S-Nitrosylation of ascorbate peroxidase is part of the signaling pathway for programmed cell death induction in plants.
Abstract
Nitric oxide (NO) is a small redox molecule that acts as a signal in different physiological and stress-related processes in plants. Recent evidence suggests that the biological activity of NO is also mediated by S-nitrosylation, a well-known redox-based posttranslational protein modification. Here, we show that during programmed cell death (PCD), induced by both heat shock (HS) or hydrogen peroxide (H2O2) in tobacco (Nicotiana tabacum) Bright Yellow-2 cells, an increase in S-nitrosylating agents occurred. NO increased in both experimentally induced PCDs, although with different intensities. In H2O2-treated cells, the increase in NO was lower than in cells exposed to HS. However, a simultaneous increase in S-nitrosoglutathione (GSNO), another NO source for S-nitrosylation, occurred in H2O2-treated cells, while a decrease in this metabolite was evident after HS. Consistently, different levels of activity and expression of GSNO reductase, the enzyme responsible for GSNO removal, were found in cells subjected to the two different PCD-inducing stimuli: low in H2O2-treated cells and high in the heat-shocked ones. Irrespective of the type of S-nitrosylating agent, S-nitrosylated proteins formed upon exposure to both of the PCD-inducing stimuli. Interestingly, cytosolic ascorbate peroxidase (cAPX), a key enzyme controlling H2O2 levels in plants, was found to be S-nitrosylated at the onset of both PCDs. In vivo and in vitro experiments showed that S-nitrosylation of cAPX was responsible for the rapid decrease in its activity. The possibility that S-nitrosylation induces cAPX ubiquitination and degradation and acts as part of the signaling pathway leading to PCD is discussed.
Nitric oxide (NO) is a gaseous and diffusible redox molecule that acts as a signaling compound in both animal and plant systems (Pacher et al., 2007; Besson-Bard et al., 2008). In plants, NO has been found to play a key role in several physiological processes, such as germination, lateral root development, flowering, senescence, stomatal closure, and growth of pollen tubes (Beligni and Lamattina, 2000; Neill et al., 2002; Correa-Aragunde et al., 2004; He et al., 2004; Prado et al., 2004; Carimi et al., 2005). In addition, NO has been reported to be involved in plant responses to both biotic and abiotic stresses (Leitner et al., 2009; Siddiqui et al., 2011) and in the signaling pathways leading to programmed cell death (PCD; Delledonne et al., 1998; de Pinto et al., 2006; De Michele et al., 2009; Lin et al., 2012; Serrano et al., 2012).
The cellular environment may greatly influence the chemical reactivity of NO, giving rise to different biologically active NO-derived compounds, collectively named reactive nitrogen species, which amplify and differentiate its ability to activate physiological and stress-related processes. Many of the biological properties of NO are due to its high affinity with transition metals of metalloproteins as well as its reactivity with reactive oxygen species (ROS; Hill et al., 2010). However, recent evidence suggests that protein S-nitrosylation, due to the addition of NO to reactive Cys thiols, may act as a key mechanism of NO signaling in plants (Wang et al., 2006; Astier et al., 2011). NO is also able to react with reduced glutathione (GSH), the most abundant cellular thiol, thus producing S-nitrosoglutathione (GSNO), which also acts as an endogenous trans-nitrosylating agent. GSNO is also considered as a NO store and donor and, as it is more stable than NO, acts as a long-distance NO transporter through the floematic flux (Malik et al., 2011). S-Nitrosoglutathione reductase (GSNOR), which is an enzyme conserved from bacteria to humans, has been suggested to play a role in regulating S-nitrosothiols (SNO) and the turnover of S-nitrosylated proteins in plants (Liu et al., 2001; Rusterucci et al., 2007).
A number of proteins involved in metabolism, stress responses, and redox homeostasis have been identified as potential targets for S-nitrosylation in Arabidopsis (Arabidopsis thaliana; Lindermayr et al., 2005). During the hypersensitive response (HR), 16 proteins were identified to be S-nitrosylated in the seedlings of the same species (Romero-Puertas et al., 2008); in Citrus species, S-nitrosylation of about 50 proteins occurred in the NO-mediated resistance to high salinity (Tanou et al., 2009).
However, while the number of candidate proteins for S-nitrosylation is increasing, the functional significance of protein S-nitrosylation has been explained only in a few cases, such as for nonsymbiotic hemoglobin (Perazzolli et al., 2004), glyceraldehyde 3-phosphate dehydrogenase (Lindermayr et al., 2005; Wawer et al., 2010), Met adenosyltransferase (Lindermayr et al., 2006), and metacaspase9 (Belenghi et al., 2007). Of particular interest are the cases in which S-nitrosylation involves enzymes controlling ROS homeostasis. For instance, it has been reported that S-nitrosylation of peroxiredoxin IIE regulates the antioxidant function of this enzyme and might contribute to the HR (Romero-Puertas et al., 2007). It has also been shown that in the immunity response, S-nitrosylation of NADPH oxidase inactivates the enzyme, thus reducing ROS production and controlling HR development (Yun et al., 2011).
Recently, S-nitrosylation has also been shown to be involved in PCD of nitric oxide excess1 (noe1) rice (Oryza sativa) plants, which are mutated in the OsCATC gene coding for catalase (Lin et al., 2012). In these plants, which show PCD-like phenotypes under high-light conditions, glyceraldehyde 3-phosphate dehydrogenase and thioredoxin are S-nitrosylated. This suggests that the NO-dependent regulation of these proteins is involved in plant PCD, similar to what occurs in animal apoptosis (Sumbayev, 2003; Hara et al., 2005; Lin et al., 2012). The increase in hydrogen peroxide (H2O2) after exposure to high light in noe1 plants is responsible for the production of NO required for leaf cell death induction (Lin et al., 2012). There is a strict relationship between H2O2 and NO in PCD activation (Delledonne et al., 2001; de Pinto et al., 2002); however, the mechanism of this interplay is largely still unknown (for review, see Zaninotto et al., 2006; Zhao, 2007; Yoshioka et al., 2011). NO can induce ROS production and vice versa, and their reciprocal modulation in terms of intensity and timing seems to be crucial in determining PCD activation and in controlling HR development (Delledonne et al., 2001; Zhao, 2007; Yun et al., 2011).
In previous papers, we demonstrated that heat shock (HS) at 55°C and treatment with 50 mm H2O2 promote PCD in tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cells (Vacca et al., 2004; de Pinto et al., 2006; Locato et al., 2008). In both experimental conditions, NO production and decrease in cytosolic ascorbate peroxidase (cAPX) were observed as early events in the PCD pathway, and cAPX decrease has been suggested to contribute to determining the redox environment required for PCD (de Pinto et al., 2006; Locato et al., 2008).
In this study, the production of nitrosylating agents (NO and GSNO) in the first hours of PCD induction by HS or H2O2 treatment in tobacco BY-2 cells and their role in PCD were studied. The possibility that S-nitrosylation could be a first step in regulating cAPX activity and turnover as part of the signaling pathway leading to PCD was also investigated.
RESULTS
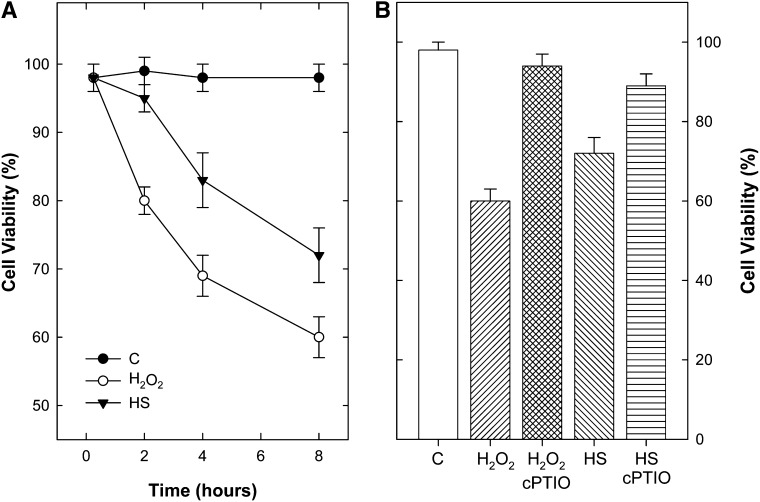
PCD was induced in tobacco BY-2 cell suspensions by 10 min of HS at 55°C or treatment with 50 mm H2O2 as reported previously (Vacca et al., 2004; de Pinto et al., 2006). The time-dependent viability of cells in the first 8 h after PCD induction is reported in Figure 1A. In both experimental conditions, cell death increased over time, although it was faster in H2O2-treated cells than in heat-shocked cells. The addition of the NO scavenger 2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) in the culture medium of tobacco BY-2 cells, before exposure to the PCD-inducing stimulus (H2O2 or HS), significantly blocked cell death (Fig. 1B), thus confirming the key role of NO in the induction of these two types of PCD (de Pinto et al., 2006; Locato et al., 2008). Therefore, NO level was measured during the first 8 h after PCD induction (Fig. 2A). Both PCD-inducing stimuli led to an increase in NO, which was already evident after 15 min. A further increase occurred in the subsequent hours, although it was much higher in the heat-shocked cells than in H2O2-treated cells.
Figure 1.
Effects of HS and H2O2 on cell viability of tobacco BY-2 cells. A, Time-dependent viability of cells treated with 50 mm H2O2 or exposed for 10 min to 55°C (HS). B, Cell viability 8 h after exposure to the PCD-inducing stimuli. Where indicated, cells were preincubated for 10 min with 500 μm cPTIO. The viability of cells was measured by trypan blue staining and examination by phase-contrast microscopy. The reported results are means ± se of five independent experiments. C, Control.
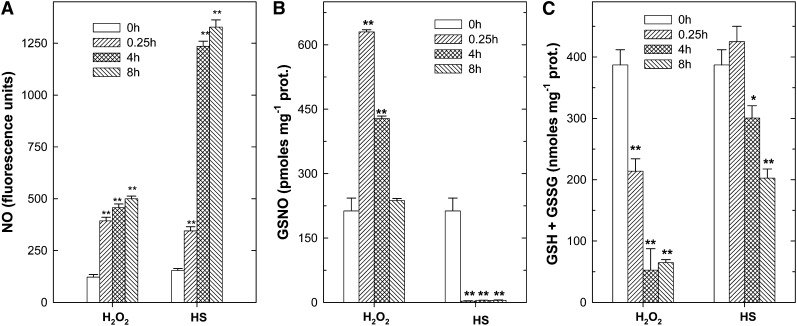
Figure 2.
Time-dependent changes in NO, GSNO, and the glutathione pool during HS- and H2O2-induced PCD. A, NO fluorescence emission of tobacco BY-2 cells was measured at the indicated times in cells loaded with 4,5-diaminofluorescein 2-diacetate before exposure to H2O2 or HS. B and C, GSNO (B) and the glutathione pool (GSH + oxidized glutathione [GSSG]; C) in tobacco BY-2 cells collected at the indicated times after exposure to PCD-inducing stimuli. The values represent means ± se of three independent experiments. *P < 0.05, **P < 0.01 (Student’s t test).
Since it is known that NO strongly reacts with thiol groups, the levels of GSNO and the glutathione pool were determined in the HS- and H2O2-treated cells. In H2O2-treated cells, GSNO increased very rapidly after the treatment and progressively decreased, with values comparable to the controls, after 8 h (Fig. 2B). On the other hand, in heat-shocked cells, GSNO levels rapidly decreased and remained very low at all the analyzed times (Fig. 2B). The glutathione pool also changed as a consequence of PCD induction: in H2O2-treated cells, the glutathione pool decreased notably 15 min after the treatment; a further decrease occurred in the following hours. In heat-shocked cells, the first decrease in the glutathione pool was observed 4 h after the treatment and was always less marked than in H2O2-treated cells (Fig. 2C).
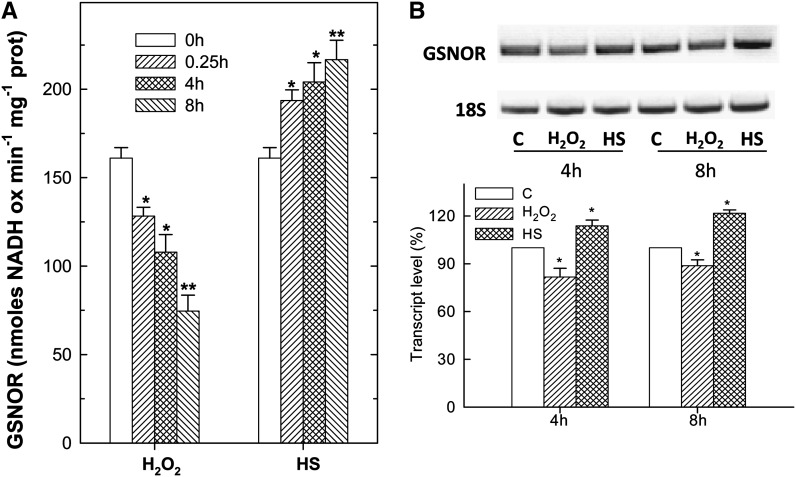
The difference in GSNO content in the two PCD conditions was in line with the behavior of GSNOR, the enzyme responsible for GSNO removal. Indeed, the activity of GSNOR decreased in H2O2-treated cells, whereas it increased in heat-shocked cells (Fig. 3A). The changes in GSNOR activity depended on changes in its expression, as shown by reverse transcription (RT)-PCR analysis (Fig. 3B).
Figure 3.
Changes in GSNOR activity and expression during HS- and H2O2-induced PCD. A, GSNOR activity was measured at the indicated times after exposure to PCD-inducing stimuli. B, Representative image and densitometric analyses of semiquantitative RT-PCR results for GSNOR at 4 and 8 h after PCD induction. Densitometric values of GSNOR transcript level are expressed as a percentage considering the values obtained for control as 100%. rRNA was used to normalize the results. The values represent means ± se of three independent experiments. *P < 0.05, **P < 0.01 (Student’s t test). C, Control.
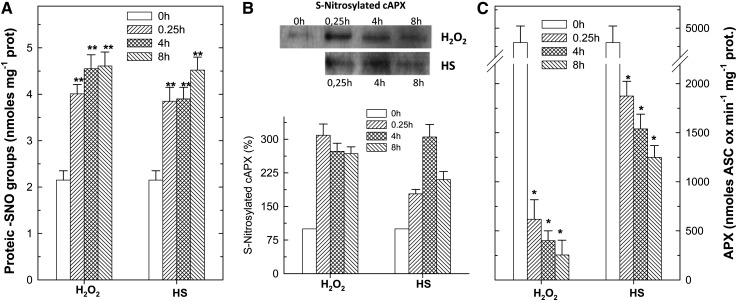
Despite the different contents of NO and GSNO, S-nitrosylated proteins quickly increased after exposure to both the PCD-inducing stimuli and remained higher than the controls for all the analyzed times (Fig. 4A). Immunoblotting with a monoclonal antibody that recognizes cAPX on S-nitrosylated proteins, isolated by the biotin-switch method (Jaffrey and Snyder, 2001) and purified with neutravidin-agarose beads, confirmed that S-nitrosylation of cAPX happened in both experimental conditions (Fig. 4B). Densitometric analysis showed that S-nitrosylation of cAPX occurred faster during the H2O2-induced PCD than during the HS one (Fig. 4B). Interestingly, a decrease in cAPX activity was found in parallel with its S-nitrosylation (Fig. 4C). The activity of cAPX decreased 15 min after exposure of the cells to both PCD-inducing stimuli and further declined in the following hours. However, this decrease was stronger in H2O2-treated cells than in HS-treated cells. The decrease in cAPX was a precocious event in the PCD signaling pathway, since it occurred when no cellular death hallmarks (cell shrinkage, micronuclei formation, and DNA laddering) were evident (data not shown).
Figure 4.
S-Nitrosylation of cAPX during PCD. A, Total protein SNO groups at different times after HS and H2O2 treatment. B, Representative image and densitometric analyses of S-nitrosylated cAPX. Densitometric values of S-nitrosylated cAPX are expressed as a percentage considering the values obtained for control as 100%. In each well, 5 μg of total nitrosylated proteins was loaded. The gels are representative of five different experiments. C, Specific activities of cAPX during H2O2- and HS-induced PCD. The values represent means ± se of five independent replicates. *P < 0.05, **P < 0.01 (Student’s t test).
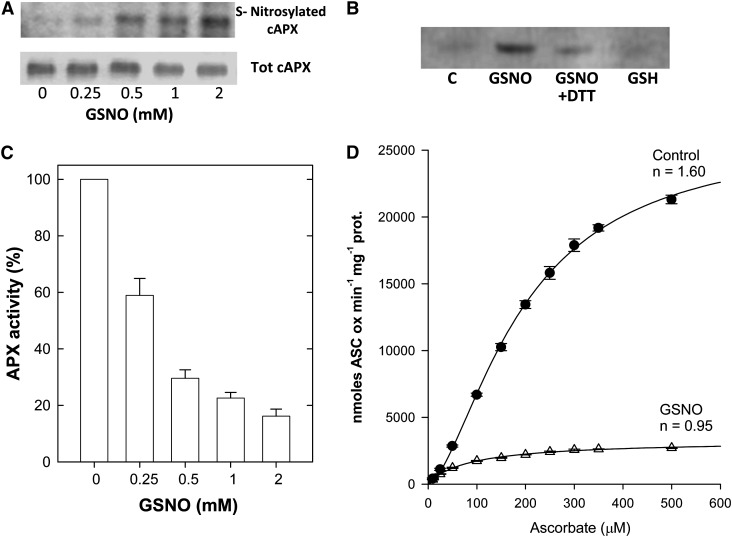
In order to confirm that S-nitrosylation was responsible for cAPX inhibition, in vitro experiments were performed. Partial purification of cAPX was obtained from nontreated tobacco BY-2 cells (Table I). The purified cAPX was exposed to a range of GSNO concentrations typically used for in vitro S-nitrosylation, and the formation of SNO-APX was monitored by the biotin-switch method. Figure 5A shows that cAPX was S-nitrosylated by GSNO in a concentration-dependent manner. Dithiothreitol (DTT) addition markedly reduced the concentration of SNO-APX formation, which was consistent with the presence of a reversible thiol modification (Fig. 5B). The absence of an effect following exposure to GSH confirmed the specificity of this posttranslation modification (Fig. 5B). The exposure to GSNO also significantly reduced the activity of the enzyme in a concentration-dependent manner (Fig. 5C). Interestingly, in vitro S-nitrosylation of partially purified cAPX from control BY-2 cells was also responsible for the alteration of enzyme kinetics from sigmoidal to hyperbolic (Fig. 5D), with a Hill coefficient changing from 1.6 ± 0.08 in control cAPX to 0.95 ± 0.07 in the S-nitrosylated enzyme.
Table I. Partial purification of cAPX from tobacco BY-2 cells.
The enrichment in cAPX was determined by the increase in its specific activity and protein level observed by western blotting.
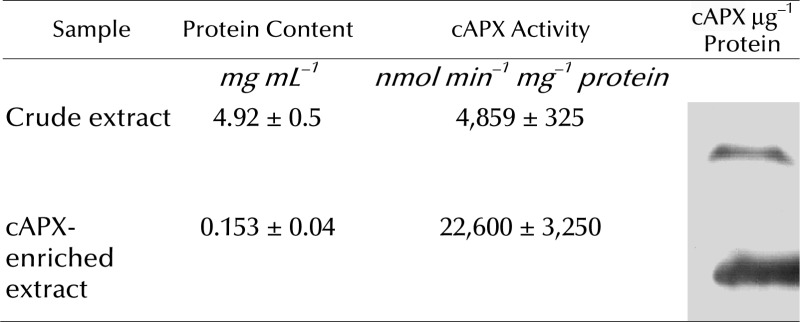 |
Figure 5.
In vitro S-nitrosylation of cAPX is responsible for its activity inhibition and kinetic changes. A, Representative images of S-nitrosylated and total cAPX after treatment with increasing GSNO concentration. B, cAPX S-nitrosylation obtained with 2 mm GSNO was reverted in the presence of 5 mm DTT and did not occur in the presence of GSH. C, Control. The gels in A and B are representative of three different experiments. C, Inhibition of cAPX activity in the presence of increasing GSNO concentrations; cAPX activity was expressed as a percentage of the activity of nontreated extracts considered as 100%. D, Changes in kinetic behavior of cAPX after in vitro S-nitrosylation. The analysis was performed on partially purified cAPX before and 30 min after treatment with 2 mm GSNO. ASC, Ascorbate. The values represent means ± se of three independent experiments. The n values are Hill coefficients.
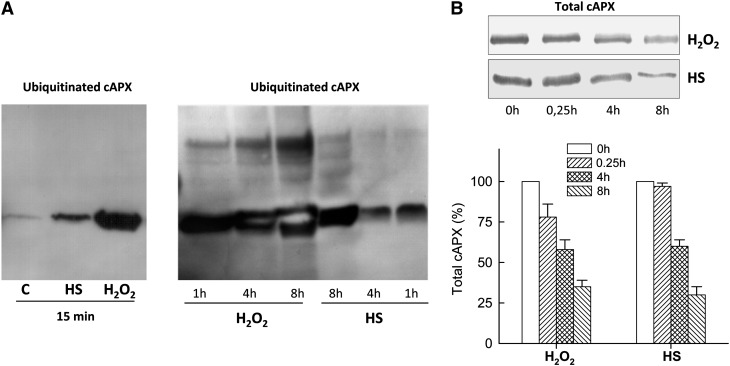
Since S-nitrosylation could be a signal for protein degradation via the ubiquitin-proteasome pathway (Kim et al., 2004; Kwak et al., 2010), we also investigated whether cAPX was ubiquitinated during PCD in tobacco BY-2 cells. Immunoblotting with anti-cAPX on enriched fractions of ubiquitinated proteins highlighted that ubiquitination of cAPX occurred during the PCD induced by the two stimuli, although not at the same time (Fig. 6A). Indeed, ubiquitinated cAPX appeared 15 min after PCD induction in both kinds of PCD but was higher in H2O2 cells. In addition, the appearance over time of bands with a higher Mr, indicative of polyubiquitinated cAPX, clearly demonstrated that ubiquitination of cAPX occurred faster in H2O2 cells than in HS cells. (Fig. 6A). In line with this, a decrease in the total level of cAPX was observed during both types of PCD but was faster in H2O2 cells than in HS cells (Fig. 6B).
Figure 6.
Ubiquitin-dependent degradation of cAPX during H2O2- and HS-induced PCD. A, Representative images of ubiquitinated cAPX at various times after PCD induction by H2O2 and HS. C, Control. B, Representative image and densitometric analysis of total cAPX during PCD. Densitometric values of total cAPX are expressed as a percentage considering the values obtained for control as 100%. All gels are representative of three different experiments. The values represent means ± se of three independent experiments.
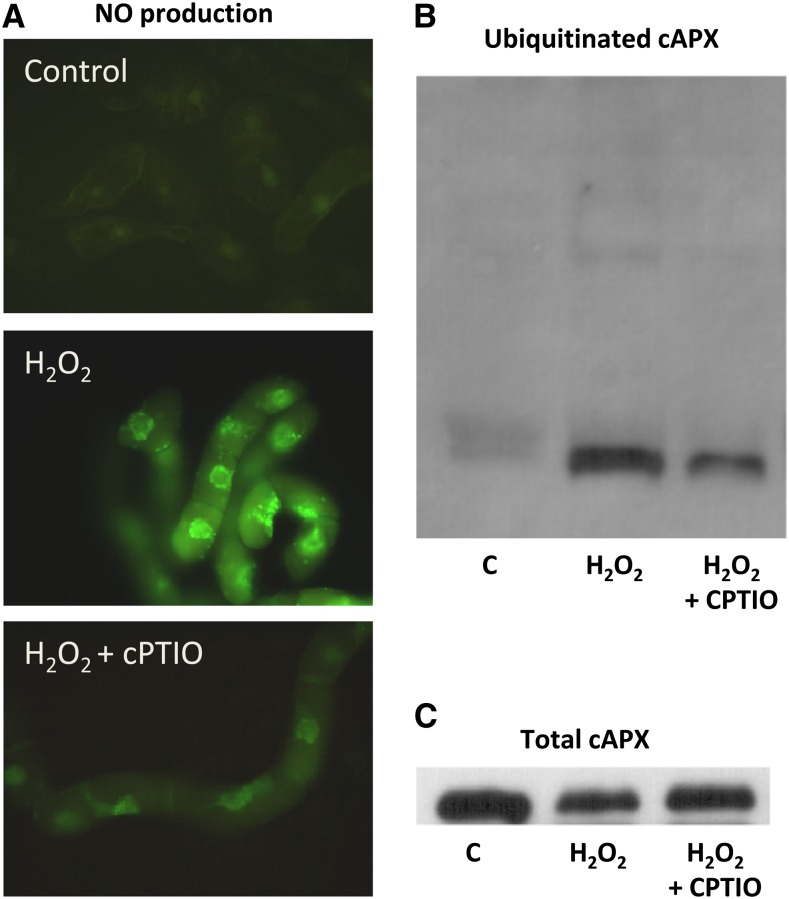
Finally, in order to verify whether cAPX ubiquitination and degradation were NO-dependent events, these analyses were performed in cells subjected to PCD stimuli in the presence of cPTIO. The cPTIO ability to scavenge NO produced during H2O2-induced PCD has been confirmed by using the specific fluorescent probe 4,5-diaminofluorescein 2-diacetate (Fig. 7A). Consistently, in the presence of cPTIO, the levels of cAPX ubiquitination and degradation were also remarkably decreased (Fig. 7, B and C). Similar results were obtained in cells undergoing PCD after HS exposure (data not shown).
Figure 7.
NO-dependent ubiquitination and degradation of cAPX during H2O2-induced PCD. A, NO production in H2O2-treated cells; where indicated, cells were preincubated for 10 min with 500 μm cPTIO. B, cAPX ubiquitination in H2O2-treated cells in the absence and presence of cPTIO. C, Total cAPX during H2O2-induced PCD in the absence and presence of cPTIO. All gels are representative of three different experiments. C, Control.
DISCUSSION
The production of a NO burst is a common event in the induction of PCD due to different stimuli. In fact, NO performs a pivotal role in HR induction (Delledonne et al., 1998; Zaninotto et al., 2006), leaf senescence (Carimi et al., 2005), Cd2+-induced PCD in suspension cell cultures (De Michele et al., 2009), and PCD induced by high light in leaves of noe1 rice lacking catalase activity (Lin et al., 2012).
We confirmed that NO plays a key role in H2O2- and HS-dependent PCD induction in tobacco BY-2 cells (de Pinto et al., 2006; Locato et al., 2008), since treatment of cells with the NO scavenger cPTIO, before exposure to the PCD-inducing stimuli, significantly blocked cell death (Fig. 1B). Our data show that the accumulation of NO occurs rapidly (within 15 min) in the PCDs induced by H2O2 and HS. However, higher levels of this molecule subsequently accumulated in heat-shocked cells over a period of 8 h (Fig. 2A). In H2O2-treated cells, the lower increase in NO was accompanied by a simultaneous increase in GSNO, while in heat-shocked cells, the level of GSNO was lower than that observed in the control cells (Fig. 2B). The decrease in GSNO observed in heat-shocked cells does not seem to be due to a temperature-dependent degradation, since very low amounts of this molecule were found at all the analyzed times, even when the cells were maintained for 8 h at their optimal temperature. The different levels of GSNO observed in the two experimentally induced PCDs may in part be explained by the changes observed in GSNOR activity and expression (Fig. 3). In fact, in heat-shocked cells, the increase in GSNOR could induce GSNO removal, whereas in H2O2-treated cells, the decrease in GSNOR could favor the accumulation of GSNO. Accordingly, an opposite correlation between GSNO content and GSNOR has been demonstrated previously in different pepper (Capsicum annuum) plant organs (Airaki et al., 2011). In addition, the different levels of NO and GSNO in the two experimentally induced PCDs are also in line with a different behavior of the glutathione pool. It is not surprising that in tobacco BY-2 cells, the glutathione pool is much higher than GSNO (Fig. 2, B and C), since the range of the two metabolites in tobacco BY-2 cells concurs with the literature (Singh et al., 1996; Airaki et al., 2011). It is known that NO release from GSNO is stimulated by high GSH concentrations (Singh et al., 1996). In heat-shocked cells, the maintenance of a high content of the glutathione pool after PCD induction might promote the decomposition of GSNO and thus contribute to the strong increase in NO occurring in cells undergoing PCD after HS exposure. On the other hand, in H2O2 cells, the more rapid and marked decrease in the glutathione pool could be a factor favoring GSNO accumulation.
It has been suggested that an increase in NO and GSNO and a decrease in NO removal are part of NO signaling (Diaz et al., 2003; Rusterucci et al., 2007). GSNO can act as a NO donor by directly transferring a NO group to a target Cys residue, thus playing a key role in regulating the biological activity of NO (Pawloski et al., 2001). However, S-nitrosylation may also be due to the direct reaction of ROS with Cys residues. Thus, despite the different contents in NO and GSNO, observed in H2O2 and HS cells, protein S-nitrosylation promptly occurs in both kinds of PCD (Fig. 4A).
S-Nitrosylation is considered to be one of the most important functional forms of NO-dependent posttranslational modification and is involved in regulating various signaling processes (Astier et al., 2011). In plants, several proteome-wide analyses show that protein S-nitrosylation might affect a large spectrum of cellular functions (Lindermayr et al., 2005; Romero-Puertas et al., 2008; Tanou et al., 2009). It has also been reported that protein S-nitrosylation occurs during abiotic stress (Ortega-Galisteo et al., 2012; Camejo et al., 2013) and is an important mediator of H2O2-induced cell death in rice leaves (Lin et al., 2012), even if this does not exclude a direct role of H2O2 in activating specific pathways leading to PCD or to stress responses.
Our data show that cAPX, a key enzyme for H2O2 removal in plant cells (Shigeoka et al., 2002; De Gara 2004), is S-nitrosylated in both H2O2- and HS-induced PCD (Fig. 4B). In fact, Lin et al. (2012) reported ascorbate peroxidase among S-nitrosylated proteins in the H2O2-induced cell death in noe1 plants. It is worth noting that S-nitrosylation of cAPX seems to be responsible for the immediate decrease in the enzyme activity observed in both HS- and H2O2-induced PCD (Fig. 4C). This is supported by in vitro results showing that the GSNO-dose-dependent S-nitrosylation of cAPX causes a parallel decrease in enzyme activity (Fig. 5). The faster S-nitrosylation of cAPX observed during H2O2-induced PCD compared with that observed in HS-induced PCD is also consistent with the more rapid and stronger decrease in cAPX activity observed in H2O2-treated cells (Fig. 4) and, as a consequence, with the different rates of PCD occurrence in the two different conditions (Fig. 1A). H2O2 itself could also contribute to cAPX inactivation (Hiner et al., 2000), further explaining the timing of PCD occurrence faster in H2O2-treated cells than in the HS-exposed ones. A decrease in antioxidants has also been reported as part of the strategies that cells exploit to activate the oxidative burst that characterizes PCDs triggered by various stimuli (for review, see de Pinto et al., 2012).
In previous papers (Vacca et al., 2004; de Pinto et al., 2006), we showed that the immediate decrease in cAPX activity, observed during PCD induced by exposure to HS or H2O2, is due to a change in the kinetics of the enzyme from sigmoidal to hyperbolic. The data from this work prove that this kinetic change is due to cAPX S-nitrosylation, since it occurs when the partially purified enzyme is in vitro trans-nitrosylated by GSNO treatment (Fig. 5D). The loss of cooperativity in the S-nitrosylated cAPX, evident from the change in the Hill coefficient (Fig. 5D), suggests that one of the two ascorbate-binding sites in the catalytic domain of the enzyme has been lost. It has been reported that pea (Pisum sativum) cAPX possesses two binding sites for ascorbate, with different affinities for the substrate (Lad et al., 2002). The high-affinity site is located in the vicinity of the Cys-32 residue (Mandelman et al., 1998). Since the Cys-32 residue is well conserved in the cAPX of various species, including tobacco, we can speculate that the NO- or GSNO-dependent S-nitrosylation of this residue could be responsible for the changes in enzyme kinetics and catalytic turnover. In Arabidopsis, for example, APX1, a cytosolic isoenzyme of ascorbate peroxidase, has been identified among S-nitrosylable proteins, with Cys-32 recognized as the site of S-nitrosylation (Fares et al., 2011).
Some evidence suggests that S-nitrosylation could represent a signal for protein degradation through the ubiquitin-proteasome pathway in animal cells (Kim et al., 2004; Kwak et al., 2010; Dunne et al., 2013). Our data show that cAPX is also promptly ubiquitinated in cells undergoing H2O2- and HS-induced PCDs (Fig. 6A). Consistently, we found that the proteasome is involved in HS-induced PCD in tobacco BY-2 cells (Vacca et al., 2007). Moreover, the faster ubiquitination of cAPX in H2O2-treated cells also correlates with the faster decrease in cAPX protein level occurring under this experimental condition in comparison with HS (Fig. 6B). Interestingly, when NO production is reduced by treatment with the NO scavenger cPTIO, cAPX ubiquitination and degradation are remarkably prevented (Fig. 7), thus indicating, to our knowledge for the first time, that also in plants S-nitrosylation acts as a signal for ubiquitin-dependent protein degradation. Further studies are required in order to verify whether the oxidative environment activating PCD in H2O2-treated cells induces other alterations in cAPX that contribute to its ubiquitination.
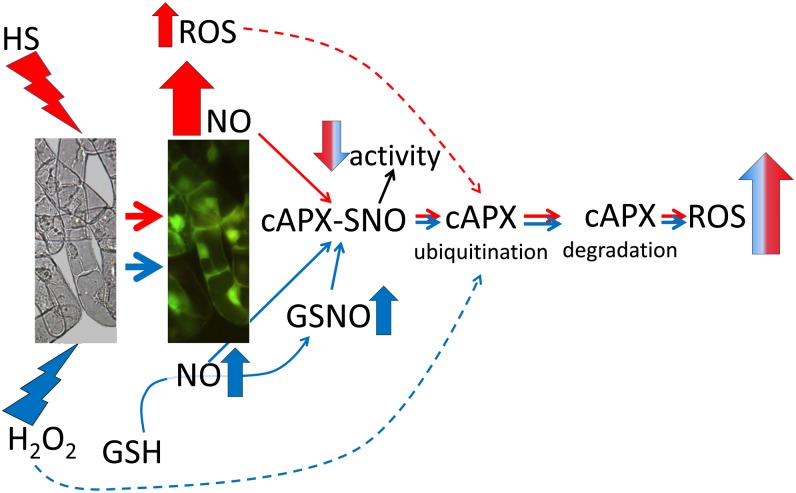
In conclusion, our results suggest that S-nitrosylation of cAPX, which takes place when no markers of cell death are evident, is part of the early signaling pathway leading to NO-dependent PCD. In vitro and in vivo evidence suggests that S-nitrosylation of cAPX is pivotal for the rapid decrease in enzyme activity, since it affects its kinetic properties. As summarized in Figure 8, the rapid decrease in cAPX activity, due to an immediate posttranslational modification, supports the hypothesis that alterations in both ROS production and ROS scavengers are critical for the oxidative burst to occur as a precocious event of plant PCD. Our data also further underline the tight relationship between NO and ROS in the control of PCD.
Figure 8.
S-Nitrosylation of cAPX is a part of PCD signaling in tobacco BY-2 cells. The diagram summarizes the results described in this paper. H2O2 treatment induces an increase of both NO and GSNO in cells; on the other hand, the HS-mediated oxidative stress determines only a high production of NO. In both cases, an increase in S-nitrosylation of cAPX occurs. S-Nitrosylation of cAPX, possibly in association with other oxidative mechanisms (dotted lines), affects cAPX activity and makes cAPX a target for ubiquitination and consequent degradation. The decrease in cAPX activity and protein level determines the high ROS production involved in PCD.
MATERIALS AND METHODS
Cell Treatments and Analyses of Cell Viability
The tobacco (Nicotiana tabacum) ‘Bright-Yellow 2’ cell suspensions were routinely propagated and cultured according to Nagata et al. (1992). For the experiments, a stationary culture (7 d) was diluted 4:100 (v/v). On day 4 of culture, cells were treated with 50 mm H2O2 (de Pinto et al., 2006) or subjected to HS by transferring to 55°C for 10 min (Vacca et al., 2004). At the indicated times, 1 mL of cell suspension was collected for cell viability and NO analyses. Cell viability was determined by trypan blue staining as described previously (de Pinto et al., 1999). For each sample 1,000 cells were scored.
For other analyses, cells were separated from the culture medium by vacuum filtration on Whatman 3MM paper (Whatman International).
NO, SNO, and Glutathione
NO was measured with the fluorescent probe 4,5-diaminofluorescein 2-diacetate according to Locato et al. (2008).
SNO groups were assayed following the method of Saville (1958) after separation of the extracts on the basis of molecular mass. Briefly, cells were homogenized in 0.1 m Tris-HCl, pH 6.8, at 4°C and centrifuged at 20,000g for 15 min. The supernatants were successively separated with Amicon Ultra 5K tubes (Millipore) in two fractions: low molecular mass (less than 5 kD) and high molecular mass (more than 5 kD). The two fractions were incubated for 5 min with an equivalent volume of 1% (w/v) sulfanilamide dissolved in 0.5 m HCl in the presence or absence of 0.2% (w/v) HgCl2. Samples were then incubated for 5 min with an equivalent volume of 0.02% (w/v) N-(1-naphthyl)-ethylenediamine dihydrochloride dissolved in 0.5 m HCl, and the absorbance of azo-dye products was read at 550 nm. SNO levels were quantified as the difference of absorbance in samples incubated in the presence and absence of HgCl2 and comparing the values with a standard curve made with GSNO (Sigma-Aldrich). SNO of the low-molecular-mass fraction were considered as GSNO. The results were normalized against the whole-cell lysate protein content measured according to Bradford (1976).
Glutathione content was measured as described by de Pinto et al. (1999).
GSNOR and cAPX Activities
Cells (0.3 g) were homogenized in liquid N2 with 4 volumes of buffer containing 50 mm Tris-HCl, pH 7.8, 0.05% (w/v) Cys, and 0.1% (w/v) bovine serum albumin. The homogenates were then centrifuged at 20,000g for 15 min, and the supernatants were used for the determination of enzymatic activities.
GSNOR (EC 1.1.1.284) activity was measured following the decrease in A340 due to the oxidation of NADH occurring for GSNO consumption. The reaction mixture contained 0.05 m Tris-HCl, pH 8.0, 0.5 mm EDTA, 0.2 mm NADH, and 0.4 mm GSNO. An extinction coefficient of 6.2 mm−1 cm−1 was used, and the activity was expressed as nmoles of NADH oxidized per minute per milligram of protein.
cAPX (EC 1.11.1.11) activity was determined following the H2O2-dependent oxidation of ascorbate at 290 nm, as described by Paradiso et al. (2012).
Semiquantitative RT-PCR
Total RNA was isolated from tobacco BY-2 cells using the RNeasy plant minikit (Qiagen) according to the supplier’s recommendation. Residual DNA was removed from the RNA samples using a DNA-free kit (Ambion). Synthesis of complementary DNA was performed from 2 mg of total RNA with 10-mm random primers (Amersham Biosciences Europe), utilizing an Omniscript Reverse Transcriptase kit (Qiagen) according to the supplier’s recommendation. PCR was performed with specific primers for GSNOR (5′-GCAAGTTCTGCAAATCAGGA-3′ and 5′-GCTCTTTGGGGTTGATGAAT-3′; accession number HQ830156.1) and 18S rybosomial RNA (rRNA; AJ236016; 5′-CATGATAACTCGACGGATCG-3′ and 5′-GAAGGCCAACGTAATAGGAC-3′). The 18S rRNA was used as an internal control in order to normalize each sample for variations in the amount of initial RNA. The products of PCR amplification produced a single band at the predicted sizes of 441 and 594 bp for GSNOR and 18S rRNA, respectively. These were analyzed on 1.5% (w/v) agarose gels containing 0.5 mg mL−1 ethidium bromide.
Immunodetection of S-Nitrosylated cAPX
Cells were homogenized in liquid N2 with four volumes of buffer containing 50 mm HEPES, pH 7.5, 5 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride. The homogenates were centrifuged at 20,000g for 15 min and the supernatants subjected to biotin switch (Jaffrey and Snyder, 2001). Briefly, Cys-free residues were blocked by incubating 2 mg of proteins with 5 μL of 20 mm methyl-methanthiosulfate (Pierce) and 25 μL of 25% (w/v) SDS for 30 min at 50°C, with frequently vortexing. Samples were subsequently precipitated with 2 volumes of ice-cold acetone and resuspended in 100 μL of buffer containing 25 mm HEPES, pH 7.7, 1 mm EDTA, and 1% (w/v) SDS. The resuspended samples were incubated for 1.5 h in the dark with 1 mm ascorbate and 1 mm N-(6-(biotinamido)hexyl-3′-(2′-pyridyldithio)propionamide (Pierce) and frequently vortexed. Purification of biotinylated proteins was obtained as described by Belenghi et al. (2007). Biotinylated samples were diluted with 2 volumes of neutralization buffer (25 mm HEPES, 1 mm EDTA, 100 mm NaCl, and 0.8% (v/v) Triton X-100, pH 7.7) supplemented with 140 μL of neutroavidin-agarose (Pierce) and incubated overnight, at 4°C, with continuous shaking. Beads were washed three times with 2 mL of buffer containing 25 mm HEPES, pH 7.7, 1 mm EDTA, 600 mm NaCl, and 0.8% (v/v) Triton X-100 and resuspended in 40 μL of SDS-PAGE loading buffer (0.225 m Tris-HCl, pH 6.8, 7.5% (w/v) SDS, 30% (v/v) glycerol, 1.5% (w/v) dithioerythritol, and 0.03% (w/v) bromphenol blue). After 5 min of incubation at 100°C and subsequent cooling, samples were centrifuged at 20,000g for 5 min. The supernatants were separated by 12.5% (w/v) SDS-PAGE and subjected to immunoblotting with the anti-cAPX monoclonal antibody as described previously (de Pinto et al., 2002).
Immunodetection of Ubiquitinated cAPX
Cells were homogenized in liquid N2 with 2 volumes of buffer containing 50 mm HEPES, pH 7.5, 5 mm EDTA, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and 10 mm N-Ethylmaleimide. The homogenates were centrifuged at 20,000g for 15 min and the supernatants used for the analysis. In particular, ubiquitinated proteins were obtained using the Ubiquitinated Protein Enrichment Kit (Calbiochem), following the manufacturer’s instructions. The obtained extracts, enriched in ubiquitinated proteins, were separated by 12.5% (w/v) SDS-PAGE and subjected to immunoblotting with the anti-cAPX monoclonal antibody as described previously (de Pinto et al., 2002). In order to detect total cAPX level, crude extracts were also subjected to immunoblotting with cAPX antibody.
In Vitro Studies of Partially Purified cAPX
cAPX was partially purified by electroelution as described by De Gara et al. (1997). Briefly, proteins were separated by native PAGE, and a gel lane was cut and stained for ascorbate peroxidase activity in order to identify the ascorbate peroxidase position on the gel (De Gara et al., 1997). The area containing cAPX isoenzymes on the remaining gel was then electroeluted with a GE 200 SixPac Gel Eluter (Hoefer Scientific Instruments) following the experimental conditions recommended by the supplier.
Aliquots of semipurified cAPX were incubated in the dark at room temperature for 30 min with different concentrations of GSNO (ranging from 0.25 to 2 mm) or with 2 mm GSH in MES buffer (pH 7.5). A cAPX aliquot treated with 2 mm GSNO was then incubated with 10 mm of the reducing agent DTT. Excess of reagents were eliminated by protein precipitation with 2 volumes of cold acetone. Proteins were then subjected to biotin switch, resuspended in SDS-PAGE loading buffer, and loaded on 12.5% (w/v) SDS-PAGE gels. S-Nitrosylated cAPX was detected with an anti-biotin antibody (Pierce).
The kinetics of control and GSNO-treated cAPX were obtained by determining its activity with 170 μm H2O2 and increasing ascorbate concentrations (10–500 μm). The value for the Hill coefficient was obtained by analysis with the Enzyme Kinetics software (SPSS).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number HQ830156.1 (GSNOR).
Glossary
- NO
nitric oxide
- PCD
programmed cell death
- ROS
reactive oxygen species
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- SNO
S-nitrosothiols
- GSNOR
S-nitrosoglutathione reductase
- HR
hypersensitive response
- H2O2
hydrogen peroxide
- HS
heat shock
- BY-2
Bright Yellow-2
- cAPX
cytosolic ascorbate peroxidase
- RT
reverse transcription
- DTT
dithiothreitol
- GSH
reduced glutathione
- cPTIO
2-(4-Carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- rRNA
rybosomial RNA
References
- Airaki M, Sánchez-Moreno L, Leterrier M, Barroso JB, Palma JM, Corpas FJ. (2011) Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol 52: 2006–2015 [DOI] [PubMed] [Google Scholar]
- Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, Jeandroz S, Durner J, Lindermayr C, Wendehenne D. (2011) S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Belenghi B, Romero-Puertas MC, Vercammen D, Brackenier A, Inzé D, Delledonne M, Van Breusegem F. (2007) Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem 282: 1352–1358 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L. (2000) Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210: 215–221 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Camejo D, Romero-Puertas MdC, Rodríguez-Serrano M, Sandalio LM, Lázaro JJ, Jiménez A, Sevilla F. (2013) Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J Proteomics 79: 87–99 [DOI] [PubMed] [Google Scholar]
- Carimi F, Zottini M, Costa A, Cattelan I, De Michele R, Terzi M, Lo Schiavo F. (2005) NO signalling in cytokinin-induced programmed cell death. Plant Cell Environ 28: 1171–1178 [Google Scholar]
- Correa-Aragunde N, Graziano M, Lamattina L. (2004) Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218: 900–905 [DOI] [PubMed] [Google Scholar]
- De Gara L (2004) Ascorbate metabolism and plant growth: from germination to cell death. In H Asard, J Smirnoff, N May, eds, Vitamin C: Its Function and Biochemistry in Animals and Plants. Bios Scientific Publisher, Oxford, pp 83–95 [Google Scholar]
- De Gara L, de Pinto MC, Arrigoni O. (1997) Ascorbate synthesis and ascorbate peroxidase activity during the early stage of wheat germination. Physiol Plant 100: 894–900 [Google Scholar]
- De Michele R, Vurro E, Rigo C, Costa A, Elviri L, Di Valentin M, Careri M, Zottini M, Sanità di Toppi L, Lo Schiavo F. (2009) Nitric oxide is involved in cadmium-induced programmed cell death in Arabidopsis suspension cultures. Plant Physiol 150: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinto MC, Francis D, De Gara L. (1999) The redox state of the ascorbate-dehydroascorbate pair as a specific sensor of cell division in tobacco BY-2 cells. Protoplasma 209: 90–97 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Locato V, De Gara L. (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35: 234–244 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Paradiso A, Leonetti P, De Gara L. (2006) Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J 48: 784–795 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Tommasi F, De Gara L. (2002) Changes in the antioxidant systems as part of the signaling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco Bright-Yellow 2 cells. Plant Physiol 130: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz M, Achkor H, Titarenko E, Martínez MC. (2003) The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett 543: 136–139 [DOI] [PubMed] [Google Scholar]
- Dunne KA, Allam A, McIntosh A, Houston SA, Cerovic V, Goodyear CS, Roe AJ, Beatson SA, Milling SW, Walker D, et al (2013) Increased S-nitrosylation and proteasomal degradation of caspase-3 during infection contribute to the persistence of adherent invasive Escherichia coli (AIEC) in immune cells. PLoS ONE 8:e68386, /10.1371/journal.pone.0068386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares A, Rossignol M, Peltier JB. (2011) Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Biophys Res Commun 416: 331–336 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Hill BG, Dranka BP, Bailey SM, Lancaster JR, Jr, Darley-Usmar VM. (2010) What part of NO don’t you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem 285: 19699–19704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiner AN, Rodríguez-López JN, Arnao MB, Lloyd Raven E, García-Cánovas F, Acosta M. (2000) Kinetic study of the inactivation of ascorbate peroxidase by hydrogen peroxide. Biochem J 348: 321–328 [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1. [DOI] [PubMed] [Google Scholar]
- Kim S, Wing SS, Ponka P. (2004) S-nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol Cell Biol 24: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YD, Ma T, Diao SY, Zhang X, Chen YM, Hsu J, Lipton SA, Masliah E, Xu HX, Liao FF. (2010) NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener 5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad L, Mewies M, Raven EL. (2002) Substrate binding and catalytic mechanism in ascorbate peroxidase: evidence for two ascorbate binding sites. Biochemistry 41: 13774–13781 [DOI] [PubMed] [Google Scholar]
- Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M. (2009) NO signals in the haze: nitric oxide signalling in plant defence. Curr Opin Plant Biol 12: 451–458 [DOI] [PubMed] [Google Scholar]
- Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, Hu B, Yang F, Loake GJ, Chu C. (2012) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Bahnweg G, Durner J. (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281: 4285–4291 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494 [DOI] [PubMed] [Google Scholar]
- Locato V, Gadaleta C, De Gara L, De Pinto MC. (2008) Production of reactive species and modulation of antioxidant network in response to heat shock: a critical balance for cell fate. Plant Cell Environ 31: 1606–1619 [DOI] [PubMed] [Google Scholar]
- Malik SI, Hussain A, Yun BW, Spoel SH, Loake GJ. (2011) GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci 181: 540–544 [DOI] [PubMed] [Google Scholar]
- Mandelman D, Jamal J, Poulos TL. (1998) Identification of two electron-transfer sites in ascorbate peroxidase using chemical modification, enzyme kinetics, and crystallography. Biochemistry 37: 17610–17617 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. (1992) Tobacco BY-2 cell-line as the HeLa cell in the cell biology of higher plants. Int Rev Cytol 132: 1–30 [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT. (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC. (2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J Exp Bot 63: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. (2007) Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87: 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso A, de Pinto MC, Locato V, De Gara L. (2012) Galactone-γ-lactone-dependent ascorbate biosynthesis alters wheat kernel maturation. Plant Biol (Stuttg) 14: 652–658 [DOI] [PubMed] [Google Scholar]
- Pawloski JR, Hess DT, Stamler JS. (2001) Export by red blood cells of nitric oxide bioactivity. Nature 409: 622–626 [DOI] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. (2004) Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA. (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Mattè A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. (2008) Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8: 1459–1469 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AM, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M. (2007) S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Espunya MC, Díaz M, Chabannes M, Martínez MC. (2007) S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol 143: 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville B. (1958) A scheme for the colorimetric determination of microgram amounts of thiols. Analyst (Lond) 83: 670–672 [Google Scholar]
- Serrano I, Romero-Puertas MC, Rodríguez-Serrano M, Sandalio LM, Olmedilla A. (2012) Peroxynitrite mediates programmed cell death both in papillar cells and in self-incompatible pollen in the olive (Olea europaea L.). J Exp Bot 63: 1479–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53: 1305–1319 [PubMed] [Google Scholar]
- Siddiqui MH, Al-Whaibi MH, Basalah MO. (2011) Role of nitric oxide in tolerance of plants to abiotic stress. Protoplasma 248: 447–455 [DOI] [PubMed] [Google Scholar]
- Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. (1996) The chemistry of the S-nitrosoglutathione/glutathione system. Proc Natl Acad Sci USA 93: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbayev VV. (2003) S-nitrosylation of thioredoxin mediates activation of apoptosis signal-regulating kinase 1. Arch Biochem Biophys 415: 133–136 [DOI] [PubMed] [Google Scholar]
- Tanou G, Job C, Rajjou L, Arc E, Belghazi M, Diamantidis G, Molassiotis A, Job D. (2009) Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J 60: 795–804 [DOI] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passarella S, Marra E, De Gara L. (2004) Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol 134: 1100–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, Valenti D, Bobba A, de Pinto MC, Merafina RS, De Gara L, Passarella S, Marra E. (2007) Proteasome function is required for activation of programmed cell death in heat shocked tobacco Bright-Yellow 2 cells. FEBS Lett 581: 917–922 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yun BW, Kwon E, Hong JK, Yoon J, Loake GJ. (2006) S-nitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot 57: 1777–1784 [DOI] [PubMed] [Google Scholar]
- Wawer I, Bucholc M, Astier J, Anielska-Mazur A, Dahan J, Kulik A, Wysłouch-Cieszynska A, Zareba-Kozioł M, Krzywinska E, Dadlez M, et al (2010) Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem J 429: 73–83 [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Mase K, Yoshioka M, Kobayashi M, Asai S. (2011) Regulatory mechanisms of nitric oxide and reactive oxygen species generation and their role in plant immunity. Nitric Oxide 25: 216–221 [DOI] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH, et al. (2011) S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268 [DOI] [PubMed] [Google Scholar]
- Zaninotto F, La Camera S, Polverari A, Delledonne M. (2006) Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol 141: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. (2007) Interplay among nitric oxide and reactive oxygen species: a complex network determining cell survival or death. Plant Signal Behav 2: 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]