) was obtained from Addgene and cloned into pTNT vector (Promega) for mRNA synthesis.
RNA synthesis
VEE plasmids were linearized with MluI digest and used as templates for RNA synthesis. The synthesis of RNA replicon was performed with the RiboMAX Large Scale RNA Production System-SP6 or T7 (Promega) kit. After the DNase treatment, the synthesized RNA was purified by ammonium acetate precipitation (2.5 M). For the 5′-Capping of RNA, we used ScriptCap m7G Capping System and ScriptCap 2′-O-Methyltransferase (Epicentre, currently available from CELLSCRIPT) to produce cap 1-capped RNA, which proceeds 100% efficiency of capping. After the 5′-Capping of RNA, RNA was purified by ammonium acetate precipitation, and then additional poly(A) tail (~150 bases) was added by Poly(A) Polymerase (Epicentre, currently available from CELLSCRIPT). RNA was purified, resuspended in the RNA Storage Solution (Ambion) at 1mg/ml concentration, and stored at −80 °C. B18R mRNA was synthesized as same as replicon RNA except for using 25% modified nucleotides (psudouridine and 5-methyl-cytidine, TriLink Biotechnologies).
Preparation of B18R conditioned medium
B18R-CM (conditioned medium) was generated by B18R mRNA (1 μg for 1 well of 6-well plate) transfection into HFFs with Lipofectamine 2000 (Invitrogen). After 3 hr, medium was changed to Advanced DMEM containing 15% FCS (ES cell qualified, Millipore) or ES culture medium. Medium was collected on next day, filtrated, and diluted with fresh culture medium to 20% final concentration.
iPS generation by replicon transfection
BJ and HFFs were passaged to 6-well plate on day 0 and cultured to 90–100% confluency (4×105 cells/well) on day 1. 1 μg RNA mixture (3:1 ratio VEE RNA to B18R mRNA) was transfected with Lipofectamine 2000. After 3 hr, transfection medium was changed to the Advanced DMEM (Invitrogen) containing 15% FCS (ES cell qualified, Millipore). ES culture medium was used from day 7. One day after the final transfection, cells were passaged to STO feeder with several dilutions. ES medium containing B18R-CM was changed everyday until iPS cell colonies were generated. Colonies were mechanically picked for isolation of clones or stained with Alkaline Phosphase Detection kit (Millipore) or manually prepared AP-staining solution containing 1 mg/ml of FastRed TR (Sigma) and 0.4 mg/ml of 1-Naphthyl phosphate (Sigma) in AP buffer (100 mM Tris, 100 mM NaCl and 50 mM MgCl2, pH 9.5).
Optimization of iPS generation with one time transfection of replicon
Cells were passaged on gelatin-coated 6-well plate on day 0 and cultured to 90–100% confluency on day 1. To minimize the interferon response, cells were started to treat with 20% B18R-CM 20 min before transfection. 1 μg RNA mixture (3:1 ratio VEE RNA to B18R mRNA for SP6 VEE RNA, or 1:1 ratio for T7 VEE RNA) was transfected with Lipofectamine 2000. After 3 hr, transfection medium was changed to the Advanced DMEM or Pluriton medium (Stemgent) containing 20% B18R-CM. On day 7, Advanced DMEM was replaced to ES culture medium. Puromycin (0.8 μg/ml) was added from day 2 to 10. Cells were passaged onto STO feeder cells on day 10 and cultured in ES culture medium. B18R-CM was supplied everyday until iPS cell colonies were generated.
qRT-PCR
Total RNAs from feeder free culture of iPSCs clones, HUES-9, BJ and HFFs were isolated with RNeasy mini kit. TaqMan RT-PCR reactions were carried out using RNA-to-Ct one-step reaction (Applied Biosystem). Primers and probes were obtained from AB TaqMan Gene Expression Assay catalog (GAPDH, Hs99999905_m1; POU5F1 Hs03005111_g1; Sox2 Hs01053049_s1; DNMT3B Hs00171876_m1; TERT Hs00972656_m1; Lin28 Hs00702808_s1; Nanog Hs02387400_g1; TDGF1 Hs02339499_g1). Data were analyzed on the 7300 real-time PCR system using the delta-delta Ct method.
Bisulfite genomic sequencing
Conversion of unmethylated cytosines into uracil of genomic DNA was performed with EZ DNA Methylation-Gold Kit (Zymo Research). The promoter region of OCT4 or NANOG was amplified by PCR (Table S3), cloned into the T-vector, and then sequenced.
Immunoblotting
Cells were lysed with 2 × RIPA buffer containing 0.3 M NaCl, 80 mM Tris-HCl (pH7.5), 0.4% SDS, 2% Triton-X 100, 2% sodium deoxycholate, 100 μg/ml phenylmethylsulfonyl fluoride (PMSF), aprotinin (5 μg/ml) and leupeptin (5 μg/ml). Equal amount of proteins (around 20 μg) were used for 9% SDS-PAGE and electroblotted onto a Nitrocellulose membrane. Membranes were incubated with primary antibodies for overnight at 4 °C after blocking with 4% Milk in PBS-T (0.05% Tween 20), and then incubated with horseradish peroxidase-conjugated anti-rabbit, goat or mouse IgG (Santa Cruz). Protein bands were visualized using the ECL reagent (SuperSignal West Pico, Thermo Scientific).
Antibodies
Antibodies used in this research are as follows; anti-OCT4 (sc-9081), anti-KLF4 (sc-20691), anti-GLIS1 (sc-67584), anti-c-MYC (sc-42), TRA-1-60 (sc-21705), SSEA1 (sc-21702), SSEA4 (sc-21704), anti-mouse (sc-2005), anti-rabbit (sc-2004) and anti-goat (sc-2020) from Santa Cruz; anti-SOX2 (AF2018) and anti-NANOG (AF1997) from R&D Systems; TRA-1-81 (09-0011) from Stemgent; AE1/AE3 (RB-9010P0), Desmin (MS-376-S0), AFP (RB-365) and GFAP (RB-087) from Labvision; NF-1 (NB-300-155) from Novus Biological; anti-alpha-Actinin (A7811) from Sigma; anti-Cardiac Troponin T (MS-295-P0) from Thermo Scientific; Alexa Flour 488 anti-mouse (A11001) for cardiomyocytes analysis, Alexa Fluor 488 anti-mouse (A11029), Alexa Fluor 488 anti-rabbit (A11034) and Alexa Fluor 488 anti-goat (A11055) for immunostaining of iPS clones from Life Technologies.
RNA Sequence
Total RNAs were isolated with RNeasy mini kit (Qiagen), and cDNA library of each cells were synthesized and analyzed as described before (Fox-Walsh et al., 2011).
Cardiomyocyte differentiation
hiPSCs were differentiated into cardiomyocytes as previously described with minor modifications (Yang et al. 2008). Briefly, iPS cells were treated with activin A, bone morphogenetic protein 4 (BMP4), basic fibroblast growth factor (bFGF), vascular endothelial growth factor (VEGF) and dickkopf homolog 1 (DKK1) in serum-free media at normoxia and video recorded on day 15. At day 16, spontaneously contractile EBs were dissociated, re-plated onto glass slides, fixed in 4% formaldehyde and were immunostained with mouse anti-α-Actinin (Sigma #A7811) or anti-Cardiac Troponin T (Thermo# MS-295-P0) that were detected with Anti-Mouse IgG Alexa Fluor 488 (Life Technologies #A11001).
Teratoma formation
iPSC clones were intramuscularly or subcutaneously injected into the hind limb muscles or dorsal flank of nude mice (approximately 10 cm dish cultured cells for 1 shot of injection). After 5 to 8 weeks of injection, tumors were dissected and fixed with 4% paraformaldehyde, embedded into paraffin, and sectioned for hematoxilin and eosin (H&E) staining or immunostaining.
Supplementary Material
Highlights.
Synthetic, single, self-replicative RNA expresses four reprogramming factors
Generation of human iPS cells by single transfection of self-replicating RNA replicon
Human iPS cells generated with RNA replicon are transgene free
Acknowledgments
We thank I. Frolov (UAB) for VEE vector and T. Kogiso for valuable input. This work was supported by grants from the California Institute for Regenerative Medicine (A.R.M., B.Y., D.C.D., S.F.D.), National Institutes of Health (N.C.C.), Department of Defense (X.F.) and the Howard Hughes Medical Institute (S.F.D.).
Footnotes
ACCESSION NUMBERS
RNA-Seq data have been submitted and can be accessed by the Gene Expression Omnibus (GEO) accession number GSE38265.
Supplemental Information includes four figures, one movie, three tables and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcamí A, Symons JA, Smith GL. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol. 2000;74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel M, Yanik MF. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS One. 2010;5:e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- Davis NL, West A, Reap E, MacDonald G, Collier M, Dryga S, Maughan M, Connell M, Walker C, McGrath K, et al. Alphavirus replicon particles as candidate HIV vaccines. IUBMB Life. 2002;53:209–11. doi: 10.1080/15216540212657. [DOI] [PubMed] [Google Scholar]
- Durbin AP, Whitehead SS. Dengue vaccine candidates in development. Curr Top Microbiol Immunol. 2010;338:129–43. doi: 10.1007/978-3-642-02215-9_10. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics. 2011;98:266–71. doi: 10.1016/j.ygeno.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Weber J, Dewannieux M, Gorelik E, Heidmann T, Hengartner H, Zinkernagel RM, Hangartner L. Recombination of retrotransposon and exogenous RNA virus results in nonretroviral cDNA integration. Science. 2009;323:393–396. doi: 10.1126/science.1167375. [DOI] [PubMed] [Google Scholar]
- González F, Boué S, Izpisúa Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Nagy K, Nagy A. Human induced pluripotent stem cells: the past, present, and future. Clin Pharmacol Ther. 2011;89:741–745. doi: 10.1038/clpt.2011.37. [DOI] [PubMed] [Google Scholar]
- Kinney RM, Johnson BJ, Welch JB, Tsuchiya KR, Trent DW. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170:19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Mochiduki Y, Okita K. Methods for iPS cell generation for basic research and clinical applications. Biotechnol J. 2012;7:789–797. doi: 10.1002/biot.201100356. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–2207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Petrakova O, Volkova E, Gorchakov R, Paessler S, Kinney RM, Frolov I. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B, Thiel V. T7 RiboMAX Express: Generation of 27kb in vitro Transcripts in Minutes. 2007 http://www.promega.com/resources/articles/pubhub/enotes/t7-ribomax-express-generation-of-27kb-in-vitro-transcripts-in-minutes/
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Ni Y, Wang J, Guo X. Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Sci Rep. 2012;2:657. doi: 10.1038/srep00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.