Abstract
The hepatitis C viral (HCV) genome is translated through an internal ribosome entry site (IRES) as a single polyprotein precursor that is subsequently cleaved into individual mature viral proteins. Non-structural protein 5A (NS5A) is one of these proteins that has been implicated in regulation of viral genome replication, translation from the viral IRES and viral packaging. We sought to identify cellular proteins that interact with NS5A and determine whether these interactions may play a role in viral production. Mass spectrometric analysis of coimmunoprecipitated NS5A complexes from cell extracts identified heat shock proteins (HSPs) 40 and 70.Weconfirmed anNS5A/HSPinteraction by confocal microscopy demonstrating colocalization of NS5A with HSP40 and with HSP70. Western analysis of coimmunoprecipitated NS5A complexes further confirmed interaction of HSP40 and HSP70 with NS5A.Atransient transfection, luciferase-based, tissue culture IRES assay demonstrated NS5A augmentation of HCV IRES-mediated translation, and small interfering RNA (siRNA)-mediated knockdown of HSP70 reduced this augmentation. Treatment with an inhibitor of HSP synthesis, Quercetin, markedly reduced baseline IRES activity and its augmentation by NS5A. HSP70 knockdown also modestly reduced viral protein accumulation, whereas HSP40 and HSP70 knockdown both reduced infectious viral particle production in an HCV cell culture system using the J6/JFH virus fused to the Renilla luciferase reporter. Treatment with Quercetin reduced infectious particle production at nontoxic concentrations. The marked inhibition of virus production by Quercetin may partially be related to reduction of HSP40 and HSP70 and their potential involvement in IRES translation, as well as viral morphogenesis or secretion.
Conclusion
Quercetin may allow for dissection of the viral life cycle and has potential therapeutic use to reduce virus production with low associated toxicity.
Hepatitis C virus (HCV) infection has a world-wide prevalence of 3% and is the main entity responsible for liver transplantation for treatment of cirrhosis in developed countries.1 In the United States, HCV is the most common chronic blood-borne infection, affecting 1.8% of the population, and it appears to be the major causative factor responsible for the recent doubling of hepatocellular carcinoma in the United States.2 Current therapy consists of pegylated interferon-alpha and ribavirin, which results in 76% to 82% sustained virological response in patients infected with genotypes 2 and 3.3 Unfortunately, 70% of patients in the United States are infected with genotype 1, for which sustained virological response is significantly lower at 42% to 46%. Generally, therapy of all genotypes can be accompanied by adverse effects, and contraindications to therapy are not infrequent.4 For these reasons, there is a need to develop additional therapies that are less toxic and result in higher sustained virological response either as adjuncts or replacement therapies.
Heat shock proteins (HSPs), including HSP40 and HSP70, are required for cell survival during stress and have classically been designated as protein chaperones required for proper protein folding, protein degradation, and reactivation of damaged proteins.5,6 More recently HSPs have been found to be highly expressed in malignancies, including hepatocellular carcinoma, which has elevated levels of HSP70 compared with benign liver and benign liver proliferations.7 Infection by viruses, including human immunodeficiency virus 1, polyomaviridae, molluscum contagiosum, influenza, hepatitis B, human T-lymphotropic virus, papillomavirus, and adenovirus, has been shown to induce HSPs and facilitate viral production.8 Two viruses actually encode their own HSP homologs. Closteroviridae encode an HSP70 homolog, and polyomaviridae encode a DNAJ homolog containing protein (T antigen) that activates HSP70 in a similar fashion to HSP40 (a DNAJ motif containing protein).
The HCV nonstructural protein 5A (NS5A) protein is a 56-kDa to 59-kDa phosphoprotein that has been found to be associated with the viral replicase complex. It has been implicated in the regulation of HCV genome replication, internal ribosome entry site (IRES)-mediated translation of the viral polypeptide, and viral assembly.9–12 NS5A also appears to regulate cell signaling through interaction with several key proteins, including GRB2, PI3K, SRC proteins, P53, BAX, and CDK1.11 Additionally, NS5A interacts with double-stranded RNA-dependent protein kinase blocking the antiviral response and apoptosis.13
Here, we report identification of HSP70 and HSP40 in complex with NS5A and implicate HSPs in HCV IRES–mediated translation. Furthermore, treatment with the HSP expression inhibitor Quercetin14,15 reduced IRES translation. Treatment of HCV infection with Quercetin in tissue culture lowered intracellular viral accumulation and infectious particle production.
Materials and Methods
Cell Culture
The cell lines 293T, Huh-7, and Huh-7.5 were maintained in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (Fisher Scientific, Pittsburgh, PA) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA), 100 U/mL penicillin-streptomycin, and 2 mM L-glutamine (Invitrogen, Carlsbad, CA). In all experiments, Quercetin was used at 50 µM.
Antibodies
Antibodies used in this study were: β-Actin (Cell Signaling Technology, Danvers, MA), HSP70 (Santa Cruz Biotech, Santa Cruz, CA), HSP40 (Abcam, Cambridge, MA), NS5A (Abcam), and FLAG (Strata-gene, La Jolla, CA)
Plasmid Constructs and Virus
An MSCV-GFPIRES- PURO (pMSCVGFP) retroviral expression vector was a gift of D. Rawlings. An NS5AFLAG-expressing plasmid (pMSCVNS5AFLAG) was generated by performing polymerase chain reaction (PCR) on the H77 strain (serotype 1a) clone of HCV with end-modified primers synthesized by Integrated DNA Technologies, Inc. (Coralville, IA): NS5A fwd 5′AAAAAAACCGGTGCCGCCATGTCCGGTTCCTGGCTAAGGGAC3 ′ and NS5A rev 5′AAAAAGATCTGCAGCACQCGQCQTCTTCCG3′. The product was digested by AgeI and BglII followed by cloning into an MSCV-FLAG-IRES-PURO retroviral vector precut with AgeI and BamHI. Construct integrity was verified by sequencing. The IRES reporter construct has been previously described.16 The HCV reporter virus pNRLFC was in vitro transcribed and electroporated into Huh-7.5 cells to generate infectious viral supernatant as previously described.17
Co-immunoprecipitation Assay
Cell cultures 293T-NS5Aflag and 293T-GFP (green fluorescing protein; GFP) were established by transfection with MSCVNS5AFLAG and MSCVGFP, respectively, using Fugene 6 (Roche, Indianapolis, IN). Cell-free extracts were obtained from 108 cells by lysis using 1% Brij 97 lysis buffer in phosphate-buffered saline supplemented with protease inhibitors (Sigma, St. Louis, MO) and treated with either 1 mM Dithiobis (succinimidylpropionate) (Sigma), 1mMDithio-bismaleimidoethane (Pierce, Rockford, IL), or dimethylsulfoxide (DMSO) at 4°C for 2 hours. The Dithiobis (succinimidyl-propionate) reaction was quenched by 50 mM Tris-HCl (pH 7.5). Triton X-100 was added to 1% to all the samples and incubated at 4°C for 30 minutes.
Cell-free extracts were immunoprecipitated with anti-flag antibody and Protein G Plus-Agarose beads (Santa Cruz Biotech) at 4°C overnight and proteins eluted with FLAG peptide (Sigma). Eight percent gels were run and analyzed by standard western analysis methods. Gels were stained with the Silver Stain Plus kit (Bio-Rad, Hercules, CA). The resulting bands of interest were then carefully excised and subjected to mass spectrometry analysis.
Mass Spectrometry
Excised silver-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis bands were destained,18 followed by digestion of gel-embedded proteins with sequencing-grade trypsin (Promega, Madison, WI), and subsequent peptide extraction using standard protocols.19 Peptide analysis was accomplished with nanoflow high-performance liquid chromatography system (Dionex-LC Packings, Sunnyvale, CA) with a nano-electrospray interface (Protana, Odense, Denmark) and an Applied Biosystems/Sciex QSTAR XL mass spectrometer (Foster City, CA).
Proteins were identified by querying the tandem mass spectrometry data against spectra predicted for human and Escherichia coli Swiss-Prot/TREMBL database entries employing Mascot software (Matrix Science, London, UK). Protein identification criteria required Mascot peptide scores with less than 5% probability of a random match and that at least two peptides were observed. All searches employed peptide and product ion tolerances of 0.3 Da.
Immunofluorescence
NS5A flag-expressing Huh-7 cells were fixed in methanol for 20 minutes, blocked 1 hour in blocking buffer BB (phosphate-buffered saline, 1% bovine serum albumin, 0.1% NP40) and incubated with either HSP40 and FLAG or HSP70 and FLAG antibodies at 1:100 overnight in BB. Slides were then washed for 5 minutes with phosphate-buffered saline 3 times then incubated for 1 hour in anti-rabbit Texas red and anti-mouse fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) followed by washing. Slides were mounted and visualized by confocal microscopy using the Zeiss LSM 5 pascal laser scanning microscope with pascal software.
IRES Reporter Assay
SiRNA directed against HSPA1A, DNAJA2, and a nontargeting control (siG-ENOME Non-Targeting siRNA Pool #2) (Dharmacon, Chicago, IL) were transfected at a final concentration of 50 nM using HiPerFect Transfection reagent (Qiagen, Valencia, CA) followed by HCV IRES reporter plasmid with either pMSCV5AFLAG or pMSCVGFP using Fugene 6. Renilla and Firefly luciferase activity determined using the Dual-Glo Luciferase Assay System (Promega).
Quantitative Real-Time Reverse-Transcription PCR
The RNeasy Mini kit (Qiagen, Valencia, CA) along with QIAshredder spin columns (Qiagen) and on-column DNase digestion (Qiagen) were used to purify RNA. Real-time PCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR System with 2 × SYBR Green Master Mix (Diagenode, Sparta, NJ) in 25-µL reactions. The real-time PCR cycling conditions were 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute each as well as a final dissociation stage of 95°C for 15 seconds and 60°C for 15 seconds. The primers for the viral genome were derived from the region encoding NS3 and were CTGGGCTTTGGTGCTTACAT and GGCTGCCAGTGGTAATTGTT. HCV RNA levels were normalized to the housekeeping gene 36B4 using the primers TGGCAGCATCTACAACCCTGAAGT and TGGGTAGCCAATCTGCAGACAGACAC.20
Viral Assays
Huh-7.5 cells were infected with viral supernatant containing HCV virus NRLFC with a viral titer of 103 focus-forming units and a multiplicity of infection of 0.1. Subgenomic replicon-harboring cells were generated as previously described.21
Cell Viability
Cell viability was determined using the MTT Cell Proliferation assay (ATCC, Manassas, VA).
Statistical Analysis
Error bars reflect the standard deviation. P values were determined by student t test.
Results
NS5A Complex Purification
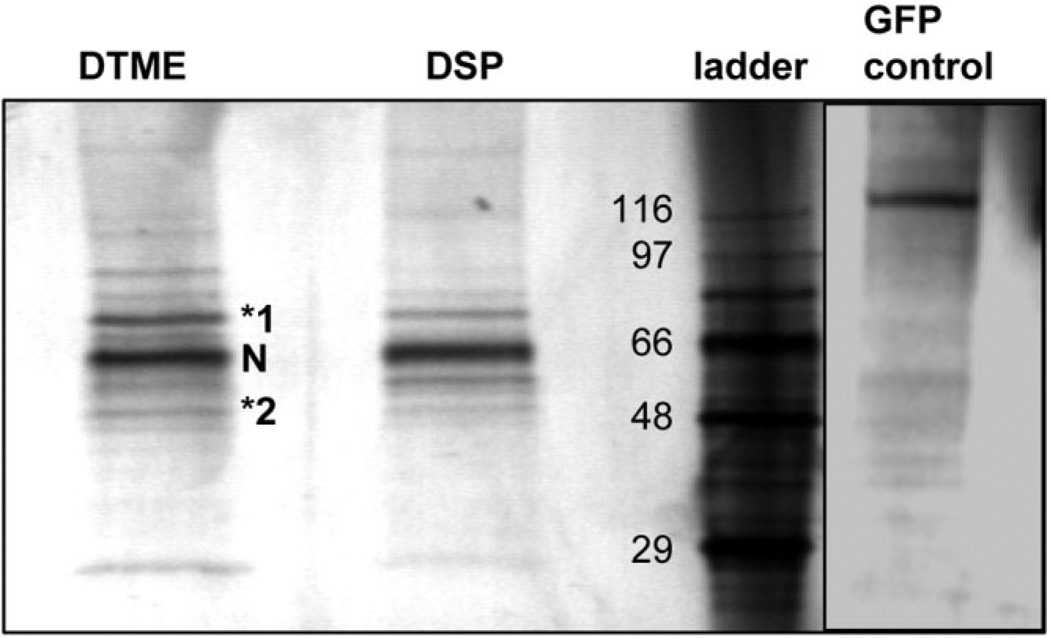
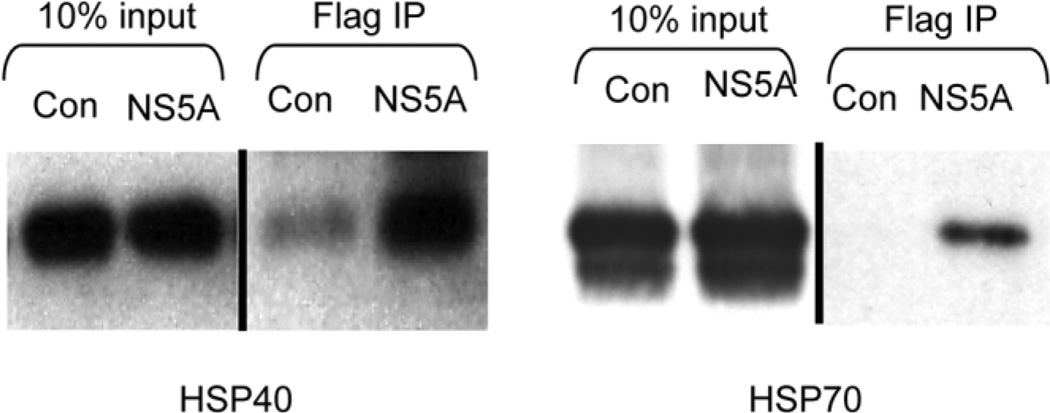
In an effort to identify novel factors that interact with NS5A, a construct expressing the Flag epitope fused to the carboxy-terminus of NS5A was generated and used to establish polyclonal 293T cell lines expressing either NS5A-FLAG or GFP. Immunoprecipitation of NS5A-FLAG in cell-free extracts revealed several co-immunoprecipitating host proteins (Fig. 1). Bands from this gel were carefully excised and analyzed by mass spectrometry. The proteins identified, with the number of corresponding unique peptide matches indicated in brackets, were as follows: [7] HSP70 (specific gene HSPA1A), [5] HSC70 (specific gene HSP8), [2] conserved between HSP70 and HSC 70, and [1] HSP40 (specific gene DNAJA2). The terms HSP70 and HSP40 will refer to the proteins specifically encoded by HSPA1A and DNAJA2, respectively, in experiments performed in the rest of this paper. Other excised bands did not yield any identifiable peptides. HSC70 is a constitutive, noninducible form of HSP70 that presumably plays a housekeeping role, and is essential for benign and tumor cell viability.22 SiRNA knockdown of HSC70 has previously been show to significantly affect cell viability, precluding examination of its effect on virus production in this study. Anti-FLAG immunoprecipitations performed on non–cross-linked extracts demonstrated interaction of NS5A with both HSP40 and HSP70 by western analysis, suggesting that co-immunoprecipitation of HSPs with NS5A was not an artifact of crosslinking (Fig. 2).
Fig. 1.
Co-immunoprecipitation of HSP40 and HSP70 with NS5A in cell-free extracts. Cell lysates from cell lines expressing either GFP or NS5A-FLAG were crosslinked with either Dithio-bismaleimidoethane or Dithiobis (succinimidylpropionate) (to improve complex stability), immunoprecipitated with anti-FLAG antibody, eluted with FLAG peptide, resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and silver stained. Bands were excised and analyzed by mass spectrometry. Bands 1 and 2 were identified as HSP70 and HSP40. N, NS5AFLAG. Protein size standards are indicated on the left of the ladder in kDa. GFP control extracts were crosslinked with Dithio-bismaleimidoethane. N = 3 with reproducible results.
Fig. 2.
NS5A co-immunoprecipitates with HSP40 and HSP70. Western analysis of NS5A-Flag immunoprecipitation probed for HSP40 and HSP70 demonstrates NS5A/HSP40 and NS5A/HSP70 complex formation. Input is shown at a lower exposure because of the abundance of HSP40 and HSP70 in these extracts. C, control GFP; N, NS5AFLAG. N = 2 with reproducible results.
NS5A Co-localizes with HSP40 and HSP70
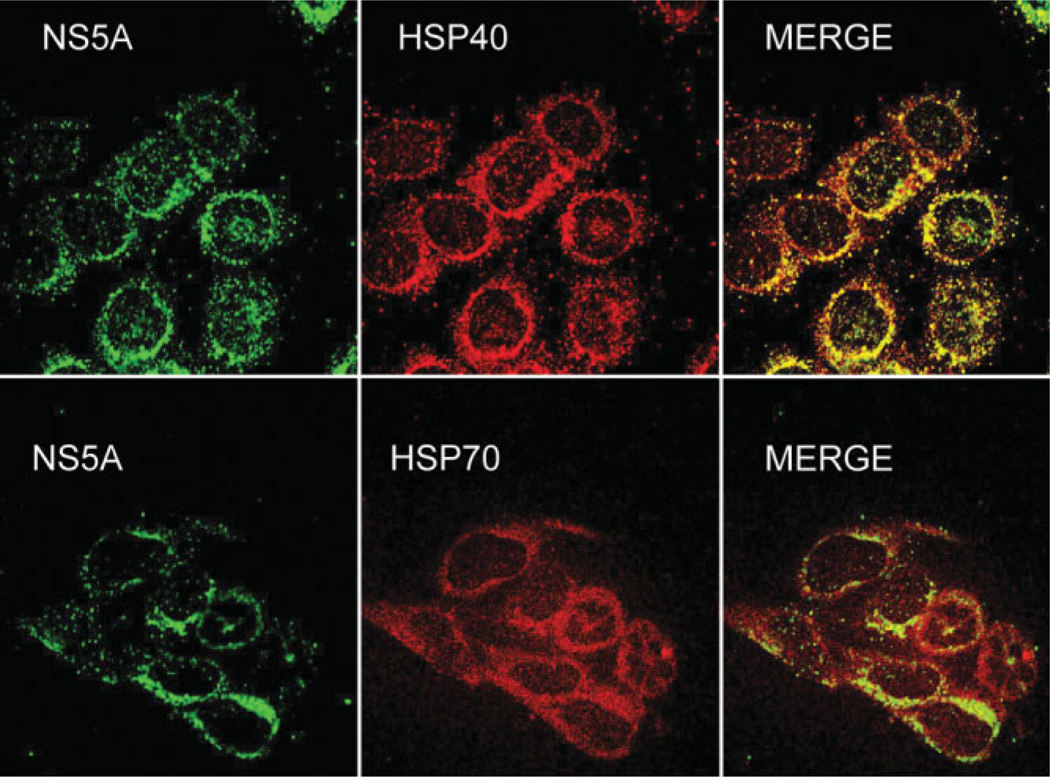
To determine whether NS5A colocalizes with HSP40 or HSP70, immunofluorescence was performed on Huh-7 cells expressing NS5A-FLAG (Fig. 3). HSP70 was more diffusely distributed in the cytoplasm but did colocalize with NS5A in the perinuclear region (consistent with endoplasmic reticulum/site of HCV replication23). HSP40 showed more overlap with NS5A because it appeared to be predominantly distributed within the perinuclear areas (also consistent with endoplasmic reticulum location). These results along with HSP co-immunoprecipitation with NS5A suggest there is an interaction of these HSPs with NS5A.
Fig. 3.
NS5A colocalizes with HSP40 and HSP70. Huh-7 cells expressing NS5AFLAG were stained with anti-Flag/anti-mouse-fluorescein isothiocyanate and either anti-HSP40 or anti-HSP70/anti-rabbit Texas red and analyzed by confocal microscopy. N = 2 with reproducible results.
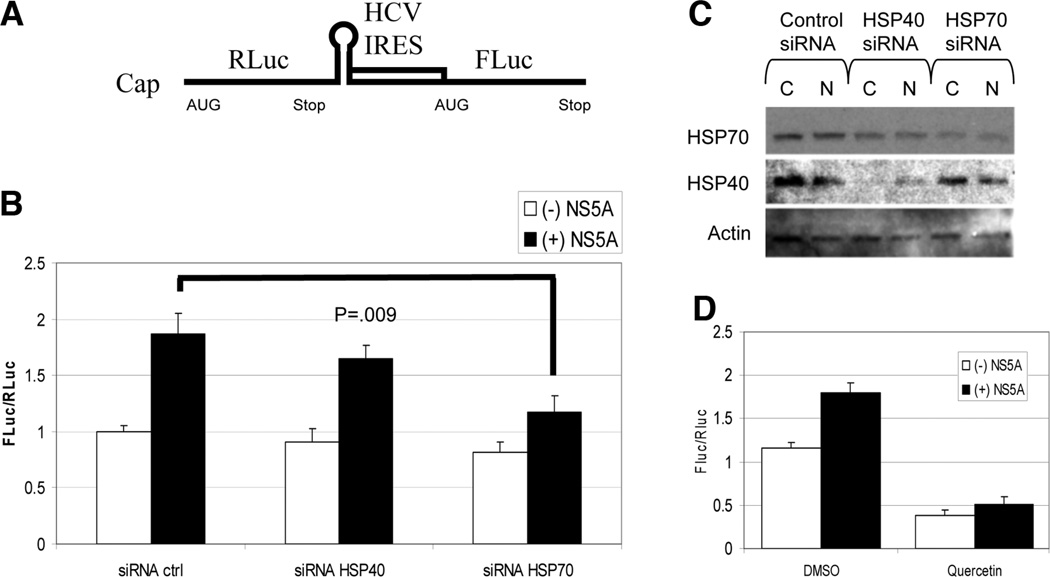
HSP70 Knockdown and Quercetin Treatment Reduces NS5A Augmentation of HCV IRES-Mediated Translation
NS5A has previously been reported to alter HCV IRES-mediated translation,24,25 and a HSP40 family member has been implicated in IRES-dependent translation in yeast.26 To determine whether HSP40 and HSP70 were involved with NS5A alteration of IRES activity, we used a cell culture– based bicistronic reporter system (Fig. 4A), where Renilla luciferase expression reflects 5′ cap-dependent translation and Firefly luciferase reflects HCV IRES-mediated translation.27 Changes in IRES translation can be measured by alterations in the ratio of Firefly to Renilla luciferase. As is evident in Fig. 4B, expression of NS5A almost doubled the IRES-dependent translation relative to 5′ cap-dependent translation compared with control. SiRNA knockdown of HSP40 (DNAJA2) showed a minimal decrease in NS5A augmented IRES/cap translation, whereas knockdown of HSP70 (HSPA1A) showed a statistically significant decrease (P = 0.009). Western analysis confirmed modest knockdown of HSP40 and HSP70 (Fig. 4C). Quercetin treatment in this system showed a marked reduction in baseline IRES translation relative to cap-dependent translation in the absence of NS5A (Fig. 4D) and also blocked NS5A augmentation. It is likely that the baseline IRES activity also requires HSPs, which are reduced with Quercetin treatment, resulting in an overall reduction in the Fluc/Rluc ratio.
Fig. 4.
(A) Schematic of the IRES assay reporter construct. A 5′ cap is present upstream of Renilla Luciferase (RLuc) followed by a stop codon, the HCV IRES, and the Firefly Luciferase (FLuc). (B) IRES reporter assay demonstrates NS5A augmented HCV IRES translation that is reduced by knockdown of HSP70. Huh-7 cells were treated with the indicated siRNAs on day 1 and transfected with control or NS5A and the IRES reporter DNA plasmids on day 2. Two days after the co-transfections, Firefly and Renilla luciferase levels were measured and displayed as the ratio of Firefly/Renilla luciferase activity. Extracts were normalized to protein concentration. N = 2 with reproducible results. (C) SiRNA knockdown of HSPs. Western analysis for HSP70 and HSP40 was performed on extracts from the transfection. C, control GFP; N, NS5AFLAG. HSP40 is reduced to 52% and HSP70 to 49% by quantitative reverse transcription PCR under similar conditions (data not shown). (D) Quercetin decreases IRES activity. Experimental conditions were the same as in part B but with and without treatment with Quercetin and in the presence or absence of NS5A. N = 3 with reproducible results.
Quercetin Inhibits Viral Protein Production Independently of Viral Genome Replication
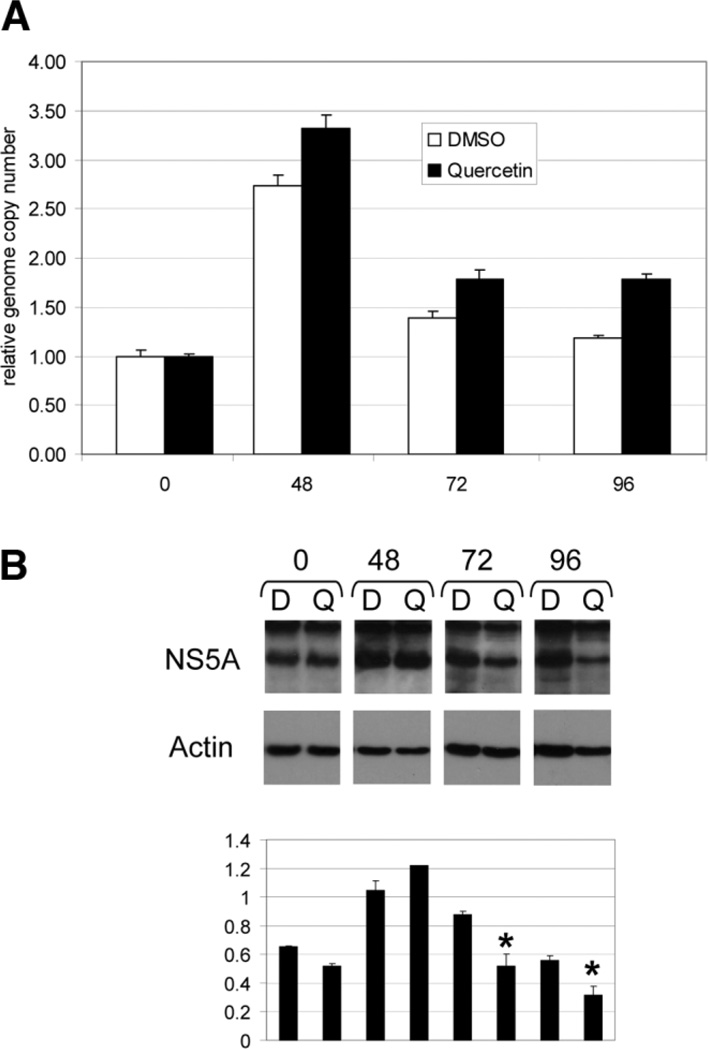
Decreased viral protein production may result from decreased viral genome production or lowered IRES translation and polypeptide production. We used a subgenomic replicon system to assess Quercetin’s effect on these two components of viral replication.21 The replicon-harboring cells were plated out at low density (approximately 30%) and treated with either Quercetin or DMSO control. HCV replicon replication is known to be dependent on cell division,28 so that when cells are initially plated from a dense population of cells, replication is low and then increases once the cells are able to commence division, as is seen here at 48 hours (Fig. 5A). Quercetin had no effect on viral genome copy number at 48, 72, and 96 hours. In fact, there appeared to be a slight increase in genome replication with Quercetin. This may be related an unknown feedback mechanism in which the replication apparatus senses low viral protein levels and so increases genomic replication. Alternatively, the kinase inhibitory properties of Quercetin may reduce hyperphosphorylation of NS5A, which has been shown to increase genomic replication when replicon systems are treated with experimental inhibitors of NS5A phosphorylation.29
Fig. 5.
(A) Quercetin treatment does not reduce viral genome replication in an HCV subgenomic replicon cell line. An Huh-7 cell line harboring a HCV subgenomic replicon was plated at low density and treated with either DMSO or Quercetin (50 µm) and RNA harvested at the hour time points indicated on the x-axis. Quantitative reverse transcription PCR was performed on DNase-treated RNA samples, and values were normalized to the 0 time point. (B) Upper panel. Quercetin lowers viral protein production in an HCV subgenomic replicon cell line. Western analysis was used to detect NS5A and actin in the same experiment performed in A. All films were exposed for the same amount of time from membranes processed at the same time. Hours are indicated at the top of the panel. D, DMSO; Q, Quercetin. N = 2 with reproducible results. Bottom panel: Ratio of NS5A signal to actin signal measured by densitometry. *P = 0.05.
In contrast to genome levels, NS5A protein levels were reduced with Quercetin treatment by 72 hours, suggesting IRES translation inhibition (Fig. 5B). Because the IRES immediately upstream of the viral coding sequence in this replicon is derived from the encephalomyocarditis virus,30 these data suggest that Quercetin also can interfere with encephalomyocarditis virus IRES activity. SiRNA knockdown experiments could not be performed because control siRNA inhibited replicon replication in this system (data not shown). SiRNA has previously been shown to activate the interferon pathway,31 which may explain the replicon replication inhibition under these conditions.
Hepatitis C Viral Replication Is Inhibited by Quercetin and Mildly Reduced by SiRNA-Mediated Knockdown of HSP70
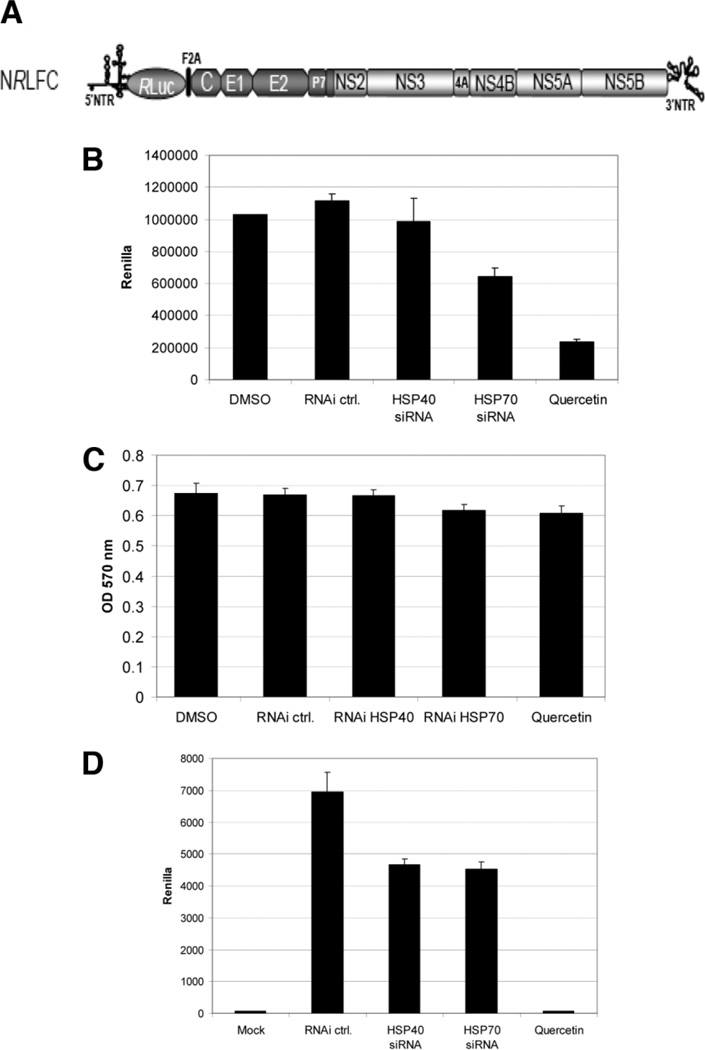
To determine the impact of HSPs on viral replication, we used a reporter virus NRLFC (Fig. 6A) in an HCV cell culture system. SiRNA-mediated knockdown of HSP40 appeared to have no effect on viral replication as measured by the Renilla reporter (Fig. 6B), whereas knockdown of HSP70 showed a modest reduction. Quercetin, however, markedly reduced luciferase activity by 80% or more, indicating decreased protein production. These treatments did not appear to affect cell viability (Fig. 6C).
Fig. 6.
(A) Schematic of the intragenotype chimeric reporter HCV viral genome NRLFC used in this study.17 The 5′ NTR, structural regions, p7, and part of NS2 are derived from the J6CF strain, and the remaining nonstructural regions are derived from JFH-1. Renilla Luciferase (RLuc) is inserted downstream of the 5′ NTR. (B) Hepatitis C viral protein production is inhibited by Quercetin and modestly reduced by HSP70 siRNA but not HSP40 siRNA. Huh-7.5 cells were transfected with siRNA 24 hours before infection or treated with Quercetin at the time of infection. Cells were infected with NRLFC, at a multiplicity of infection of 0.1. Renilla luciferase activity was assayed on the cells at 72 hours after infection. (C) SiRNA and Quercetin are not toxic to Huh-7.5 cells. Huh-7.5 cells were treated with siRNA or Quercetin for 72 hours and MTT assays performed to determine cell viability. (D) Hepatitis C virus production is inhibited by Quercetin treatment and mildly reduced by HSP knockdown. Huh-7.5 cells were infected with NRLFC virus at a multiplicity of infection of 0.1. Cells were treated with siRNA or Quercetin. Supernatants were harvested 2 days after infection and used to infect naïve Huh-7.5 cells to determine the amount of infectious virus production. Renilla luciferase assays were performed on cells 48 hours after this second infection. N = 2 with reproducible results.
Hepatitis C Viral Secretion (Viral Egress or Viral Release) Is Reduced by siRNA Knockdown of HSP40 and HSP70 and Blocked by Quercetin
To address the possibility that HSP40 and HSP70 may play a role in production of infectious virus particles, supernatants from the previous replication experiments were tested for infectivity. In this case. siRNA knockdown of both HSP40 and HSP70 modestly reduced virus production (Fig. 6D). Quercetin, however, strikingly blocked virus production to almost below detection by this assay. Titration of Quercetin showed 29% reduction at 0.5 µM, 90% reduction at 5 µM, and near 100% reduction at 50 µM (data not shown).
Discussion
Recently the HCV protein NS5A has been implicated in the regulation of HCV genome replication, IRES-mediated translation of the viral polypeptide, and viral assembly.24,32 We have identified a NS5A/HSP complex composed of NS5A, HSP40, and HSP70 that may impact one or more of these stages of the viral life cycle. A potential interaction of NS5A with HSP70 (same specific family member, HSPA1A) has previously been reported by the Rice laboratory, where HSP70 was identified through a yeast two-hybrid screen with NS5A bait.33 Although no additional assays were performed to confirm the interaction in that study, siRNA knockdown of HSP70 significantly impaired virus production as well as viral RNA levels. Here we have presented additional evidence of an NS5A/HSP40 and NS5A/HSP70 interaction through identification of the complex through an alternative proteomic-based screen, coimmunoprecipitation, and colocalization by immunofluorescence in the perinuclear area where the virus replicates. In our laboratory, exogenous expression of tagged NS5A is very low relative to other tagged proteins, lowering the possibility that misfolding is the reason for its interaction with HSPs. This most likely represents a complex composed of all three proteins together, because HSP40 is well known to interact with and augment HSP70 activity.34 Although we have shown there to be a NS5A/HSP complex, we certainly cannot exclude the possibility that these HSPs are also interacting with other viral products.
The major role of HSP40 and HSP70 may not be in genome replication but rather in subsequent stages in the viral life cycle, because Quercetin treatment of subgenomic replicon-harboring cells did not affect viral RNA replication. HSP70 is implicated in NS5A-mediated IRES translation, as demonstrated by decreased IRES activity with HSP70 knockdown in the IRES reporter system. A yeast HSP40 homolog, and indirectly HSP70 by virtue of their existence as a complex, have previously been shown to be important for cellular IRES specific translation in yeast.26 The reduction in the IRES activity with knockdown of HSP40 is minimal, and with HSP70 is modest compared with Quercetin treatment and may be related to several factors. These proteins are highly abundant, rendering significant knockdown difficult. Furthermore, there are eight family members of HSP7035 and 41 of HSP40,36 clearly raising the possibility that loss of one member could be functionally complemented by one of the many others. Compensatory increased expression of numerous HSPs also has been shown to occur during heat shock with knockout of HSP70 in Drosophila.37 In mice, up-regulation of one family member of HSP70 has been seen with knockout of another HSP70 family member.38 We have further found knockdown of HSP40 results in increased HSP70 mRNA, and conversely, knockdown of HSP70 increases HSP40 mRNA expression (data not shown). The stronger impact Quercetin has on the virus compared with HSP knockdown can be explained by the fact that most of the HSP40 and HSP70 family members are more globally reduced by Quercetin.
Quercetin treatment of the subgenomic replicon system also reduced NS5A protein levels, demonstrating its effect on IRES translation, albeit an exogenous one (encephalomyocarditis virus). Our data suggest that Quercetin can also reduce baseline HCV IRES translation in the absence of NS5A expression as well as NS5A-augmented translation, as seen by the marked reduction of the ratio of IRES/cap reporter gene activity in the bicistronic reporter assay. Taken together, the IRES assay and the replicon studies suggest that Quercetin can inhibit viral IRES translation and could potentially be used to treat HCV as well as other viral infections. Quercetin has been shown to inhibit several kinases,39,40 and we cannot exclude this activity as a factor in its ability to reduce virus production. However, similar, yet modest, effects imparted by HSP knockdown support the argument that its HSP synthesis inhibitory properties are responsible for these effects.
In addition to the potential role HSPs and possibly an HSP/NS5A complex may play in IRES translation, we show that HSPs are important for viral production using an HCV cell culture reporter system. Viral replication was modestly reduced by HSP70 knockdown and more strongly by Quercetin treatment. The lack of significant effect by HSP40 knockdown and the modest reduction by HSP70 knockdown may be explained by similar mechanisms given for the IRES assay results. The decrease in infectious particle production appeared more dramatic than with viral replication, because HSP40 knockdown also affected it, and Quercetin further reduced levels to the lower limits of assay detection. This stronger effect raises the possibility that the HSPs also may play a role in viral assembly or secretion. Given the known role of HSPs as protein chaperones, it is reasonable to suggest these HSPs are important for several stages of the viral life cycle, including IRES translation, viral assembly, and secretion. Further studies to determine the role HSPs alone and in complex with NS5A in assembly and secretion are beyond the scope of this study and are currently being investigated.
The most striking finding in this study is the marked reduction in viral production imparted by HSP synthesis inhibitor Quercetin. HSP synthesis inhibition may be an attractive therapeutic avenue, because it does not block a viral-encoded protein directly but rather a cellular cofactor. This may reduce the chance of treatment-related viral escape mutants. The low toxicity and pharmacokinetics of Quercetin are well known,41,42 and it has been approved for other uses in clinical trials in the United Kingdom and the United States,41,43,44 rendering it an immediately viable candidate for treatment of HCV infection.
Acknowledgment
The authors thank T. Wakita for the JFH-1 plasmid.
Supported by the Cure Digestive Diseases Research Center (S.W.F.), Stein Oppenheimer Endowment Award (S.W.F.), National Institutes of Health (NIH) grant 1K22CA120147-01A1 (S.W.F.), and NIH grant AI 072180 (A.D.).
Abbreviations
- BB
blocking buffer
- DMSO
dimethylsulfoxide
- GFP
green fluorescing protein
- HCV
hepatitis C virus
- HSP
heat shock protein
- IRES
internal ribosome entry site
- MSCV
murine stem cell virus
- NS5A
nonstructural protein 5A
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma: an epidemiologic view. J Clin Gastroenterol. 2002;35(5) Suppl 2:S72–S78. doi: 10.1097/00004836-200211002-00002. [DOI] [PubMed] [Google Scholar]
- 3.Gambarin-Gelwan M, Jacobson IM. Optimal dose of peginterferon and ribavirin for treatment of chronic hepatitis C. J Viral Hepatol. 2008;15:623–633. doi: 10.1111/j.1365-2893.2008.01018.x. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Patel K. Future therapy of hepatitis C. Hepatology. 2002;36(5) Suppl 1:S245–S252. doi: 10.1053/jhep.2002.36795. [DOI] [PubMed] [Google Scholar]
- 5.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 6.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 7.Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology. 2007;45:725–734. doi: 10.1002/hep.21531. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan CS, Pipas JM. The virus-chaperone connection. Virology. 2001;287:1–8. doi: 10.1006/viro.2001.1038. [DOI] [PubMed] [Google Scholar]
- 9.Girard S, Vossman E, Misek DE, Podevin P, Hanash S, Brechot C, et al. Hepatitis C virus NS5A-regulated gene expression and signaling revealed via microarray and comparative promoter analyses. Hepatology. 2004;40:708–718. doi: 10.1002/hep.20371. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, Staschke K, De Francesco R, Tan SL. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation- dependent viral RNA replication? Virology. 2007;364:1–9. doi: 10.1016/j.virol.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 11.Reyes GR. The nonstructural NS5A protein of hepatitis C virus: an expanding, multifunctional role in enhancing hepatitis C virus pathogenesis. J Biomed Sci. 2002;9:187–197. doi: 10.1007/BF02256065. [DOI] [PubMed] [Google Scholar]
- 12.Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008;82:1073–1083. doi: 10.1128/JVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale M, Jr, Blakely CM, Kwieciszewski B, Tan S-L, Dossett M, Tang NM, et al. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa N, Hirayoshi K, Kudo H, Takechi H, Aoike A, Kawai K, et al. Inhibition of the activation of heat shock factor in vivo and in vitro by flavonoids. Mol Cell Biol. 1992;12:3490–3498. doi: 10.1128/mcb.12.8.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosokawa N, Hirayoshi K, Nakai A, Hosokawa Y, Marui N, Yoshida M, et al. Flavonoids inhibit the expression of heat shock proteins. Cell Struct Funct. 1990;15:393–401. doi: 10.1247/csf.15.393. [DOI] [PubMed] [Google Scholar]
- 16.Dasgupta A, Das S, Izumi R, Venkatesan A, Barat B. Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses. FEMS Microbiol Lett. 2004;234:189–199. doi: 10.1016/j.femsle.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 17.Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, et al. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog. 2008;4:e1000182. doi: 10.1371/journal.ppat.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumner LW, Wolf-Sumner B, White SP, Asirvatham VS. Silver stain removal using H2O2 for enhanced peptide mass mapping by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:160–168. doi: 10.1002/rcm.555. [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 20.Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 21.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 22.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–582. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Yan W, Coito C, Li Y, Gale M, Jr, Katze MG. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J Gen Virol. 2003;84:535–543. doi: 10.1099/vir.0.18658-0. [DOI] [PubMed] [Google Scholar]
- 25.Kalliampakou KI, Kalamvoki M, Mavromara P. Hepatitis C virus (HCV) NS5A protein downregulates HCV IRES-dependent translation. J Gen Virol. 2005;86:1015–1025. doi: 10.1099/vir.0.80728-0. [DOI] [PubMed] [Google Scholar]
- 26.Raychaudhuri S, Fontanes V, Banerjee R, Bernavichute Y, Dasgupta A. Zuotin, a DnaJ molecular chaperone, stimulates cap-independent translation in yeast. Biochem Biophys Res Commun. 2006;350:788–795. doi: 10.1016/j.bbrc.2006.09.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesan A, Dasgupta A. Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements. Mol Cell Biol. 2001;21:2826–2837. doi: 10.1128/MCB.21.8.2826-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, et al. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol. 2004;78:13306–13314. doi: 10.1128/JVI.78.23.13306-13314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 31.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 32.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, et al. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 36.Valle JW, Dangoor A, Beech J, Sherlock DJ, Lee SM, Scarffe JH, et al. Treatment of inoperable hepatocellular carcinoma with pegylated liposomal doxorubicin (PLD): results of a phase II study. Br J Cancer. 2005;92:628–630. doi: 10.1038/sj.bjc.6602394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettencourt BR, Hogan CC, Nimali M, Drohan BW. Inducible and constitutive heat shock gene expression responds to modification of Hsp70 copy number in Drosophila melanogaster but does not compensate for loss of thermotolerance in Hsp70 null flies. BMC Biol. 2008;6:5. doi: 10.1186/1741-7007-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan L, Huang H, Lian L. [Surgical procedure of gynecologic malignances in patients over 70 years] Zhonghua Fu Chan Ke Za Zhi. 2001;36:614–617. [PubMed] [Google Scholar]
- 39.Glossmann H, Presek P, Eigenbrodt E. Quercetin inhibits tyrosine phosphorylation by the cyclic nucleotide-independent, transforming protein kinase, pp60src. Naunyn Schmiedebergs Arch Pharmacol. 1981;317:100–102. doi: 10.1007/BF00506266. [DOI] [PubMed] [Google Scholar]
- 40.Shrivastava A, Manna SK, Ray R, Aggarwal BB. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J Virol. 1998;72:9722–9728. doi: 10.1128/jvi.72.12.9722-9728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferry DR, Smith A, Malkhandi J, Fyfe DW, deTakats PG, Anderson D, et al. Phase I clinical trial of the flavonoid quercetin: pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin Cancer Res. 1996;2:659–668. [PubMed] [Google Scholar]
- 42.Harwood M, Danielewska-Nikiel B, Borzelleca JF, Flamm GW, Williams GM, Lines TC. A critical review of the data related to the safety of quercetin and lack of evidence of in vivo toxicity, including lack of genotoxic/ carcinogenic properties. Food Chem Toxicol. 2007;45:2179–2205. doi: 10.1016/j.fct.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Padayatti S. Investigating the use of quercetin on glucose absorption in obesity, and obesity with type 2 diabetes. [Accessed August 7, 2009]; Available at: http://clinicaltrials.gov/ct2/show/NCT00065676. [Google Scholar]
- 44.Shoskes D, Lapierre C, Cruz-Correa M, Muruve N, Rosario R, Fromkin B, et al. Beneficial effects of the bioflavonoids curcumin and quercetin on early function in cadaveric renal transplantation: a randomized placebo controlled trial. Transplantation. 2005;80:1556–1559. doi: 10.1097/01.tp.0000183290.64309.21. [DOI] [PubMed] [Google Scholar]