Summary
We have developed a single-molecule imaging technique that uses quantum dot-labeled peptide-major histocompatibility complex (pMHC) ligands to study CD4+ T cell functional sensitivity. We found that naive T cells, T cell blasts and memory T cells could all be triggered by a single pMHC to secrete tumor necrosis factor-α (TNF-α) and interleukin-2 (IL-2) cytokines with a rate of ~1,000, ~10,000 and ~10,000 molecules/min respectively and that additional pMHCs did not augment secretion, indicating a digital response pattern. We also found that a single pMHC localized to the immunological synapse induced the slow formation of a long-lasting T cell receptor (TCR) cluster, consistent with a serial engagement mechanism. These data show that scaling up CD4+ T cell cytokine responses involves increasingly efficient T cell recruitment rather than greater cytokine production per cell.
Introduction
CD4+ T helper cells play a critical role in adaptive immunity. They modulate the functions of other important immune cells, such as B cells, macrophages and CD8+ cytotoxic T cells through cytokine secretion. A critical first step in the activation of CD4+ T cells is the specific recognition of cognate peptide-major histocompatibility complex (pMHC) ligands displayed on antigen-presenting cell (APC) surfaces by their αβ T cell receptors (TCRs) (Davis et al., 1998). Antigen recognition triggers a variety of intracellular signaling events, including protein tyrosine kinase activation, calcium flux, secretory machinery repolarization, synapse formation and cytokine secretion (Huse et al., 2007; Ueda et al., 2011).
Upon recognition of cognate pMHCs, naive CD4+ T cells typically produce a potent T cell growth factor, interleukin 2 (IL-2) which is necessary for the proliferation, development and function of different T cell subsets including helper, cytotoxic and regulatory T cells (Ruscetti et al., 1977). Naive CD4+ T cells also produce other cytokines such as tumor necrosis factor-alpha (TNF-α) (Priyadharshini et al., 2010). Activated naive CD4+ T cells differentiate into unique subsets of effector CD4+ T cells and secrete various cytokines to mediate adaptive immune responses. After the clearance of antigens, the majority of effector CD4+ T cells that participate in the primary immune response undergo apoptosis. Only a small fraction survives to become long-lived memory T cells. Naive and memory T cells differ in many aspects, but it is generally agreed that memory T cell responses require less antigen and respond more quickly and efficaciously (Dutton et al., 1998).
Cytokine secretion is one of the main functions of CD4+ T cells and typically involves the simultaneous engagement of two directionally distinct pathways, with one set of cytokines including IL-2 being directed into the synapse and another group including TNF-α being released multidirectionally (Huse et al., 2006). For CD8+ cytotoxic T cell blasts, we have shown that one pMHC can trigger calcium signaling and that three or more pMHCs can lead to functional cell killing (Purbhoo et al., 2004). Although CD4+ T cell blasts show a similar signaling sensitivity as CD8+ T cell blasts (Irvine et al., 2002), little is known about their functional sensitivity. Furthermore the characteristics of naive and memory CD4+ T cells are even less defined.
An efficient transduction of early signals into functional responses might be particularly important during the early stages of the immune response when APCs may present only a limited number of non-self pMHCs. We have previously shown that T cell signaling sensitivity can be regulated by miR-181a during T cell development (Li et al., 2007), so understanding the functional sensitivity of CD4+ T cells at different differentiation stages could provide important insights into T cell signaling and the intercellular communication among different immune cells, in which CD4+ T cells often play a central role.
In the present study we set out to define the functional sensitivity of individual CD4+ T cells by using a combination of single-molecule imaging techniques and single-cell cytokine secretion assays. Specifically we have used quantum dot (QD)-labeled pMHCs to monitor the relationship between ligand number in the immunological synapse and CD4+ T cell functional responses. This represents a substantial improvement over our previous work using phycoerythrin as a label, since this fluorophore bleaches very rapidly and only allows a “snapshot” of pMHCs at a single time point (Irvine et al., 2002; Purbhoo et al., 2004). In addition, single-cell cytokine secretion assays using real-time cytokine-reporter systems allow us to measure the rate and magnitude of cytokine production of individual cells over time. We used these two techniques to investigate whether and how the quantity of pMHC regulates a single T cell functional response.
Results
Labeling pMHCs with QDs on the APC surface
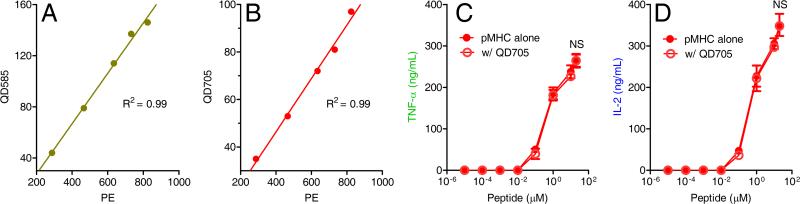
Whereas previously we have used phycoerythrin-streptavidin (PE-SA) to investigate the precise relationship between the number of agonist pMHCs and T cell signaling responses (Irvine et al., 2002), phycoerythrin is unstable and prone to photobleaching. Moreover, its emission spectrum overlaps with the auto-fluorescence of most APCs (Fig. S1A). These inherent limitations make it difficult to obtain precise single-molecule information at the APC membrane and prevent the tracking of long-term events such as cytokine secretion. To overcome these limitations, we replaced PE-SA with very bright, photostable and nano-sized (16-21.5 nm) quantum dot-streptavidin conjugates (QD-SA) to label biotin-moth cytochrome C (MCC) peptides bound to I-Ek. This biotin-MCC peptide was designed to prevent proteolysis by incorporating D-amino acids in the linker region (see methods section). After pulsing the APCs with biotin-MCC peptide, we found a linear correlation between the peptide concentration in solution and the number of biotin-MCC–I-EK molecules on the APC surface, as shown by staining with PE-SA, QD585-SA or QD705-SA (Fig. S1 B-D), consistent with previous studies using PE-SA labeling (Ebert et al., 2008; Irvine et al., 2002). We have also shown previously that, when used at saturating concentrations, PE-SA is able to bind biotinylated pMHCs presented on the APC surface at a 1:1 ratio (Irvine et al., 2002) and here we were able to show that QD-SA had the same labeling efficiency, as shown in Figures 1A and B for both QDs and also in Figure S1E for a direct comparison between QD705-SA and PE-SA. Furthermore, we found only negligible nonspecific labeling when unpulsed APCs were incubated with different concentrations of QD705-SA (Fig. S1F).
Figure 1. Detection of specific pMHC complexes on the APC surface using quantum dot labeling.
(A and B) Plots depict the mean fluorescence intensities (MFI) of MCC–I-Ek complexes on APCs as detected by PE-SA comparing QD585-SA (A) or QD705-SA (B) labeling using flow cytometry. (C and D) Dose response of TNF-α (C) and IL-2 (D) secretion measured by ELISA following 2-hour incubation of T cell blasts in response to APCs pulsed with a different concentrations of biotin-MCC peptide either labeled (open circles) or not labeled (filled circles) with QD705. No statistical differences (NS, t test) were found between labeled and unlabeled samples for TNF-α (C) and IL-2 (D) production in response to different concentrations of biotin-MCC peptide. Data are presented as mean ± SEM of three independent experiments (see also Figure S1).
To assess T cell sensitivity, we carefully kept the density of pMHCs very low (<100 pMHCs/APC) so that most pMHCs would be distant from each other (Kimachi et al., 1997), thus limiting cross-linking caused by the tetravalent nature of QD-SA. A typical APC surface area is ~1400 μm2. When loaded with less than 100 peptides, each pMHC can occupy an average area >14 μm2, which is at least 70000- or 40000-fold larger than a single QD projection area of ~2×10−4 (QD585) or 3×10−4 (QD705) μm2. Another reason cross-linking is unlikely is that there is an enormous molar excess of QD-SA in solution versus the cell surface pMHC's (>60000:1) and streptavidin-biotin interactions are essentially irreversible. Since the QDs used here are larger (diameter 16 or 21.5 nm) than the reported narrowest part of the synaptic interface (~13 nm) (Choudhuri et al., 2005; Ueda et al., 2011), one concern is that the QDs might affect pMHC mobility and T cell activation. To address this point we measured the diffusion of QD-labeled pMHCs on the B cell surface and tested the potential inhibitory effect of QD labeling to T cell activation. We first measured the diffusion of single pMHCs labeled with either Alexa-555-SA or QD-SA and found that both labeling methods yielded similar diffusion coefficients (Fig. S1 G-H). Since Alexa-555 is a very small dye (~1 kDa), the diffusion data showed that QD labeling had negligible effects on pMHC mobility. We further compared the cytokine secretion of T cell blasts following stimulation with APCs pulsed with increasing concentrations of biotin-MCC peptide labeled or not with QD705 at 2 (Fig. 1 C-D), 4 (Fig. S1 I-J) and 24 hour (Fig. S1 K-L) incubation times. Measurements using an enzyme-linked immunosorbent assay (ELISA) showed that labeling with QD705 affected neither TNF-α nor IL-2 secretion at all incubation times tested.
A single pMHC triggers cytokine secretion in CD4+ T cell blasts
The brightness of QDs allowed us to easily identify single QDs coated onto a glass coverslip (Fig. S2 A-B, top rows). However, one known limitation of QDs is their propensity to “blink” on and off which could cause a miscounting of pMHCs at the T cell-APC interface. Fortunately, we found that QD blinking was greatly suppressed when labeling pMHCs on an APC surface (Fig. S2 A-B, bottom rows and Movie S1 and S2). This is consistent with data showing that QD “blinking” is affected by changes in the local environment (Fomenko and Nesbitt, 2008). Similar suppression of QD blinking was seen in tracking neural synaptic vesicles (Zhang et al., 2009). The natural suppression of QD blinking on the APC surface allowed us to track single QD-labeled pMHCs with high spatiotemporal resolution (Movie S2). We compared QD blinking on the glass coverslip versus the APC membrane for ten consecutive frames (Fig. S2 A-B, left) and plotted the single QD intensity time traces along with the frame sequence (Fig. S2 A-B, right). The data demonstrated that our microscopy system enabled us to easily identify single QDs and also faithfully detect single pMHCs presented on a live APC surface in three-dimensions (3D) (Movie S3).
We first used this labeling method to monitor calcium signaling responses, since they occur within seconds of TCR engagement. In agreement with previous results (Irvine et al., 2002), we found that even one cognate pMHC could trigger [Ca2+]i increase in T cell blasts (Fig. S2 C-D). We then assessed the functional sensitivity of CD4+ T cell blasts by measuring cytokine secretion as a function of pMHC number. We stimulated CD4+ T cell blasts with APCs pulsed with very low concentrations of biotin-MCC peptide in vitro and measured the production of two representative cytokines: TNF-α and IL-2. TNF-α is initially synthesized as a transmembrane protein that is released from the plasma membrane by metalloprotease cleavage. Upon the addition of the metalloprotease inhibitor TAPI-0 to the culture medium, newly synthesized TNF-α was retained on the T cell surface so that it was detectable with a fluorescent anti-TNF-α antibody (Huse et al., 2006). To capture secreted IL-2 at the T cell surface we used a bifunctional antibody that binds CD45 on one arm and IL-2 on the other arm. The bound IL-2 is then detected by a specific fluorescent anti-IL-2 antibody (Manz et al., 1995). In agreement with our previous study (Huse et al., 2006), we found that CD4+ T cells forming conjugates with cognate APCs released TNF-α and IL-2 after 2 hours of incubation at 37°C and 5% CO2. We imaged these T cell-APC conjugates using a 3D fluorescence microscopy system and found that for some conjugates, even a single pMHC was capable of activating cytokine secretion. Figure 2 shows representative responses of CD4+ T cell blasts to single pMHC molecules. A T cell was found to stop, form a stable synapse with the APC and then secreted both TNF-α (Fig. 2A) and IL-2 (Fig. 2B). Single pMHCs were clearly visible at the T cell-APC interface (Fig. 2, also see 3D Movie S4). These T cell responses were specific to cognate pMHC, as T cells that contacted APCs in areas without labeled pMHC did not produce TNF-α (Fig. 2A) or IL-2 (Fig. S2 E-G), nor were cytokines produced when T cells were incubated with unpulsed APCs (Fig. S2 H-I). One possible problem that might affect the results measured here would be if there were some “dark pMHC complexes”, that is agonist ligands that had somehow lost the biotin tag on the peptide and/or the QD label through protease activity. Therefore, to minimize this, we substituted “D” amino acids that are highly resistant to most proteases (Miller et al., 1995) for all of the linker residues on the peptide beyond the MCC core nine amino acid sequence (Irvine et al., 2002). In addition, if this was insufficient or if some other mechanism besides protease activity could result in “dark pMHCs”, then we should see some T cell-APC conjugates secreting cytokines in the absence of QDs at the synapse. Therefore we examined 156 cytokine-positive T cell-APC conjugates selected at random and in every case there was one or more QD-labeled pMHCs at the synapse. Thus “dark pMHC complexes” are not a factor in these experiments. In summary, these results show that it is feasible to measure the functional sensitivity of CD4+ T cells by combining a single-molecule imaging technique with a single-cell cytokine secretion assay and that a single pMHC is sufficient to trigger cytokine secretion in CD4+ T cell blasts.
Figure 2. A single pMHC stimulates T cell blasts to secrete cytokines.
Shown are the DIC images of T cell-APC conjugates after 2 hours of incubation. TNF-α (A, shown as green) and IL-2 (B, shown as blue) secretion was detected by fluorescent antibody staining in response to a single pMHC presented at the T cell-APC interface as measured by 3D single-molecule fluorescence imaging. 3D reconstructions of the QD705-labeled pMHC fluorescence at the T cell-APC interface were viewed from the top of the T cell-APC contact sites (top view) or from the cell interfaces from the T cell looking at the APC (side view). The fluorescence signal along a line scan drawn through the cellular interface indicates the position of a single pMHC in the 3D reconstruction. The single peak proves the presence of a single pMHC and indicates its location (Xie et al., 2008). The fluorescence image of cytokine secretion (green for TNF-α and blue for IL-2) was overlaid with that of QD705 (red dot) at the focal plane to show the activation of a single T cell by a single pMHC. Scale bar (white), 5 μm. Data are representative of 115 independent measurements (see also Figure S2 and movie S1-S4).
T cell blasts respond similarly to pMHCs labeled with different QDs
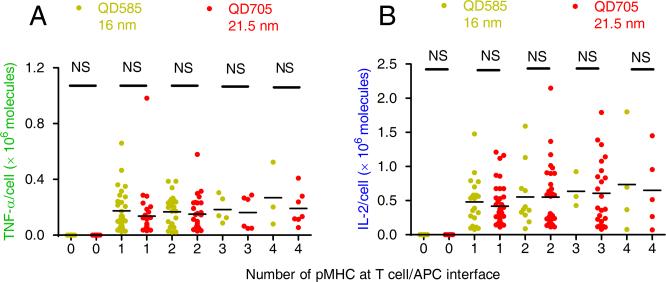
Using this approach, we also quantified TNF-α and IL-2 cytokine production in response to a low number of pMHCs labeled by QD585 or QD705. After a 2-hour incubation, we found that T cells formed conjugates with APCs and ~30% of those T cells secreted cytokines in a typical experiment, while the remaining T cells did not exhibit detectable responses and they either did not encounter pMHCs (Fig. 2A and Fig. S2 E-G, non-responding T cells) or did not respond to the pMHCs at the synapse (Fig. S3A). The amount of cytokines secreted by each responding T cell blast was quantified using standard fluorescent calibration beads (Fig. S3 B-C). The number of pMHCs at the interface of T cell-APC was carefully checked using 3D single-molecule microscopy (Fig. S3D). Figure 3 shows the cytokine response plotted against the number of pMHCs at the T cell-APC interface. T cell blasts secreted cytokines in response to only a few pMHCs, emphasizing the high functional sensitivity of the responding cells. Here we also find that cytokine secretion was not affected by QD size, as shown by the fact that QD585 (a diameter of 16 nm) and QD705 (a diameter of 21.5 nm) labeled pMHCs induced similar cytokine responses (Fig. 3 A-B). This confirmed our previous ELISA measurements in bulk T cells (Fig. 1 C-D) and extended this result to single T cell responses. We concluded that T cell-APC synapses are fluid enough to accommodate these relatively large labels without attenuating T cell sensitivity or responses.
Figure 3. Dose response of cytokine secretion by T cell blasts.
Dose response of TNF-α (A) and IL-2 (B) cytokine secretion by T cell blasts in response to a low number of cognate pMHCs at the T cell-APC interface. pMHCs were labeled with either QD585 (yellow dots) or QD705 (red dots). No statistical differences (NS, t test) were found between QD585 and QD705 labeling for stimulating TNF-α (A) or IL-2 (B) production in response to different number of pMHCs. pMHC number was carefully defined using the single-molecule imaging technique. Cytokine secretion by single T cells was quantified using standard fluorescence beads (see Methods and Fig. S3 B-C). Each dot represents an individual measurement of pMHC number at the T cell-APC interface and the corresponding amount of secreted cytokine. The mean values are indicated by black dashes. Data are representative of 12 independent experiments (see also Figure S3).
Naive and memory T cells also recognize a single pMHC
We next investigated the functional sensitivity of purified naive and memory T cells (Fig. S4 A-D). Based on our preliminary studies, we analyzed the functional sensitivities of these T cells following a 6-hour and a 3-hour incubation respectively. As expected, naive and memory T cells did not secrete cytokine in the absence of pMHC stimulation (Fig. S4E). Most importantly our data show that a single pMHC could activate naive (Fig. 4A) and memory (Fig. 4B) T cells and induce TNF-α and IL-2 secretion. Similar to T cell blasts, we also found that many naive and memory T cells do not respond to pMHCs at the synapse (Fig. S4F) as reported by many investigators (Grakoui et al., 1999; Naramura et al., 1998). The percentage of responding T cells in each category is given by Figure 5 C-D. We then directly compared the cytokine secretion kinetics of responding naive T cells, T cell blasts and memory T cells. Since T cells need time to warm up (from 4°C to 37°C), settle onto a glass surface, engage with APCs, search for antigens and prepare for cytokine secretions, we normalized the cytokine production using their actual secretion times estimated from time-lapse experiments. Here we found that naive T cells produced only ~10% as much cytokine as T cell blasts while memory T cell secretion was comparable to the blast cells. In addition, we found that the production of IL-2 was generally higher than that of TNF-α for all three T cells (Fig. 4 C-D), especially at the longer time points (Fig. S1 I-L). Taken together the above results showed that all three types of T cells measured here have a similar functional sensitivity, but that memory T cells and blasts produce much more cytokine than naive T cells.
Figure 4. A single pMHC stimulates naive and memory T cells to secrete cytokines.
(A and B) Shown are the images of naive (A) and memory (B) T cell-APC conjugates after 6 hours and 3 hours incubation respectively. TNF-α (shown as green) and IL-2 (shown as blue) secretion was detected by fluorescent antibody staining in response to a single pMHC (yellow dot) present at the interface of T cell-APC conjugates measured by 3D single-molecule fluorescence imaging. A single pMHC labeled with QD585 was visualized at the T cell-APC interface at the overlaid images. (C and D) Normalized TNF-α (C) and IL-2 (D) cytokine secretion of naive T cells (cyan dots), T cell blasts (purple dots) and memory T cells (red dots) in response to different number of pMHCs at the T cell-APC interface. Scale bar (white), 5 μm. Each dot represents an individual measurement. The mean values are indicated as black dashes. Statistical significance of the differences between naive T cells and T cell blasts or memory T cells is shown (***, P < 0.0001; **, P < 0.01; *, P < 0.05; t-test) (see also Figure S4).
Figure 5. Digital cytokine secretion of T cells in response to a low number of pMHCs.
(A and B) Naive T cells, T cell blasts, and memory T cells display a digital cytokine secretion pattern for TNF-α (A) and IL-2 (B) in response to a low number of pMHCs. Data in A and B are presented as mean ± SEM of 3 to 30 measurements. (C and D) The percentages of naive T cells, T cell blasts, and memory T cells secreting TNF-α or IL-2 following stimulation by a defined number of pMHCs at the T cell-APC interface are presented. 80 to 269 T cell-APC conjugates were analyzed for each condition. The arrow indicates the trend of percentage increase with increasing pMHC number. pMHCs were labeled with QD585 (see also Figure S5).
All three types of CD4+ T cells display a digital cytokine secretion pattern
Having shown that a single pMHC is able to trigger CD4+ T cell cytokine secretion, we further investigated how pMHC number regulates single T cell responses. By plotting the single-cell dose responses of TNF-α and IL-2 secretion against the pMHC number in the synapse, we found that cytokine secretion displayed a digital pattern for naive T cells, T cell blasts and memory T cells since the quantities of secreted cytokines were independent of the pMHC number, at least within the range of pMHC's (1-4) measured here (Fig. 5 A-B). The digital response was independent of the QD label since we found similar results with QD705 (Fig. S5 A-B). This is consistent with our observation that increasing antigen density alone did not increase cytokine secretion in single responding naive T cells to the amount of single responding T cell blasts or memory T cells. However, we also found that cytokine dose-responses were not digital at the population level (Fig. 1 C-D). To understand this apparent discrepancy, we examined the relationship between antigen number and T cell responding frequency at the single cell level. We found that increasing the pMHC density resulted in a higher percentage of responding T cells (Fig. 5 C-D), and that the percentage was proportional to the pMHC number (Fig. S5 C-I). These results reconcile a digital response of individual T cells with the scalability of cytokine release in the cell population. Thus the immune system calibrates immune response by increasing or decreasing the number of cells that are activated, not by modulating the cytokine output of individual cells.
Naive T cells and T cell blasts secrete cytokines with different kinetics
To better characterize and compare the cytokine secretion between naive T cells and T cell blasts after stimulation, we also used time-lapse microscopy to measure the cytokine secretion kinetics of a T cell stimulated by a single pMHC (Fig. 6, pMHC information is not shown to avoid redundancy). Here we sequentially imaged T cell cytokine secretion every 5 minutes (T cell blasts) or 10 minutes (naive cells) for 3 hours (T cell blasts) or 8 hours (naive cells). For T cell blasts, TNF-α and IL-2 were detected about 40 minutes after T cell-APC conjugate formation (Fig. 6 A-B, top rows). Naive T cells required much longer to start cytokine secretion and TNF-α and IL-2 staining did not begin until about 150 minutes and 250 minutes respectively after cell conjugate formation in representative T cells (Fig. 6 A-B, bottom rows). Since quantities of released cytokines were proportional to the time for both T cell blasts and naive T cells, we fitted the data to a zero-order kinetic model. This model fitted the TNF-α (Fig. 6C) and IL-2 (Fig. 6D) secretion kinetics for both naive and T cell blasts equally well, indicating that the cytokine release rates obtained using this methods were accurate. The rates were 2,200 ± 600 (TNF-α) or 1,200 ± 500 (IL-2) molecules/min for naive T cells and 14,000 ± 2,000 (TNF-α) or 17,000 ± 5,000 (IL-2) molecules/min for T cell blasts. Consistent with our normalized cytokine secretion data at single time points (Fig. 4 C-D), we found that the cytokine release rate of T cell blasts was ~10-fold higher than that of naive T cells (Fig. 6E).
Figure 6. Kinetics of live cell cytokine secretion by T cell blasts and naive T cells in response to stimulation of a single pMHC.
(A and B) Representative real-time cytokine secretions comparing a T cell blast and a naive T cell. The DIC signal was overlaid with cytokine fluorescence signal of TNF-α (A) and IL-2 (B). (C and D) The fluorescence signals were converted into the amounts of secreted cytokines and plotted against the time following the T cell-APC conjugate formation for T cell blasts and naive T cells. Cytokine release rates were obtained by fitting the data with a zero-order kinetics model for T cell blasts and naive T cells, and the goodness of fit were indicated by the R2 values. The slopes represent the rates of cytokine secretion of TNF-α, 18,000 (blast) and 2,400 (naive) molecules/min (C) and of IL-2, 23,000 (blast) and 1,600 (naive) molecules/min (D). (E) Comparison of cytokine release rates between T cell blasts and naive T cells. Data are presented as mean ± SEM of three independent measurements (see also Figure S6 and movie S5-S7).
In parallel experiments, we tracked the diffusion of single QDs or pMHCs in different conditions: QDs covalently bound on a glass surface, QD-pMHC at the synapse either in the absence or in the presence of anti-pMHC blocking antibody (Reay et al., 2000), and QD-pMHC on an APC surface in the absence of T cells (Fig. S6 A-C, and Movie S5-S7). We found that single pMHCs were tightly trapped at the synapse for a prolonged time, while unengaged pMHCs on the APC surface were much more mobile. The diffusion coefficient of single pMHCs engaged with TCRs at the T cell-APC interface was similar to that of QDs immobilized on the glass surface and greatly increased after adding an anti-pMHC blocking antibody (Fig. S6 A-C). Blocking of the TCR-pMHC interaction resulted in reduced T cell-APC adhesion as indicated by the observation that cells began to fall apart (Fig. S6D) as we have observed previously (Huppa et al., 2003). To further investigate the stability of pMHC at the immunological synapse during T cell activation we performed time-lapse experiments to track single pMHCs at the T cell-APC interface while monitoring cytokine secretion by the conjugated T cells. Our data show that pMHC stably resided at the synapse during the T cell-APC interaction and that the continuous presence of a single pMHC was sufficient for cytokine production by individual T cells (Fig. S6E). These observation are consistent with previous findings showing that pMHCs are stable within the central supramolecular activation cluster of the immunological synapse (Grakoui et al., 1999; Monks et al., 1998) and that continuous TCR-pMHC engagement is required for T cell cytokine production (Huppa et al., 2003).
Individual pMHCs trigger the formation of sustained TCR clusters
To characterize the mechanisms by which a few pMHCs are able to trigger the observed T cell digital cytokine release, we investigated TCR recruitment at the immunological synapse during the engagement with single pMHCs. To facilitate the contacts between T cell blasts and APCs, cell mixtures were spun down and incubated for 1, 10, 30, 60 and 120 minutes. Microscopic analysis of T cell-APC conjugates showed that a single pMHC triggered the generation of a post-triggering TCR cluster that surrounded that particular pMHC. These TCR clusters were detectable after a 30-minute incubation (Fig. 7A), though their sizes and patterns were relatively small and heterogeneous. TCR clusters became clearer after a 60-minute incubation (Fig. 7B) and were sustained even after 120-minute incubation (Fig. 7C). Analysis of many TCR clusters co-localizing with single pMHCs at different time points showed that TCR clusters continuously evolved with time and become larger and more stable at longer time points (Fig. 7D). Previous work with lipid bilayers has shown that with high densities of pMHC, TCR microclusters form within seconds, and then migrate quickly toward the center of the synapse (Campi et al., 2005; Dustin and Groves, 2012; Lillemeier et al., 2010; Schamel et al., 2005; Yokosuka et al., 2005). In contrast, here we found that the TCR clusters induced by single pMHCs formed slowly over tens of minutes. Their kinetics of formation, stability and cytoskeleton dependence (Fig. S7) were reminiscent of the post-triggering TCR-CD3 clusters in the cSMAC, especially those formed at low pMHC densities (0.2-0.6 μm-2) (Grakoui et al., 1999; Krummel et al., 2000; Purtic et al., 2005; Varma et al., 2006).
Figure 7. A single pMHC triggers the formation of a sustained post-triggering TCR cluster.
(A-C) Alexa488-H57 Fab-labeled 5C.C7 TCRs (green) interacting with a single pMHC labeled with QD705 (red) on APC surface at 30 (A), 60 (B) and 120 (C) minute incubation times. 3D fluorescence microscopy was used to detect single pMHCs and to visualize TCR cluster formation. The fluorescence images of pMHC were overlaid with those of TCR cluster or DIC images to demonstrate the co-localization between single pMHC and TCR cluster or the localization of pMHC at T cell-APC interface. Representative images of at least 30 cells for each incubation time as shown. (D) Quantification of the TCR number within individual cluster at different time-points. The intensities of individual clusters were measured within a 9 × 9 pixel square (2 μm2) surrounding each single pMHCs. TCR numbers were estimated based on the average value of the intensity of a single Alexa488-H57 Fab. Data are also presented as mean ± SEM (see also Figure S7).
In summary, we have demonstrated here that CD4+ naive T cells, T cell blasts and memory T cells were able to secrete cytokines in response to even one pMHC. Furthermore we found that cytokine production in three stages of T cell maturation was independent of ligand number beyond the first pMHC and thus, they displayed a digital response pattern.
Discussion
The functional sensitivity of individual CD4+ T cells has been difficult to define using standard approaches. Starting with the method developed by Irvine et al. (Irvine et al., 2002), we have devised a single-molecule imaging technique using QD-labeled pMHCs to precisely quantify the functional sensitivity of CD4+ T cells. QDs as fluorescent probes have several attractive characteristics including excellent photo and chemical stability, bright and uniform quantum fluorescence, broad excitation and narrow emission spectra, and nanometer size (Chang et al., 2008; Pinaud et al., 2010). This enabled us to easily identify and visualize single QD-labeled pMHCs on APC surfaces and within the immunological synapses using standard epi-fluorescence microscopy. We found that QDs could effectively label pMHCs on APC surface at a 1:1 ratio and “dark pMHC complexes” (those that lost their QD labels) are not a factor in our analyses. We further demonstrated that the presence of QDs had negligible interference to the stimulatory function and mobility of labeled pMHCs on the APC surface, despite being somewhat larger than the closest sections of the two membranes. Our results are not unexpected because TCRs are able to overcome the steric hindrance of large surface molecules, such as CD45 and LFA-1 (Choudhuri et al., 2005; Springer, 1990), when binding to cognate pMHCs and forming immunological synapses (Freiberg et al., 2002; Grakoui et al., 1999; Johnson et al., 2000; Monks et al., 1998). We also used in situ cytokine-reporter methods to monitor cytokine secretion. While the amount of IL-2 release might be underestimated since not all cytokine molecules are likely to be captured by a cell-bound approach, much of it seems to be, as only the T cells engaged with pMHCs appeared IL-2 positive and not adjacent T cells. Therefore, the QD-labeling technology and cytokine-reporter methods could be broadly useful for studying T cell immunology and related questions in cell biology as well.
Naive T cells, T cell blasts and memory T cells differ in many aspects. Defining the quality and quantity of their responses to an antigenic challenge is an important and incompletely understood subject in T cell biology. Previous cytokine release assays using populations of T cells have shown that IL-2 release is more pronounced in T cell blasts and memory T cells than in naive T cells (Bruniquel and Schwartz, 2003; Kimachi et al., 1997; Sojka et al., 2004). Here we quantified these differences in cytokine secretion at the single-molecule and single-cell level. In contrast to conventional ensemble experiments in which populations of molecules and cells show deterministic and average behaviors, our single-cell measurements captured the inherently stochastic cellular biochemical reactions triggered by single-molecules. Our single-cell analysis showed T cell heterogeneity in response to pMHC stimulation, which is consistent with those found in fluorescence reporter mouse models (Naramura et al., 1998) and in synapse formation analyses (Grakoui et al., 1999). Our data also showed simultaneous IL-2 and TNF-α production at early time points with a time resolution of minutes, versus previous work found sequential cytokine release on an hourly timescale (Han et al., 2012). Unexpectedly, our study showed that naive T cells, T cell blasts and memory T cells were equally sensitive, and that a single pMHC was capable of stimulating cytokine production in all three cells, though the responses of T cell blasts and memory cells were faster and resulted in much more cytokine secretion than naive T cells. Our single-cell results are consistent with previously reported population studies showing that naive T cells and T cell blasts appear to be equally sensitive to peptide loaded APCs but that naive T cells require a 15-fold higher concentration of pMHC to produce the same amount of IL-2 and to proliferate (Kimachi et al., 1997). However they do vary somewhat from comparisons of naive and memory T cells in terms of IL-2 secretion, as we found a 10-fold difference while Croft et al. found the differences varied between 10-fold and nil (Croft et al., 1994). This discrepancy can be explained by the fact that the work cited measured the final cytokine concentration (unit: units/mL) after 24-36 hours of anti-CD3 stimulation, while we measured the cytokine secretion kinetics (unit: molecules/min) within the first tens of minutes stimulated by a few pMHCs (1-4 pMHCs). It has been shown that naive T cells have slower but more sustained secretion kinetics than memory T cells (Sojka et al., 2004) and so this would diminish the differences between naive and memory cells that we observed here. Our single-molecule and single-cell results now provide a definitive answer to the question of the sensitivity of naive T cells, T cell blasts and memory T cells. Though individual naive T cells exhibit delayed activation kinetics and produce lower amounts of cytokines, they have the same extraordinary low activation thresholds of T cell blasts and memory T cells. Since CD4+ T cells do not store cytokines for immediate release (Huse et al., 2006; Moqbel and Coughlin, 2006), these similarities and differences must reflect the intrinsic properties of these cells in their molecular machinery including antigen recognition, signal transduction, chromatin remodeling, gene transcription, and protein translation.
With respect to previous measurements of T cell functional sensitivity, Sykulev et al. first suggested that the 2C CD8+ T cell line only requires one pMHC to kill a target cell (Sykulev et al., 1996) and Purbhoo et al. later showed that the killing of CD8+ T cell blasts (including 2C) requires three or more pMHCs (Purbhoo et al., 2004). Ebert et al. later found that two pMHCs are needed to induce thymocyte death (Ebert et al., 2008). These data suggest that there are different ligand thresholds for different functional responses in different T cells. One modulator of CD4+ T cell sensitivity is miR-181a (Li et al., 2007), which represses the expression of a number of phosphatases in the TCR signaling pathway. To date, all of the work on the functional sensitivity of CD4+ T cells has been at the population level, using peptide titration into populations of T cells (Demotz et al., 1990; Harding and Unanue, 1990). While our results corroborate the relationships previously found among naive T cells, T cell blasts and memory T cells, particularly that of Grey and colleagues (Kimachi et al., 1997), their estimated functional sensitivity of CD4+ T cells to absolute numbers of ligands (50-400 pMHC) is substantially different from our conclusion here that only a single pMHC is necessary. This disparity shows the advantages of combining single-cell imaging with counting individual pMHCs within a synapse and with single-cell readouts of cytokine production. The observation that single pMHC stably resides in the synapse suggests that the size of QD's used here may prevent the movement of labeled pMHCs across the tight synaptic borders. It was previously shown that particles larger than 15 nm are increasingly segregated from the synapse on the basis of their size (Alakoskela et al., 2011). It is tempting to speculate that, QD-labeled pMHCs (16-21.5 nm) can be included in a synapse if they are trapped during initial spreading process, but may be limited in their ability to diffuse in and out afterwards by steric effects.
Furthermore we have shown here that, at least at low antigen densities, a CD4+ T cell did not “count” beyond 0 or 1 in terms of the number of pMHC present at the immunological synapse when deciding whether to secrete cytokines or not, thus displaying a digital secretion pattern. We also found that the density of cognate pMHCs influenced the frequency of productive T cell-APC encounters and the responding probability of encountered T cells, explaining how the T cell response could be upregulated with higher doses of antigens. This view is also compatible with the observation that competition between T cells for pMHCs displayed on the APC surface influences the frequency of IL-2-producing cells in vivo (Sojka et al., 2004). Digital signaling of this type (binary) has been suggested in the expression of particular genes (Walters et al., 1995) and transcription factors (Fiering et al., 1990), and has been mathematically modeled for signal transduction pathways including mitogen-associated protein kinase (Bhalla and Iyengar, 1999), extracellular signal-regulated kinase (Altan-Bonnet and Germain, 2005), Ras (Das et al., 2009), and c-Jun N terminal kinase (Lopez, 2010) signaling. Our data provided the most direct evidence of digital signaling in T cells, and that it is an all-or-nothing phenomenon both at the single-molecule and single-cell level. Thus this is a characteristic of CD4+ T cells from the initial antigen engagement to the final cytokine production. These results shed light on the mechanisms by which CD4+ T cells calibrate their responses to the degree of antigenic stimulation and should aid modeling of these responses.
Our work also has shown that one pMHC present at the immunological synapse triggers the formation of a post-triggering TCR cluster, raising the question of how hundreds of receptors might be clustered by one ligand. We have previously shown that pMHC engagement results in the coupling of TCRs to the actin cytoskeleton followed by their migration into the center of the immunological synapse, while unbound TCRs stay in the periphery of the synapse (Xie et al., 2012). Since in situ TCR-pMHC interactions have very fast binding kinetics (Huang et al., 2010; Huppa et al., 2010), which enable many TCRs to rapidly engage a given immobilized pMHC, these rapid TCR binding events might stabilize the position of the TCR by promoting their association to actin cytoskeleton. Therefore TCR-CD3 complexes may not diffuse away from the synaptic area and could coalesce within clusters. It is tempting to speculate that the formation of TCR clusters in response to individual pMHC might be instrumental for the remarkable sensitivity of T cell responses. Unlike a covalently bound pMHC that enhances its stimulatory potency by delivering continuous stimulatory signals (Xie et al., 2012), a conventional pMHC has only a ~110 ms binding half-life with a TCR (Huppa et al., 2010). Thus a T cell needs to accumulate many short-lived TCR-pMHC encounters to be functionally activated. A particular TCR nanocluster or protein island (Lillemeier et al., 2010) encountering a single cognate pMHC in the close confines of a synapse would facilitate serial engagement and/or multiple rounds of rebinding (Huppa et al., 2010; Valitutti et al., 1995). Furthermore, continuous binding by a subset of TCRs in a synapse to a cognate pMHC may serve to confine these TCR–CD3 complexes within a close-contact zone where they are protected from dephosphorylation by CD45 and CD148. This might result in long-lived phosphorylation of TCR–CD3 immunoreceptor tyrosine-based activation motifs, leading to the recruitment and activation of downstream signaling molecules, consistent with the kinetic segregation model (Choudhuri et al., 2005; Davis and van der Merwe, 2006; James and Vale, 2012; van der Merwe and Dushek, 2010). Along with the involvement of weakly stimulatory endogenous co-agonist pMHCs abundantly presented by the APCs (Krogsgaard et al., 2005), this process could result in the sustained and amplified signaling required for the low threshold and digital cytokine responses in T cells.
In conclusion, our study highlights the importance of conducting quantitative single-molecule and single-cell experiments in order to understand the fundamental processes underlying T cell activation. The results show a remarkable functional sensitivity in CD4+ T cells, a digital response to pMHC agonists, and the gradual formation of post-triggering TCR clusters even when only one or a few ligands are present. This gives us important insights into how individual T cells are employed to generate a scalable response to pathogens.
Experimental Procedures
Mice, cells and peptide
5C.C7 mice were bred and maintained in the Research Animal Facility at Stanford University Department of Comparative Medicine Animal Facility (protocol 3540) in accordance with guidelines of the US National Institutes of Health. 5C.C7 naïve T cells (Fig. S4A and S4C), T cell blasts (Irvine et al., 2002) and memory T cells (Fig. S4B) were respectively prepared. The B-cell lymphoma cell lines CH27 and A20 were used as APCs. Biotin-MCC (88–103) peptide was custom synthesized and purified by Elim Biopharm. The biotin-MCC consisted of a biotin flexible linker and a peptide sequence, biotin–AHX-SGGGSGGGANERADLIAYLKQATK. Underlined residues were synthesized as d-stereoisomers. APCs were pulsed with biotin-MCC peptide and agonist pMHCs on APC surface were fluorescently labeled and measured by flow cytometry. ELISA assays were performed to test the stimulation function of biotin-MCC pulsed APCs. Details on the cells, reagents, flow cytometry and ELISA assays are described in the Supplemental Experimental Procedures.
Imaging analysis of T cell functional sensitivity
APCs were pulsed with biotin-MCC (Irvine et al., 2002) and labeled with 10-20 nM QD585 or QD705. T cells were either pretreated with 1 μM TAPI-0 (for TNF-α) or incubated with 10 μl IL-2 cytokine catch reagent (for IL-2). The APCs and T cells were mixed and incubated for 6 hours (naive T cells), 2 hours (T cell blasts) and 3 hours (memory T cells), respectively. The cytokines at T cell surface were detected by anti-TNF-α allophycocyanin or anti-IL-2 allophycocyanin. The cells were then imaged with a 3D Zeiss microscopy imaging system. For each cell conjugate formed by a T cell and an APC, we acquired a differential interference contrast (DIC) image at the focal plane, a z-stack DIC image, a z-stack allophycocyanin image, and two consecutive z-stacks of QD images. Each z-stack is composed of 21 1-μm or 26 0.8-μm sections visualized across the whole APC (Ebert et al., 2008; Irvine et al., 2002; Purbhoo et al., 2004). The same z-stack DIC and allophycocyanin images were collected for the quantum allophycocyanin calibration beads (Bangs Laboratories) during each T cell sensitivity experiment for fluorescence calibration and cytokine release quantification (Fig. S3 B-C). Details are described in the Supplemental Experimental Procedures.
For live-cell imaging, APCs were attached to a 4-well Lab-Tek chambered coverglass and T cells were pretreated with either TAPI-0 or IL-2 cytokine catch reagent. After adding anti-TNF-α allophycocyanin or anti-IL-2 allophycocyanin, we started the time-lapse imaging at multiple positions and acquired images every 10 minutes for 8 hours (naive T cells) or every 5 minutes for 3 hours (T cell blasts). Details are described in the Supplemental Experimental Procedures.
To detect TCR clusters, pMHCs were labeled with QD705 and TCRs were labeled with anti-TCR Alexa488-H57 Fabs (Campi et al., 2005). APCs and T cells were mixed and incubated in the presence of 10 μg/mL Alexa488-H57 Fabs. In some experiments, 5C.C7 T cell blasts were pretreated with 10 μM cytochalasin D (Campi et al., 2005; Huppa et al., 2010). Cells were fixed and examined using the 3D Zeiss microscopy imaging system. Details are described in the Supplemental Experimental Procedures.
We carefully counted the number of specific pMHCs in the immunological synapse by examining the z-stack of QD images across the APC for two consecutive time points (Fig. S3D). Data were confirmed by 3D reconstruction of the T cell-APC interface using Metamorph software (Molecular Devices) at top view and side view of T cell-APC conjugates (Fig. 2). The intensities of allophycocyanin-labeled antibody staining on T cells and quantum allophycocyanin calibration beads were quantified using a custom-coded Matlab program (MathWorks) and Metamorph software. Details are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Naive, blasting and memory T cells are equally sensitive to antigenic stimulation
A single pMHC triggers CD4+ T cell cytokine secretion
CD4+ T cell cytokine secretion displays a digital signaling pattern
A single pMHC induces the formation of a TCR cluster
Acknowledgements
We thank P. Lund, C. Meyer, H. McGuire and M. Mamedov for reading the manuscript, Y. Xia for help on data analysis, L. Klein, A. Girvin and J. Huppa for training and advice on microscopy, E. Newell and F. Tynan for help on protein purification, F. Wang for sharing reagents, N. Prado for technical support, Y. Wei for helping ELISA experiment, M. Huse for showing the utility of the cytokine capture method, and members of the Davis and Chien laboratories for helpful discussions. This work has been supported by funding from the Howard Hughes Medical Institute (M.M.D.) and the US National Institutes of Health (R01 AI022511 to M.M.D.) and by the Association pour la Recherche sur le Cancer (Grant SL220100601347 to S.V.) and Institut National du Cancer (S.V.). Author contributions: J.H. and M.M.D. designed the research; J.H. developed experimental methods, performed research and analyzed data; M.B. measured diffusion on A20 B cells and provided technical support on microscopy; X.Z. prepared memory T cells and performed ELISA measurements; M.B. and X.Z. contributed equally to this work. Q.L. provided technical training to J.H.; S.V., J.X. and Y.-H.C. contributed important ideas; J.H. wrote the paper; and it was edited by S.V. and M.M.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alakoskela JM, Koner AL, Rudnicka D, Kohler K, Howarth M, Davis DM. Mechanisms for size-dependent protein segregation at immune synapses assessed with molecular rulers. Biophys J. 2011;100:2865–2874. doi: 10.1016/j.bpj.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–387. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. 2005;202:1031–1036. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YP, Pinaud F, Antelman J, Weiss S. Tracking bio-molecules in live cells using quantum dots. J Biophotonics. 2008;1:287–298. doi: 10.1002/jbio.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436:578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- Croft M, Bradley LM, Swain SL. Naive versus memory CD4 T cell response to antigen. Memory cells are less dependent on accessory cell costimulation and can respond to many antigen-presenting cell types including resting B cells. J Immunol. 1994;152:2675–2685. [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. 2006;7:803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- Demotz S, Grey HM, Sette A. The minimal number of class II MHC-antigen complexes needed for T cell activation. Science. 1990;249:1028–1030. doi: 10.1126/science.2118680. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Groves JT. Receptor signaling clusters in the immune synapse. Annu Rev Biophys. 2012;41:543–556. doi: 10.1146/annurev-biophys-042910-155238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- Ebert PJ, Ehrlich LI, Davis MM. Low ligand requirement for deletion and lack of synapses in positive selection enforce the gauntlet of thymic T cell maturation. Immunity. 2008;29:734–745. doi: 10.1016/j.immuni.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- Fomenko V, Nesbitt DJ. Solution control of radiative and nonradiative lifetimes: a novel contribution to quantum dot blinking suppression. Nano Lett. 2008;8:287–293. doi: 10.1021/nl0726609. [DOI] [PubMed] [Google Scholar]
- Freiberg BA, Kupfer H, Maslanik W, Delli J, Kappler J, Zaller DM, Kupfer A. Staging and resetting T cell activation in SMACs. Nat Immunol. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci U S A. 2012;109:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding CV, Unanue ER. Quantitation of antigen-presenting cell MHC class II/peptide complexes necessary for T-cell stimulation. Nature. 1990;346:574–576. doi: 10.1038/346574a0. [DOI] [PubMed] [Google Scholar]
- Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4:749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- Huse M, Klein LO, Girvin AT, Faraj JM, Li QJ, Kuhns MS, Davis MM. Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity. 2007;27:76–88. doi: 10.1016/j.immuni.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat Immunol. 2006;7:247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, Bromley SK, Dustin ML, Thomas ML. A supramolecular basis for CD45 tyrosine phosphatase regulation in sustained T cell activation. Proc Natl Acad Sci U S A. 2000;97:10138–10143. doi: 10.1073/pnas.97.18.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimachi K, Croft M, Grey HM. The minimal number of antigen-major histocompatibility complex class II complexes required for activation of naive and primed T cells. Eur J Immunol. 1997;27:3310–3317. doi: 10.1002/eji.1830271230. [DOI] [PubMed] [Google Scholar]
- Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Agonist/endogenous peptide-MHC heterodimers drive T cell activation and sensitivity. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JM. Digital kinases: A cell model for sensing, integrating and making choices. Commun Integr Biol. 2010;3:146–150. doi: 10.4161/cib.3.2.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz R, Assenmacher M, Pfluger E, Miltenyi S, Radbruch A. Analysis and sorting of live cells according to secreted molecules, relocated to a cell-surface affinity matrix. Proc Natl Acad Sci U S A. 1995;92:1921–1925. doi: 10.1073/pnas.92.6.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Comparison of the Proteolytic Susceptibilities of Homologous L-Amino-Acid, D-Amino-Acid, and N-Substituted Glycine Peptide and Peptoid Oligomers. Drug Development Research. 1995;35:20–32. [Google Scholar]
- Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Moqbel R, Coughlin JJ. Differential secretion of cytokines. Sci STKE. 2006;2006:pe26. doi: 10.1126/stke.3382006pe26. [DOI] [PubMed] [Google Scholar]
- Naramura M, Hu RJ, Gu H. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 1998;9:209–216. doi: 10.1016/s1074-7613(00)80603-2. [DOI] [PubMed] [Google Scholar]
- Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nat Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- Priyadharshini B, Welsh RM, Greiner DL, Gerstein RM, Brehm MA. Maturation-dependent licensing of naive T cells for rapid TNF production. PLoS One. 2010;5:e15038. doi: 10.1371/journal.pone.0015038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- Purtic B, Pitcher LA, van Oers NS, Wulfing C. T cell receptor (TCR) clustering in the immunological synapse integrates TCR and costimulatory signaling in selected T cells. Proc Natl Acad Sci U S A. 2005;102:2904–2909. doi: 10.1073/pnas.0406867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay PA, Matsui K, Haase K, Wulfing C, Chien YH, Davis MM. Determination of the relationship between T cell responsiveness and the number of MHC-peptide complexes using specific monoclonal antibodies. J Immunol. 2000;164:5626–5634. doi: 10.4049/jimmunol.164.11.5626. [DOI] [PubMed] [Google Scholar]
- Ruscetti FW, Morgan DA, Gallo RC. Functional and morphologic characterization of human T cells continuously grown in vitro. J Immunol. 1977;119:131–138. [PubMed] [Google Scholar]
- Schamel WW, Arechaga I, Risueno RM, van Santen HM, Cabezas P, Risco C, Valpuesta JM, Alarcon B. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J Immunol. 2004;172:6136–6143. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- Springer TA. Adhesion receptors of the immune system. Nature. 1990;346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- Ueda H, Morphew MK, McIntosh JR, Davis MM. CD4+ T-cell synapses involve multiple distinct stages. Proc Natl Acad Sci U S A. 2011;108:17099–17104. doi: 10.1073/pnas.1113703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2010;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DI. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci U S A. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Huppa JB, Newell EW, Huang J, Ebert PJ, Li QJ, Davis MM. Photocrosslinkable pMHC monomers stain T cells specifically and cause ligand-bound TCRs to be ‘preferentially’ transported to the cSMAC. Nat Immunol. 2012;13:674–680. doi: 10.1038/ni.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, Dustin ML, Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Tsien RW. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science. 2009;323:1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.