Abstract
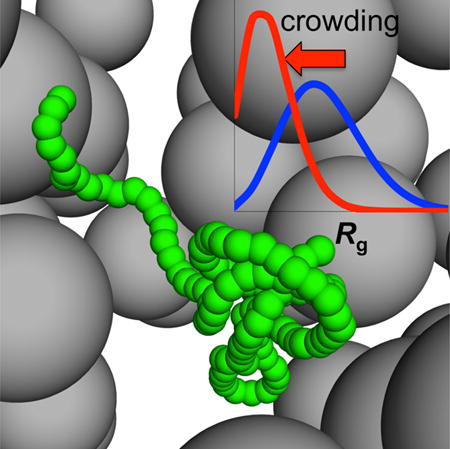
Due to their conformational malleability, intrinsically disordered proteins (IDPs) are particularly susceptible to influences of crowded cellular environments. Here we report a computational study of the effects of macromolecular crowding on the conformational ensemble of a coarse-grained IDP model, by using two approaches. In one, the IDP is simulated along with the crowders; in the other, crowder-free simulations are postprocessed to predict the conformational ensembles under crowding. We found significant decreases in the radius of gyration of the IDP under crowding, and suggest repulsive interactions with crowders as a common cause for chain compaction in a number of experimental studies. The postprocessing approach accurately reproduced the conformational ensembles of the IDP in the direct simulations here, and holds enormous potential for realistic modeling of IDPs under crowding, by permitting thorough conformation sampling for the proteins even when they and the crowders are both represented at the all-atom level.
Keywords: Intrinsically disordered proteins, macromolecular crowding, postprocessing approach
It is now accepted that a considerable proportion of proteins are disordered and play essential roles in signaling and regulation.1-12 The complex, crowded environments inside cells may significantly influence the conformational, thermodynamic, and kinetic properties of proteins.13 These influences are expected to be particularly strong for intrinsically disordered proteins (IDPs), due to their conformational malleability and propensity to oligomerize and aggregate and to bind with different cellular targets.14 While some studies15-18 have shown that IDPs preserve their disordered state under crowded conditions, these and other studies19-22 have found that the conformations of IDPs (and unfolded proteins) become more compact. Compression of the polymer PEG by Ficoll as a crowding agent23 and of unfolded RNA by PEG24 has also been observed, suggesting a common cause for the compaction in size. Here we report a computational study of the effects of macromolecular crowding on the conformational ensemble of an IDP.
Two different computational approaches have been used to study proteins under crowding. In the direct simulation approach, a test protein is simulated along with crowders;25-32 to calculate thermodynamic or kinetic properties, one has to obtain the full free energy surface that covers two end states (such as the folded and the unfolded states of a protein). To overcome the enormous computational cost of such calculations, the protein has generally been represented at a coarse-grained level, and the crowders are often treated with an even simpler representation, such as a sphere or a few linked beads. Simulations with an all-atom representation of test and crowder proteins have recently been carried out, although the relatively short simulation times limited the conformation sampling largely to near the folded state.32
We have introduced an alternative approach known as postprocessing,33-34 whereby the conformational ensemble of a protein sampled in the absence of crowders is transformed to that under crowding, through weighting by the Boltzmann factor of the excess chemical potential, Δμ, arising from protein-crowder interactions. Similar ideas have been exploited by others.21, 35 To implement the postprocessing approach, we have developed methods to accelerate the calculation of the excess chemical potential.33-34, 36 In essence, the postprocessing approach uses crowder-free simulations to predict crowding-induced changes in free energy differences and rate constants between end states,33-34, 36-42 as well as free energy surfaces of protein folding and conformational transition under crowding.39, 43
The postprocessing approach may be especially suited for studying IDPs under crowding. These proteins may not have well-defined basins of attraction and can access enormous conformational space. Sampling this conformational space is already challenging for simulations in dilute solution.44-49 The addition of crowders in these simulations may necessitate reduced representations for the IDP and the crowders. The postprocessing approach allows the more realistic representations to be preserved in modeling the effects of crowding.
The accuracy of the postprocessing approach depends on adequate sampling in the crowder-free simulations of the test protein, so as to cover all the conformational regions that would be important when crowders are present. Under-sampling of new low-energy regions that emerge in the presence of crowders is always a concern.32, 43 Previously we have validated the postprocessing approach against direct simulations for the effects of crowding on the flap open-to-closed population ratio of the HIV-1 protease dimer38 and for the folding free energy surfaces of three small proteins under crowding.43
Below we compare the conformational ensembles of an IDP under crowding, either predicted by the postprocessing approach or obtained from direct simulations with crowders present. To accommodate the direct-simulation approach, the IDP has a coarse-grained representation, with one bead for each residue.26, 50 The residues in the protein chain are connected by springs with an equilibrium length of 3.8 Å and a spring constant of 70 kcal/mol/Å2, and interact via a Morse potential
| (1) |
where rij is the distance between two residues, r0 = 9.5 Å, α = 0.707 Å−1, D0 = 0.207 kcal/mol, and ξ is a constant introduced to scale the strength of the intra-protein interactions. A higher ξ is expected to strengthen the attraction between the residues and thus result in more compact conformations. In the direct simulations, the crowders are soft spheres with a nominal radius of rc = 30 Å, with the interaction center located on the spherical surface with a radius of 23 Å; the crowders interact with a repulsive inverse r12 potential:26
| (2) |
where rαβ is the center-center distance between two crowders, and ε = 0.6 kcal/mol. When two crowders are at contact (i.e., rαβ = 60 Å), the interaction energy is ε, or roughly the thermal energy kBT at T = 300 K. Each protein residue and each crowder also interact with an inverse r12 potential:
| (3) |
We carried out Brownian dynamics simulations of a 99-residue IDP alone and along with the crowders, using a modified UHBD program as done previously.26, 38 Periodic boundary condition was imposed, with a cubic unit cell of 510 Å side length. The cutoff distances were 24.5 Å for the Morse potential and 75 Å for the inverse r12 potentials. For the simulations with crowders, 71 to 575 crowders were included to produce crowder volume fractions (ø) ranging from 0.06 to 0.49 (Table 1), the same as the crowding conditions in our previous study of the flap open-to-closed transition of the HIV-1 protease dimer.38 Five values of the scaling constant ξ for the intra-protein Morse potential were studied: 1, 0.8, 0.7, 0.6, and 0.5. For each combination of ø and ξ, conformation sampling consisted of 36 repeat trajectories that started from different random number seeds and lasted 30 μs each. The timestep was 50 fs; snapshots in the last 20 μs were saved at 10 ns intervals for analysis. For implementing the postprocessing approach, we also carried out crowder-only simulations, with the unit cell now enlarged to a side length of 1000 Å and the numbers of crowders increased in proportion to produce ø values matching those in the simulations of the protein-crowder mixtures.
Table 1. Results for Rg;rms (in Å) Obtained from Direct Simulations.
|
ø ξ |
0.00 | 0.06 | 0.12 | 0.18 | 0.24 | 0.31 | 0.37 | 0.43 | 0.49 |
|---|---|---|---|---|---|---|---|---|---|
| 1.0 | 19.2±1.3 | 19.2±1.0 | 19.0±1.0 | 19.0±1.0 | 18.7±1.1 | 18.5±1.1 | 18.5±1.5 | 18.2±0.7 | 17.9±0.5 |
| 0.8 | 30.1±5.8 | 27.7±3.5 | 26.8±3.8 | 26.5±4.3 | 25.4±3.2 | 23.1±1.4 | 22.3±1.7 | 21.4±1.4 | 20.4±0.9 |
| 0.7 | 45.2±7.6 | 45.3±7.4 | 41.7±5.9 | 39.4±6.6 | 36.7±5.7 | 32.2±4.9 | 29.3±3.9 | 25.3±2.3 | 23.1±1.6 |
| 0.6 | 52.0±4.7 | 51.4±4.4 | 49.1±5.6 | 48.5±5.9 | 45.6±5.9 | 41.9±4.8 | 36.7±5.0 | 32.7±3.8 | 28.1±2.8 |
| 0.5 | 52.3±3.8 | 52.2±4.0 | 50.7±3.7 | 50.4±3.8 | 48.6±4.4 | 45.3±3.9 | 42.8±4.0 | 37.1±4.1 | 34.2±5.2 |
Both without and with the crowders, the IDP sampled a wide variety of conformations, with the radius of gyration (Rg) spanning a broad range (Fig. 1). To demonstrate that the conformation sampling was adequate, in Fig. 2 we compare the histograms of Rg at ξ = 0.5 obtained from simulations starting from two very different initial conformations, one compact with Rg = 13 Å and the other extended with Rg= 93 Å.51 For both the crowder-free simulations (ø = 0) and the simulations at ø = 0.31, the histograms of Rg from the different initial conformations agree very well, indicating convergence of the simulation results. Additional results demonstrating convergence for ξ = 0.7 and ø = 0 are shown in Fig. S1. All subsequent results are from simulations with the compact initial conformation for the protein.
Figure 1.
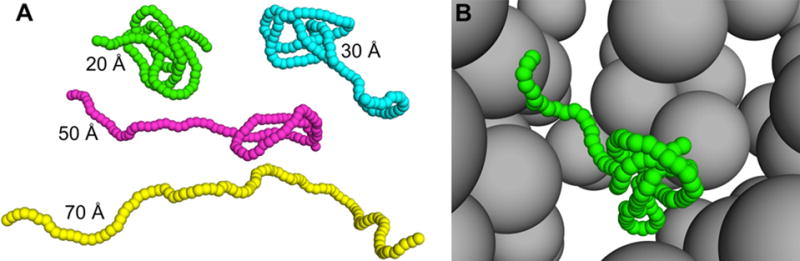
Illustrative conformations of the IDP sampled without and with crowders present (ξ = 0.8). (A) Four conformations in crowder-free simulations. The values of Rg are listed. (B) A snapshot from a simulation at ø = 0.18. The IDP has Rg= 27 Å.
Figure 2.
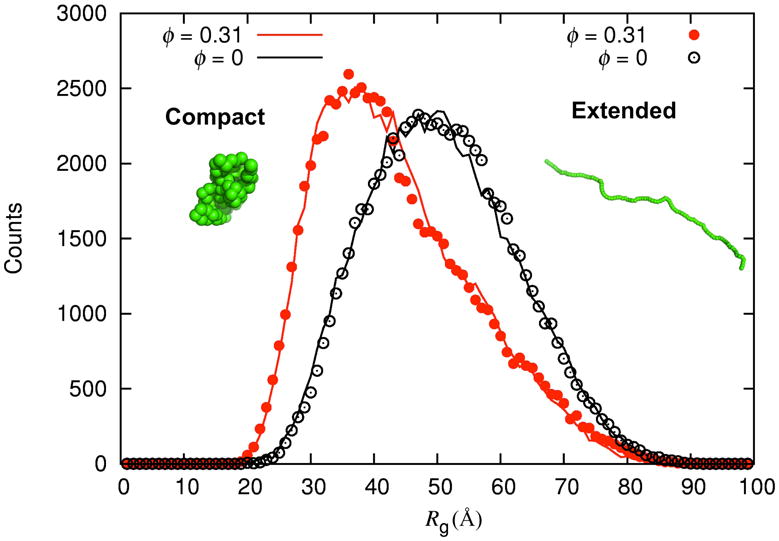
Histograms of Rg from four sets of trajectories (ξ = 0.5). Each set consists of 36 repeat trajectories. Two sets are for ø = 0 and two sets are for ø = 0.31. In each set, the trajectories were started either from a compact or an extended conformation for the IDP.
Comparing the histograms in Fig. 2 for ø = 0 and ø = 0.31, one sees that the peak position shifts toward a lower Rg value, meaning that the IDP becomes more compact under crowding. The corresponding results at ξ = 0.7, displayed in Fig. 3, further illustrate that the shift in peak position toward lower Rg values can be accompanied by a significant narrowing of the distribution of Rg. Despite the overall compaction under crowding, the histograms in Figs. 2 and 3 show that the same range of Rg values are spanned with and without crowders, suggesting that the accessible conformational spaces may largely overlap under crowder-free and crowding conditions.
Figure 3.
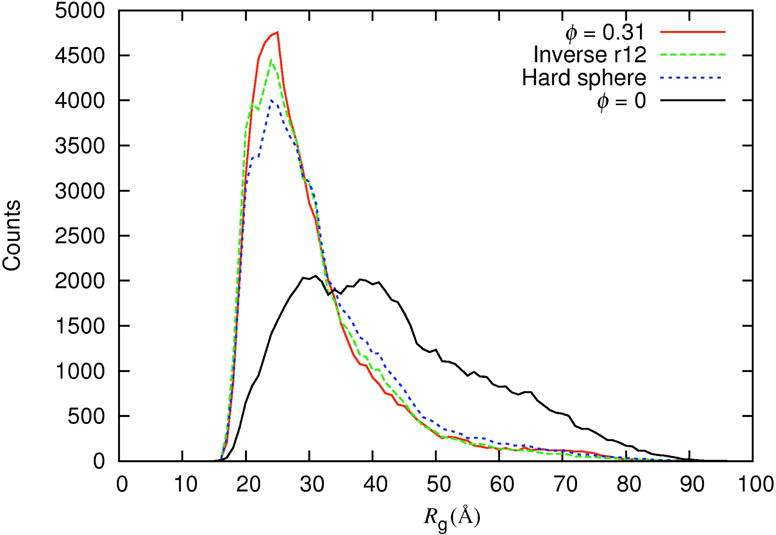
Histograms of Rgfrom direct simulations at ø = 0.31 and predicted by postprocessing the crowder-free simulations (with ø= 0). Hard sphere: Δμ calculated by approximating the crowders as hard spheres; Inverse r12: Δμ calculated by exact treatment of the inverse r12 form of protein-crowder interactions. Results are for ξ = 0.7.
From the direct simulations, the root-mean-square Rg (Rg;rms) values, along with their standard deviations among 36 repeat trajectories,52 for the various combinations of ø and ξ are obtained and listed in Table 1. These results show that, at a given ø, the protein chain becomes more and more extended as the intra-protein Morse interactions are weakened. Conversely, at a given ξ, the protein chain becomes more and more compact as the level of crowding is increased. Interestingly, the extent of the crowding-induced compaction is maximal at an intermediate ξ. At ξ = 0.7, Rg;rms is reduced by nearly 50% when the crowders are present at a volume fraction of 0.49. In comparison, under this crowding condition, the reduction in Rg is only 6.5% at ξ = 1 and 34.6% at ξ = 0.5. In addition, at intermediate ξ values (0.7 and 0.8), the standard deviations of Rg are lower under crowding than in the crowder-free condition, corresponding to the narrowing of the distribution of Rg shown in Fig. 3. Explanations for these intriguing observations at intermediate ξ will be presented below.
Based on studies of proteins unfolded by denaturants and/or low pH, a polypeptide chain with 99 residues is expected to have an Rg;rms ∼35 Å in dilute solution.53 An IDP probably has a comparable size. This expected size is produced by our model with intermediate ξ values (0.7 and 0.8) at ø = 0. Coincidentally, as just noted, it is at the intermediate ξ values that crowding exerts maximal compaction as well as narrowing of the distribution of Rg.
The magnitudes of crowding-induced compaction obtained in our simulations are similar to those observed in some recent experimental studies. In FRET measurements of Mikaelsson et al.,22 200 g/l Dextran 20 produced ∼10% decrease in donor-acceptor distance in urea-denatured ribosomal protein S16. Similarly, in SAXS measurements of Kilburn et al.,24 20% (w/v) PEG1000 produced a 16% decrease in the Rg of unfolded RNA. In comparison, the compaction for our IDP with ξ = 0.8 is 12% at ø = 0.18. Interestingly, there seems to be experimental evidence for crowding-induced narrowing of the distribution of Rg in the study of Mikaelsson et al.22 These authors fitted their FRET data with a Gaussian distribution for the donor-acceptor distance, and found that the width of the distribution decreased in the presence of 200 g/l Dextran 20.
We now use the above results from the direct simulations to benchmark the accuracy of the postprocessing approach, which uses the crowder-free simulations to predict the conformational ensembles of the IDP under the various levels of crowding. This entails weighting each crowder-free conformation by a factor exp(−Δμ/kBT).38 When the interactions between the test protein and crowders are hard-core repulsion only, the excess chemical potential can be calculated as Δμ = –kBTlnp, where p is the fraction of attempts to place the test protein into the crowded solution that do not result in protein-crowder clash. For spherical crowders, we developed an efficient method for calculating p, by mapping the covolume of a crowder and the protein onto a grid centered on the crowder.33 In these calculations, averages are taken over both conformations of the IDP and configurations of the crowders. Following our previous study,38 we first approximated the repulsive inverse r12 potentials as hard-core repulsion and applied the covolume-based method to calculate Δμ (using crowder configurations generated in previous hard-sphere simulations). Up to ø = 0.25, the postprocessing approach with the hard-sphere treatment accurately predicts the Rg;rms values from the direct simulations (Fig. 4). At ø = 0.31, it yields small but discernible underestimation of the crowding-induced compaction. We also used the crowder configurations generated by the crowder-only simulations here (with crowder-crowder interactions governed by the potential of eq (2)) but then treated the crowders as hard spheres in calculating Δμ. The predicted Rg;rms values were unchanged.
Figure 4.
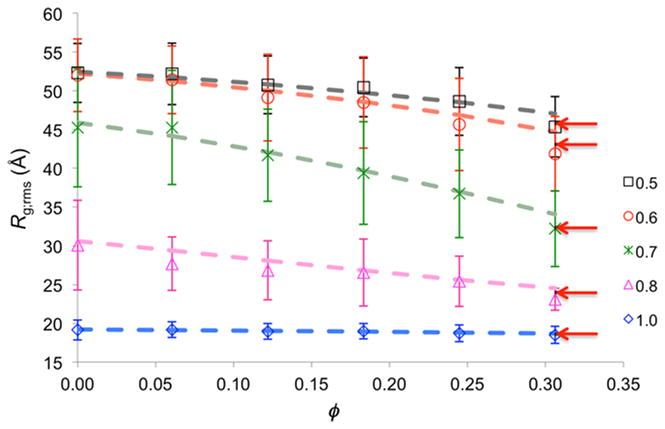
Rg;rms values obtained from direct simulations (symbols with error bars representing standard deviations among 36 repeat trajectories) and predicted by the postprocessing approach, with Δμ calculated either by approximating the crowders as hard spheres (dashed curves) or by exact treatment of the inverse r12 form of protein-crowder interactions (red arrows at ø = 0.31).
To understand the slight overestimation of Rg;rms at high ø, we note that, in the hard-sphere treatment, the interaction between a residue and a crowder is turned off at their nominal contact distance, even though the interaction energy there still roughly equals kBT. Therefore the crowders and the residues can approach each other to their contact distance more easily than they should, leading to slight underestimation of Δμ. This underestimation is worse for the more extended conformations (Fig. 3) and for the more crowded conditions. The former is because the more extended conformations have more residues exposed to the crowders; the latter is because, with more crowders around, the chance for them to approach the IDP increases.
We have just developed a new method for calculating Δμ that is based on fast Fourier transform (FFT) and can handle any form of protein-crowder interactions.36 The basic idea is to express the protein-crowder interaction energy as a correlation function and then evaluate this correlation function via FFT. Using the crowder configurations generated by the crowder-only simulations here and applying the FFT-based method to treat the inverse r12 form of protein-crowder interactions, the predictions for the Rg;rms values at ø = 0.31 are improved, and are now in good agreement with those obtained from the direct simulations (Fig. 4). The predicted histogram of Rg also agrees well with that from the direct simulations (Fig. 3), suggesting that the conformational ensemble under crowding is predicted well by the postprocessing approach when the protein-crowder interactions are properly treated when calculating Δμ. However, at an even higher level of crowding (ø = 0.49), the crowder-free simulations here under-sampled the most compact conformations (Fig. S2), resulting in a small overestimate of Rg;rms (35.7 Å predicted versus 34.2 ± 5.2 Å from the direct simulations, both at ξ = 0.5). More extensive sampling in the crowder-free simulations can be obtained by specialized techniques such as umbrella sampling or replica exchange.
The postprocessing approach provides a conceptual framework via which the crowding-induced maximal compaction and narrowing of the distribution of Rg at intermediate ξ can be easily understood. Recall that this approach yields the conformational ensemble under crowding through weighting each crowder-free conformation by a factor exp(–Δμ/kBT). Consider two different scenarios for the crowder-free conformational ensemble, one consists of similar conformations, resulting in a narrow distribution of Rg, while the other consists of vastly different conformations that span broad distribution of Rg. In the first scenario, reweighting by exp(–Δμ/kBT) will not produce a significant compaction, since all the crowder-free conformations would have similar weighting factors. In the second scenario, the more extended crowd-free conformations would have smaller weighting factors whereas the more compact crowd-free conformations would have larger weighting factors; the net result is significant compaction. The crowder-free conformations of our IDP have the broadest distribution of Rg at ξ = 0.7 (Fig. S3), thus explaining why crowding results in maximal compaction at this intermediate ξ.
That the broadest distribution of crowder-free Rg occurs at ξ = 0.7 requires some explanation. At ξ = 1, residue-residue attraction arising from the Morse potential keeps the IDP compact. As ξ is reduced, the protein chain can access more open conformations, leading to broadening of the distribution of Rg. However, as ξ is reduced further and further, the chain mostly samples highly extended conformations. With Rg bounded from above (at ∼100 Å for the fully extended conformation), the distribution of Rg again starts to narrow (compare the histograms of Rg at ξ = 0.6 and 0.5 in Fig. S3). Note that the standard deviations of crowder-free Rg is the largest at ξ = 0.7 (Table 1), consistent with the broadest distribution at this ξ.
Compaction under crowding can manifest either as a shift in the histogram of Rg toward lower Rg values (Fig. 2) or as a shift accompanied by a narrowing of the distribution of Rg (Fig. 3). A lower bound of Rg also exists, when the protein chain becomes extremely compact. As the histogram of Rg shifts toward lower Rg under crowding, the left envelope of the histogram may reach the lower bound of Rg. Then only the right envelope can shift leftward, resulting in a narrowed distribution.
We have demonstrated here that, from crowder-free simulations, the postprocessing approach can faithfully predict the conformational ensembles of IDPs under crowding. Note that, by varying ξ, we produced protein chains spanning a wide range of crowder-free Rg;rms values, both below and above the crowder radius; the postprocessing approach works equally well in both cases. While the demonstration carried out here uses simplified representations for the IDP and for the crowders, the postprocessing approach is now ready to model proteins and crowders both represented at the all-atom level.36 Using direct simulations with crowders included, presently achieving exhaustive conformation sampling for IDPs at such a level of realism seems impractical. The postprocessing approach may provide a viable solution. What is perhaps particularly attractive about this approach is that all the previously published simulations of IDPs in dilute solution44-49 can be postprocessed to predict results in crowded solution.
Additional technical advantages of the postprocessing approach are also worth noting. First, in implementing this approach, all locations in the crowded solution are probed for possible protein-crowder interactions.33, 36 This is in contrast to the situation with direct simulations, where the test protein can be trapped in interactions with one or two particular crowder molecules, especially when attractive components are included in the interactions. Second, the same crowder-free simulations can be used to predict conformational ensembles for many different crowding conditions (as illustrated here) and for different types of protein-crowder interactions. In particular, our FFT-based method can treat both repulsive and attractive protein-crowder interactions.36
Minton54 proposed both an equivalent hard sphere model and a Gaussian cloud model for treating unfolded protein chains. Applying these models to our IDP, we found that they significantly overestimated the magnitudes of crowding-induced compaction (Fig. S4). While simplified theoretical models can be very useful for predicting qualitative trends, predictions that have the potential to reach quantitative agreement with experimental studies of crowding effects will likely require an atomistic representation for the proteins and crowders.
Similar to what is found here for an IDP, decreases in Rg have been reported in previous computational studies for unfolded proteins under crowding.25, 29 The present study, especially with the help of the postprocessing approach, reinforces the notion that repulsive interactions with crowder molecules may be a common cause for the compaction of proteins and other polymer chains observed in experimental studies.15-16, 19-24 Crowders could also stabilize partially structured intermediates for IDPs.18 Based on thorough conformation sampling in dilute solution, the postprocessing approach holds enormous potential in realistically modeling this and other rich effects of crowding on IDPs.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM088187.
Footnotes
Supporting Information Four figures further demonstrating the convergence of simulation results (Fig. S1), presenting Rg results at ø = 0.49 (Fig. S2) and at ø = 0 (Fig. S3), and showing the predictions of simple theoretical models (Fig. S4); and a table listing the standard deviations of Rg in the combined distributions (Table S1). This material is available free of charge via the Internet http://pubs.acs.org.
References
- 1.Wright PE, Dyson HJ. Intrinsically Unstructured Proteins: Re-Assessing the Protein Structure-Function Paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic Disorder Protein Function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN. Natively Unfolded Proteins: A Point Where Biology Waits for Physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tompa P. Intrinsically Unstructured Proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 5.Dyson HJ, Wright PE. Intrinsically Unstructured Proteins and Their Functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically Disordered Proteins in Human Diseases: Introducing the D2 Concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 7.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and Structure of Inherently Disordered Proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ. Linking Folding and Binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babu MM, Van Der Lee R, De Groot NS, Gsponer J. Intrinsically Disordered Proteins: Regulation and Disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Dyson HJ. Expanding the Proteome: Disordered and Alternatively Folded Proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou HX. Intrinsic Disorder: Signaling via Highly Specific but Short-Lived Association. Trends Biochem Sci. 2012;37:43–48. doi: 10.1016/j.tibs.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tompa P. Intrinsically Disordered Proteins: A 10-Year Recap. Trends Biochem Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Zhou HX, Rivas G, Minton AP. Macromolecular Crowding Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou HX. Influence of Crowded Cellular Environments on Protein Folding, Binding, and Oligomerization: Biological Consequences and Potentials of Atomistic Modeling. FEBS Lett. 2013;587:1053–1061. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mcnulty BC, Tripathy A, Young GB, Charlton LM, Orans J, Pielak GJ. Temperature-Induced Reversible Conformational Change in the First 100 Residues of Alpha-Synuclein. Protein Sci. 2006;15:602–608. doi: 10.1110/ps.051867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szasz C, Alexa A, Toth K, Rakacs M, Langowski J, Tompa P. Protein Disorder Prevails Under Crowded Conditions. Biochemistry. 2011;50:5834–5844. doi: 10.1021/bi200365j. [DOI] [PubMed] [Google Scholar]
- 17.Schlesinger AP, Wang Y, Tadeo X, Millet O, Pielak GJ. Macromolecular Crowding Fails to Fold a Globular Protein in Cells. J Am Chem Soc. 2011;133:8082–8085. doi: 10.1021/ja201206t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cino EA, Karttunen M, Choy WY. Effects of Molecular Crowding on the Dynamics of Intrinsically Disordered Proteins. Plos One. 2012;7:E49876. doi: 10.1371/journal.pone.0049876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel R, Westphal AH, Huberts DH, Nabuurs SM, Lindhoud S, Visser AJ, Van Mierlo CP. Macromolecular Crowding Compacts Unfolded Apoflavodoxin Causes Severe Aggregation of the Off-Pathway Intermediate During Apoflavodoxin Folding. J Biol Chem. 2008;283:27383–27394. doi: 10.1074/jbc.M802393200. [DOI] [PubMed] [Google Scholar]
- 20.Hong J, Gierasch LM. Macromolecular Crowding Remodels the Energy Landscape of a Protein by Favoring a More Compact Unfolded State. J Am Chem Soc. 2010;132:10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen D, Jeffries CMJ, Hammouda B, Trewhella J, Goldenberg DP. Effects of Macromolecular Crowding on an Intrinsically Disordered Protein Characterized by Small-Angle Neutron Scattering With Contrast Matching. Biophys J. 2011;100:1120–1128. doi: 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikaelsson T, Aden J, Johansson LB, Wittung-Stafshede P. Direct Observation of Protein Unfolded State Compaction in the Presence of Macromolecular Crowding. Biophys J. 2013;104:694–704. doi: 10.1016/j.bpj.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Coeur C, Teixeira J, Busch P, Longeville S. Compression of Random Coils Due to Macromolecular Crowding: Scaling Effects. Phys Rev E. 2010;81:061914. doi: 10.1103/PhysRevE.81.061914. [DOI] [PubMed] [Google Scholar]
- 24.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular Crowding Stabilizes Folded RNA Structure by the Excluded Volume Effect. J Am Chem Soc. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung MS, Klimov D, Thirumalai D. Molecular Crowding Enhances Native State Stability and Refolding Rates of Globular Proteins. Proc Natl Acad Sci U S A. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minh DD, Chang CE, Trylska J, Tozzini V, Mccammon JA. The Influence of Macromolecular Crowding on HIV-1 Protease Internal Dynamics. J Am Chem Soc. 2006;128:6006–6007. doi: 10.1021/ja060483s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittal J, Best RB. Dependence of Protein Folding Stability Dynamics on the Density Composition of Macromolecular Crowders. Biophys J. 2010;98:315–320. doi: 10.1016/j.bpj.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, Cheung MS. Structure Function And Folding of Phosphoglycerate Kinase Are Strongly Perturbed by Macromolecular Crowding. Proc Natl Acad Sci U S A. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christiansen A, Wang Q, Samiotakis A, Cheung MS, Wittung-Stafshede P. Factors Defining Effects of Macromolecular Crowding on Protein Stability: An In Vitro/In Silico Case Study Using Cytochrome c. Biochemistry. 2010;49:6519–6530. doi: 10.1021/bi100578x. [DOI] [PubMed] [Google Scholar]
- 30.Rosen J, Kim YC, Mittal J. Modest Protein-Crowder Attractive Interactions Can Counteract Enhancement of Protein Association by Intermolecular Excluded Volume Interactions. J Phys Chem B. 2011;115:2683–2689. doi: 10.1021/jp200625k. [DOI] [PubMed] [Google Scholar]
- 31.Kang M, Roberts C, Cheng YH, Chang CEA. Gating Intermolecular Interactions in Ligand-Protein Association: Coarse-Grained Modeling of HIV-1 Protease. J Chem Theory Comput. 2011;7:3438–3446. doi: 10.1021/ct2004885. [DOI] [PubMed] [Google Scholar]
- 32.Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. Reduced Native State Stability in Crowded Cellular Environment Due to Protein-Protein Interactions. J Am Chem Soc. 2013;135:3696–3701. doi: 10.1021/ja3126992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin S, Zhou HX. Atomistic Modeling of Macromolecular Crowding Predicts Modest Increases in Protein Folding Binding Stability. Biophys J. 2009;97:12–19. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin S, Zhou HX. Generalized Fundamental Measure Theory for Atomistic Modeling of Macromolecular Crowding. Phys Rev E. 2010;81:031919. doi: 10.1103/PhysRevE.81.031919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mcguffee SR, Elcock AH. Diffusion Crowding & Protein Stability in a Dynamic Molecular Model of the Bacterial Cytoplasm. Plos Comput Biol. 2010;6:E1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin S, Zhou HX. FFT-Based Method for Modeling Protein Folding and Binding Under Crowding: Benchmarking on Ellipsoidal and All-Atom Crowders. J Chem Theory Comput. 2013 doi: 10.1021/ct4005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batra J, Xu K, Qin S, Zhou HX. Effect of Macromolecular Crowding on Protein Binding Stability: Modest Stabilization Significant Biological Consequences. Biophys J. 2009;97:906–911. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin S, Minh DDL, Mccammon JA, Zhou HX. Method to Predict Crowding Effects by Postprocessing Molecular Dynamics Trajectories: Application to the Flap Dynamics of HIV-1 Protease. J Phys Chem Lett. 2010;1:107–110. doi: 10.1021/jz900023w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H, Qin S, Zhou HX. Effects of Macromolecular Crowding on Protein Conformational Changes. Plos Comput Biol. 2010;6:E1000833. doi: 10.1371/journal.pcbi.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjong H, Zhou HX. The Folding Transition-State Ensemble of a Four-Helix Bundle Protein: Helix Propensity as a Determinant Macromolecular Crowding as a Probe. Biophys J. 2010;98:2273–2280. doi: 10.1016/j.bpj.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillip Y, Harel M, Khait R, Qin S, Zhou HX, Schreiber G. Contrasting Factors on the Kinetic Path to Protein Complex Formation Diminish the Effects of Crowding Agents. Biophys J. 2012;103:1011–1019. doi: 10.1016/j.bpj.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin S, Cai L, Zhou HXA. Method for Computing Association Rate Constants of Atomistically Represented Proteins Under Macromolecular Crowding. Phys Biol. 2012;9:066008. doi: 10.1088/1478-3975/9/6/066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin S, Mittal J, Zhou HX. Folding Free Energy Surfaces of Three Small Proteins Under Crowding: Validation of the Postprocessing Method by Direct Simulation. Phys Biol. 2013;10:045001. doi: 10.1088/1478-3975/10/4/045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Liu Z. Kinetic Advantage of Intrinsically Disordered Proteins in Coupled Folding-Binding Process: A Critical Assessment of the “Fly-Casting” Mechanism. J Mol Biol. 2009;393:1143–1159. doi: 10.1016/j.jmb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Cino EA, Wong-Ekkabut J, Karttunen M, Choy WY. Microsecond Molecular Dynamics Simulations of Intrinsically Disordered Proteins Involved in the Oxidative Stress Response. Plos One. 2011;6:E27371. doi: 10.1371/journal.pone.0027371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knott M, Best RBA. Preformed Binding Interface in the Unbound Ensemble of an Intrinsically Disordered Protein: Evidence From Molecular Simulations. Plos Comput Biol. 2012;8:E1002605. doi: 10.1371/journal.pcbi.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Ganguly D, Chen J. Residual Structures Conformational Fluctuations and Electrostatic Interactions in the Synergistic Folding of Two Intrinsically Disordered Proteins. Plos Comput Biol. 2012;8:E1002353. doi: 10.1371/journal.pcbi.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittal J, Yoo TH, Georgiou G, Truskett TM. Structural Ensemble of an Intrinsically Disordered Polypeptide. J Phys Chem B. 2013;117:118–124. doi: 10.1021/jp308984e. [DOI] [PubMed] [Google Scholar]
- 49.Das RK, Pappu RV. Conformations of Intrinsically Disordered Proteins Are Influenced By Linear Sequence Distributions Of Oppositely Charged Residues. Proc Natl Acad Sci USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tozzini V, Mccammon JAA. Coarse Grained Model for the Dynamics of Flap Opening in HIV-1 Protease. Chem Phys Lett. 2005;413:123–128. [Google Scholar]
- 51.The compact conformation had C-alpha positions of an HIV-1 protese monomer. The extendedconformation was selected as the snapshot with the largest Rgfrom a simulation starting from the compact conformation.
- 52.The variations in Rg;rmsamong the 36 repeat trajectories, when scaled up by a factorof 2, mirror closely the standard deviations of Rgin the combined distributions. The latter standard deviations are listed in Table S1.
- 53.Zhou HX. Dimensions of Denatured Protein Chains From Hydrodynamic Data. J Phys Chem B. 2002;106:5769–5775. [Google Scholar]
- 54.Minton AP. Models for Excluded Volume Interaction Between an Unfolded Protein and Rigid Macromolecular Cosolutes: Macromolecular Crowding and Protein Stability Revisited. Biophys J. 2005;88:971–985. doi: 10.1529/biophysj.104.050351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


