Abstract
Sensitive and quantitative assessment of changes in circulating tumor cells (CTCs) can help in cancer prognosis and in the evaluation of therapeutics efficacy. However, extremely low occurrence of CTCs in the peripheral blood (approximately one CTC per billion blood cells) and potential changes in molecular biomarkers during the process of epithelial to mesenchymal transition (EMT) create technical hurdles to the enrichment and enumeration CTCs. Recently, efforts have been directed toward development of antibody-capture assays based on the expression of the common biomarker - the epithelial cell adhesion molecule (EpCAM) of epithelium-derived cancer cells. Despite some promising results, the assays relying on EpCAM capture have shown inconsistent sensitivity in clinical settings and often fail to detect CTCs in patients with metastatic cancer. We have addressed this problem by the development of an assay based on hybrid magnetic/plasmonic nanocarriers and a microfluidic channel. In this assay cancer cells are specifically targeted by antibody-conjugated magnetic nanocarriers and are separated from normal blood cells by a magnetic force in a microfluidic chamber. Subsequently, immunofluorescence staining is used to differentiate CTCs from normal blood cells. We demonstrated in cell models of colon, breast and skin cancers that this platform can be easily adapted to a variety of biomarkers, targeting both surface receptor molecules and intracellular biomarkers of epithelial-derived cancer cells. Experiments in whole blood showed capture efficiency greater than 90% when two cancer biomarkers are used for cell capture. Thus, the combination of immunotargeted magnetic nanocarriers with microfluidics provides an important platform that can improve the effectiveness of current CTC assays by overcoming the problem of heterogeneity of tumor cells in the circulation.
Keywords: gold shell/magnetic core nanoparticles, circulating tumor cells, immunomagnetic assay, microfluidic chip
Detection of disseminated tumor cells or tumor biomarkers in human fluids such as blood, urine, and saliva can provide an opportunity to develop an accessible tool for cancer detection and monitoring.1–3 In particular, accurate quantitation of cancer cells in the bloodstream can help to determine prognosis and monitor the effectiveness of cancer therapy.4–6 However, the challenge of detecting circulating tumor cells (CTCs) is their rare occurrence, estimated as one to few CTCs among millions of leukocytes and billions of erythrocytes.
Several antibody-based capture assays have been introduced to detect and to count CTCs7–9 and they all rely on one common biomarker - epithelial cell surface marker (EpCAM) - expression on disseminated tumor cells. The single approved system, CellSearch (Veridex LLC), utilizes ferrofluids conjugated with anti-EpCAM antibodies to immunomagnetically enrich CTCs that express EpCAM.7 Microfluidic chambers with anti-EpCAM antibody-coated microposts have also been applied to capture CTCs from whole blood.9 However, due to tumor heterogeneity and epithelial to mesenchymal transition (EMT), subpopulations of metastatic tumor cells often do not express this specific epithelial surface antigen or express it at very low levels,10,11 thereby limiting the value of EpCAM based assays for CTC detection. Thus, EpCAM-dependent assays have limited capability to detect CTCs from “normal-like” subtype of breast cancers which lack such expression.11,12 Furthermore, in a recent prospective multicenter clinical study CTCs were detected in only 61% of metastatic breast cancer patients.4 Consequently, new approaches are required for an effective, highly sensitive, and specific detection of CTCs in whole blood. Our hypothesis is that a versatile platform that can target multiple clinically relevant cancer biomarkers may significantly improve CTC capture and, thus, provide a more accurate determination of the CTCs prevalence in cancer patients.
A key component of our approach to a versatile CTC assay is built around recent progress in development of core-shell nanostructures which have been used in a wide range of applications such as drug delivery, imaging and cellular trafficking.13–16 Notably, bimetallic nanoparticles containing a magnetic core and a plasmonic gold shell enable novel imaging approaches and photothermal therapy.17–19 Furthermore, the gold shell facilitates conjugation of biological molecules to the nanoparticle surface for molecular targeting. Previously, we introduced directional antibody conjugation method through the Fc portion that leaves the antigen binding sites on the Fab moiety available for targeting; this approach improves molecular specificity of the conjugates.20,21 In addition, nanoparticles with relatively small diameters (less than 10 nm) provide a number of unique advantages in molecular targeting such as reducing non-specific interactions, minimizing possible steric hindrance and increasing permeability in a biological environment such as cells and tissues.22,23
Here, we describe a CTC assay that is based on advances in the synthesis of immunotargeted magnetic nanocarriers in combination with a microfluidic device (Fig. 1). The nanocarriers are based on gold shell/iron oxide core nanoparticles conjugated with monoclonal antibodies that are specific to common biomarkers of CTCs. The very thin gold shell of ca. 1nm provides a convenient surface for antibody conjugation and the magnetic core is used for efficient magnetic force separation of the labeled cancer cells from normal cells in whole blood. We demonstrated versatility of the proposed platform for detection and enumeration of rare cells in capturing experiments of phenotypically different cancer cells including breast, colon and skin cancers.
Figure 1.
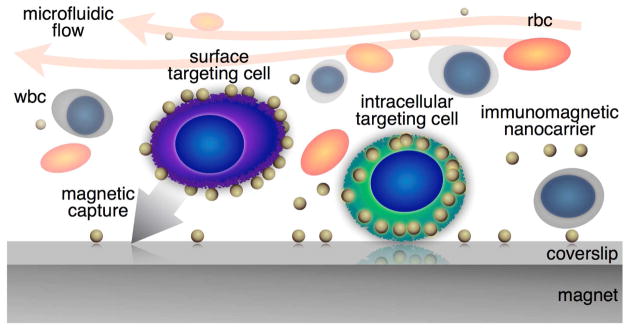
Conceptual cartoon of the versatile immunomagnetic nanocarrier platform in microfluidics for capturing circulating tumor cells in whole blood.
RESULTS AND DISCUSSION
Gold shell/iron oxide core nanoparticles
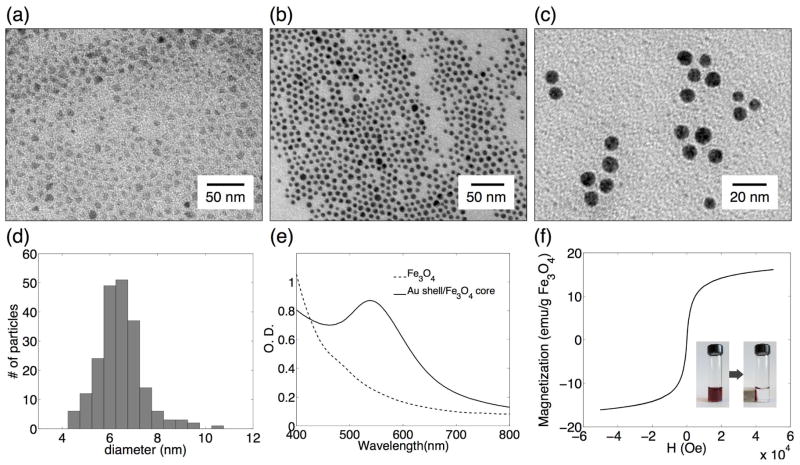
Parameters of an optimal immunomagnetic nanocarrier to detect CTCs in blood include monodispersity, high-stability in aqueous phase, and ease of conjugation with targeting antibodies. In this study, highly uniform core/shell Fe3O4/Au nanoparticles were synthesized via thermal decomposition of iron(III) acetylacetonate in a mixture of oleylamine and oleic acid followed by reduction of gold acetate in the presence of the iron oxide seeds.24 Transmission electron microscopy (TEM) of both Fe3O4 and core/shell Fe3O4/Au nanoparticles dispersed in organic solvent shows spherical, uniform nanocrystals (Fig. 2a, 2b). The core/shell nanoparticles were transferred into aqueous phase by mixing the particles in hexane with alpha-cyclodextrin (α-CD) molecules dissolved in water. α-CD is cyclic oligosaccharides containing six glucopyranose units whose hydrophobic cavities can form complexes with organic molecules and hydroxyl groups on rims provide hydrophilic properties.25 Therefore, the interaction between α-CD and oleic acid on nanoparticle surface stabilizes nanoparticles during phase transfer. The α-CD modified core/shell nanoparticles were readily dispersed in water with no detectable aggregation (Fig. 2c). The core/shell nanoparticles in water phase had a narrow size distribution with the mean diameter of 6.2 ± 0.8 nm that was determined from TEM measurements of more than 200 particles (Fig. 2d).
Figure 2.
Characterization of magnetic core/shell nanocarriers. TEM images of Fe3O4 nanoparticles in hexane before (a) and after (b) coating with gold shell; gold shell/magnetic core nanoparticles after transfer into aqueous phase (c). Gold shell/Fe3O4core nanoparticle size distribution (6.2 ± 0.8 nm) as determined from TEM image analysis of more than 200 particles (d). UV-V is spectrum of oleic acid and oleylamine stabilized Fe3O4 nanoparticles (dashed) and gold shell/magnetic core particles in hexane (solid) (e). Magnetization hysteresis at 300 K of gold shell/magnetic core nanoparticles (f); the insert: separation of nanoparticles from a colloidal suspension using a magnetic field gradient created by a simple permanent magnet.
The uniform gold coating is evident from the darker appearance of the core/shell nanoparticles as compared to the Fe3O4 precursors in TEM images (Fig. 2a and 2b). In addition, the UV-Vis absorption spectrum of Fe3O4/Au core/shell nanoparticles shows a distinctive absorption band at 533 nm that is associated with the surface plasmon resonance of the gold shell (Fig. 2e); this plasmon resonance determines red color of the core/shell nanoparticle suspension. Size comparison of Fe3O4 and Fe3O4/Au core/shell nanoparticles using TEM images showed that the thickness of the gold layer is approximately 1.1 nm. Magnetic properties of the core/shell nanoparticles were characterized using SQUID magnetometry upon cycling the field between −50 K Oe to 50 K Oe at 300 k. The maximum magnetization value is 16.13 emu/g, and neither coercivity nor remanence was observed indicating superparamagnetic property of the nanoparticles (Fig. 2f). The nanoparticles can be quickly separated from a colloidal suspension using a magnetic field gradient created by a simple permanent magnet as can be seen in the insert in Fig. 2f.
Molecular targeting
For molecular specific targeting of cancer biomarkers the core/shell nanoparticles were conjugated with monoclonal antibodies. Monoclonal antibodies are widely utilized probes due to their high binding constants and availability for a large number of established biomarkers.26 Our conjugation strategy relies on directional covalent attachment of antibodies to gold nanoparticles through Fc moiety with the antigen binding sites on the Fab portion directed outward from the gold surface, and therefore available for targeting.20,21 The conjugation is carried out using a heterobifunctional polyethylene glycol (PEG) linker terminated at one end by a hydrazide moiety, and by a di-thiol group at the other end. First, the carbohydrate moiety on the antibody’ Fc region is oxidized to an aldehyde group using sodium periodate; thereby allowing for preferential oxidation of orthodiols through a dehydration reaction. Although tyrosine and serine amino acids contain hydroxyl groups, they are not on nearest neighbor carbons and therefore cannot be oxidized by sodium periodate.27,28 Then, the oxidized antibodies interact with hydrazide-PEG-thiol linkers in a reaction where the hydrazide portion of the linker reacts with the aldehyde formed during the oxidation reaction of antibody carbohydrate to form a stable linkage (Fig. 3). The linker modified antibodies interact with gold surface of core/shell nanoparticles through the linker’s thiol groups. Subsequently, mPEG-thiol molecules are added to passivate any remaining bare gold surfaces thereby increasing the biocompatibility and reducing potential nonspecific interactions (Fig. 3). Attachment of antibodies through the Fc region can be expected to diminish non-specific interactions between nanoparticle conjugates and Fc receptors of blood cells such as macrophages.
Figure 3.
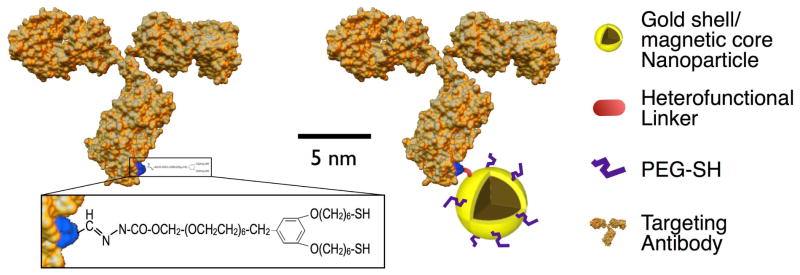
Schematic of an antibody molecule modified using a hetero-functional linker (left) and animmunomagnetic nanocarrier (right).
Molecular specificity of immunomagnetic nanocarriers
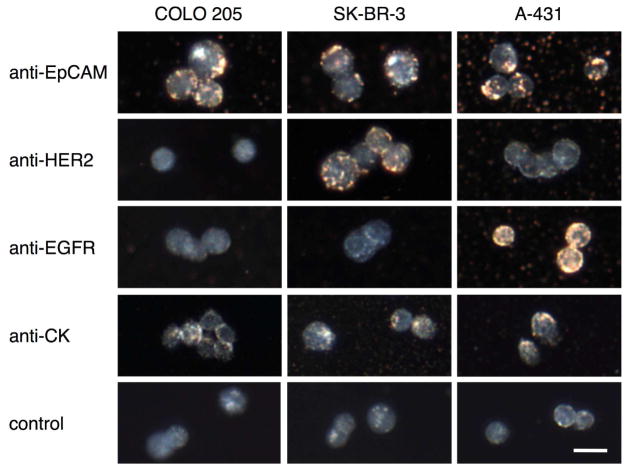
The molecular specificity was demonstrated in three cell lines with known phenotypes: 1) COLO 205 - a model of colorectal cancer which expresses a high level of epithelial cell adhesion molecules (EpCAM), is positive for cytokeratin (CK) expression and is negative for both epidermal growth factor receptor-2 (HER2) and epidermal growth factor receptor-1 (EGFR); 2) SK-BR-3 - a breast cancer model which is EpCAM+/HER2+/EGFR−/CK+; and 3) A-431 - a model of skin cancer with the following expression profile EpCAM+/HER2−/EGFR+/CK+.11,29 Each cancer cell line was labeled with immunomagnetic nanocarriers targeted to either EpCAM, HER2, EGFR, or CK. The specificity of labeling was characterized by comparing the binding of the immunotargeted nanoparticles to cells with varying biomarker expression using dark-field microscopy (Fig. 4). In dark field images, a yellow-orange color indicates specific binding of the nanoparticles to cancer cells whereas a grey-bluish color corresponds to the endogenous scattering of unlabeled cells.30–34 As can be seen in Figure 4 the labeling pattern of cancer cells correlates very well with their known expression profiles demonstrating molecular specificity of the immunomagnetic nanocarriers. Indeed, all cells showed good labeling with anti-EpCAM nanoparticles while only HER2+ SK-BR-3 cells and EGFR+ A-431 cells were labeled with anti-HER2 and anti-EGFR nanoparticles, respectively. Unbound nanocarries can be seen in some of the images; this is due to residual nanoparticles after a washing step. In addition to labeling of cytoplasmic membrane proteins - HER2, EGFR and EpCAM, we also demonstrated successful targeting of cytokeratin that is an intracellular biomarker of epithelial cells (Fig. 4). The intracellular labeling is facilitated by the small size of the immunomagnetic nanoparticles. In addition, the cells were permeabilized using a procedure that is common in immunostaining of intracellular proteins. The ultra-small immunotargeted nanoparticles permitted passage through the permeabilized cell membrane and specific interactions with intracellular molecules. The ability to target a variety of intracellular molecular biomarkers opens new opportunities for the capture of CTCs; since, prevalent and universal biomarkers inside the cells can reduces the variability that results from the heterogeneous levels of surface antigen expression.11,12
Figure 4.
Darkfield reflectance images of cancer cells labeled with immunomagnetic nanocarriers. Columns correspond to cancer cells with the following expression profiles: COLO 205 (EpCAM+/HER2−/EGFR−/CK+), SK-BR-3 (EpCAM+/HER2+/EGFR−/CK+), and A-431 (EpCAM+/HER2−/EGFR+/CK+). Rows show labeling results obtained with (from top to bottom): anti-EpCAM, anti-HER2, anti-EGFR, anti-CK and no nanocarriers. The yellow-orange color in the darkfield images is associated with binding of the nanocarriers; unlabeled cells have a grey-bluish appearance.
Capture efficiency
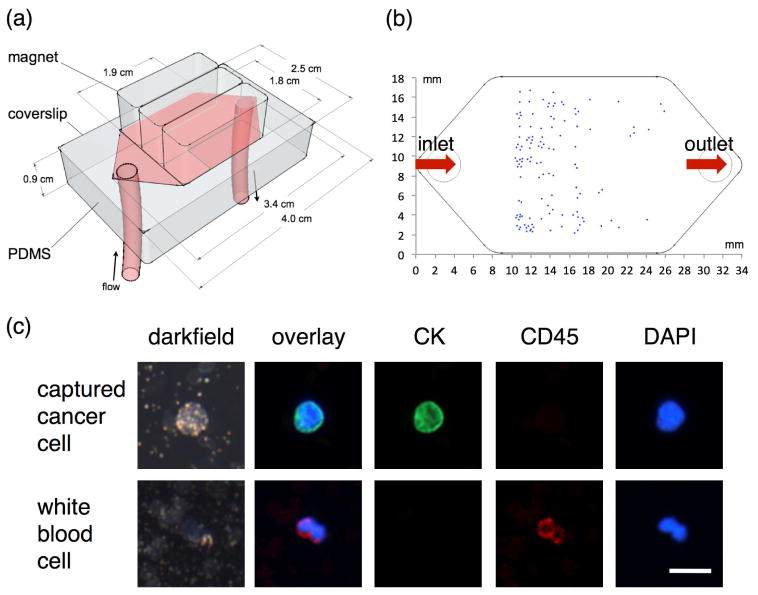
The efficiency of the immunomagnetic nanocarriers for the capture of cancer cells was examined with a microfluidic magnetic chip that we previously developed (Fig. 5a).8 In this device a magnetic field gradient is generated by a permanent magnet that is placed on top of a 20 × 30 × 0.5 mm microfluidic chamber.8 In an example, shown in Figure 5, 2.5 milliliters of whole blood from a normal volunteer was spiked with ~100–200 COLO 205 cells and anti-EpCAM magnetic nanoparticles were added to label the cells. Then, the sample was passed through the microfluidic chamber at a continuous rate of 2.5 ml/hour using a syringe pump. No additional purification or isolation steps were carried out prior to introduction of the blood sample to the chamber. The captured cancer cells were identified using fluorescent staining which allows distinguishing cancer cells with the epithelial tissue phenotype from the much larger population of nucleated white blood cells. The staining scheme that has been widely used in CTCs capture and enumeration experiments includes anti-CK, anti-CD45 and DAPI stains which are specific for epithelial cells, white blood cells and all nucleated cells, respectively.4 Figure 5c shows an example of staining results for a captured cancer cell and a white blood cell where cancer cells can be easily identified by the positive CK and negative CD-45 staining, while white blood cells are CK negative and CD45 positive. Most of the captured cancer cells were found where the first maximal magnetic field gradient exists (Fig. 5b).35 For this design, these capture sites are located around 10 mm away from the inlet of the microfluidic chamber. Cancer cells with less nanoparticle loading travelled a longer path in the microchamber and were captured at regions farther away from the inlet. In this assay coordinates of captured cancer cells can be recorded to facilitate subsequent specific molecular characterization analyses such as fluorescence in situ hybridization (FISH) or hyperspectral microscopic imaging (HMI) which can identify and select a broad spectrum of molecular moieties for better delineation of the true status of the captured cells. No false positive cells were observed in experiments with normal blood without spiked cancer cells.
Figure 5.
Design of a microfluidic channel for immunomagnetic capture, detection and characterization of CTCs (a). An example of distribution of captured COLO 205 cancer cells targeted with anti-EpCAM nanocarriers (b). Fluorescence and darkfield images of a captured COLO 205 cell (DAPI+/CK+/CD45−) and a white blood cell (DAPI+/CK−/CD45+); the cells were labeled using cytokeratin (CK), CD45 and DAPI stains (c).
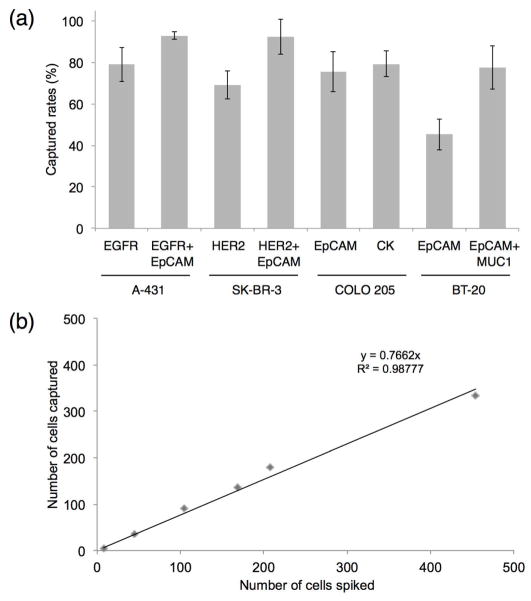
The combination of the microfluidic magnetic chip and the immunomagnetic nanocarriers provides flexibility in capturing rare cancer cells using different extracellular and intracellular biomarkers with high capturing efficiency (Fig. 6 and Table 1). To demonstrate this concept we carried out a series of spiked experiments in the whole blood where A-431 skin cancer cells, SK-BR-3 breast cancer cells and COLO 205 colon cancer cells were captured using cytoplasmic membrane protein targeted nanocarriers - anti-EGFR, anti-HER2 and anti-EpCAM, respectively, according to an expression profile of each cell type (Fig. 6a). In addition, the COLO 205 cells were also captured using nanocarriers targeted to the intracellular biomarker – cytokeratin (CK). Notably, the capture efficiency exhibited by the cytokeratin targeting nanocarrier is the same as the efficiencies of the nanocarriers targeting cell surface proteins. The capture yield was 70–80% in cases where a single nanocarrier was used that is comparable to the FDA-approved CellSearch system and the recently described microchip system with antibody-coated microposts.7,9
Figure 6.
Cancer cell capture and enumeration. (a) Capture efficiency in spike experiments in 2.5 ml whole blood samples from a normal volunteer where COLO 205 (colon), SK-BR-3 (breast), A-431 (skin) or BT-20 (breast) cells were captured using immunomagnetic nanocarriers targeted to cancer biomarkers which are listed in the parentheses; each experiment was repeated at least 3 times. Note a significant increase in the capture efficiency when a combination of nanocarriers is used for detection of A431, SK-BR-3 and BT-20 cells; this increase is especially pronounced in the case of a low EpCAM expressing BT-20 cells. (b) Number of captured cells as a function of spiked COLO 205 cells in 2.5 ml whole blood samples; the number of spiked cells was varied from 5 to 500 cells.
Table 1.
Capture efficiency in spike experiments in whole blood samples from a normal volunteer.
| Nanocarriers against antibody | Cell line | Capture yield (%) |
|---|---|---|
| EGFRa | A-431 (skin) | 79 ± 8 |
| EGFR+EpCAMb | A-431 | 93 ± 2 |
| HER2c | SK-BR-3 (breast) | 69 ± 7 |
| HER2+EpCAM | SK-BR-3 | 93 ± 10 |
| EpCAM | COLO 205 (colon) | 75 ± 9 |
| CKd | COLO 205 | 79 ± 6 |
| EpCAM | BT-20 (breast) | 45 ± 8 |
| EpCAM+MUC1e | BT-20 | 78 ± 10 |
anti-Epidermal Growth Factor Receptor 1,
anti-Epithelial Cell Adhesion Molecule,
anti-Epidermal Growth Factor Receptor 2,
anti-Cytokeratin, and
anti-Mucin 1.
However, our approach to CTC assay allows straightforward multiplexing of immunotargeted nanocarriers to various cancer cell antigens, thereby increasing CTC detection.36 For example, using anti-EGFR and anti-EpCAM nanocarriers simultaneously for detection of A-431 cells increases the capture yield from 79% in the case of anti-EGFR nanoparticles alone to 93% for the combination, similarly combining anti-HER2 and anti-EpCAM nanoparticles improves the capture yield of SK-BR-3 cells from 69% obtained in the case of HER2 targeting alone to 93% for the combination (Figure 6a and Table 1).
We also explored the concept of using multiple nanocarriers to improve capture in cancers which express EpCAM antigen poorly or not at all. First, we manually reproduced CellSearch capture assay in spiked experiments in whole blood using the basal-like subtype of breast cancer cell line, BT-20, which expresses relative low EpCAM; the assay showed the capture efficiency of only ca. 44%. The same recovery rate was obtained using our anti-EpCAM immunomagnetic nanocarriers (Fig. 6a). However, combining anti-EpCAM and anti-MUC1 nanocarriers together improved the capture efficiency from 45% to 78%. The nanocarriers in the conmbination were applied in 1:1 ratio and the concentration of each nanoparticle was the same as in experiments with a single nanocarrier.
Thus, utilizing multiple nanocarriers opens the route to significantly improve capture of CTCs with various expression profiles including capture of cancer cells that express EpCAM weakly on their surface.4 We also demonstrated that cell capture efficiency exhibits a linear behavior (the R2 value for the linear regression fit is 0.98777) in spiking experiments with a number of COLO 205 colon cancer cells ranging from 5 to 500 in whole blood (Fig. 6b). For the group with only 5 cells, the capture yield was 86%, whereas an average ca. 77% capture yield was obtained from the linear graph shown on Fig. 6b.
In addition to the multiplexing capabilities demonstrated above the advantages of our platform include the use of whole blood in CTC detection that eliminates multiple pre-processing steps including plasma replacement, centrifugation and sample transfer between tubes which are commonly used in other assays such as CellSearch. We also demonstrated the possibility of fluorescent staining inside the microfluidic chamber following cell capture. This in-channel procedure provides an efficient washing and fluorescent labeling due to short diffusion distances thus saving the amount of fluorescent reagents used, improving uniformity of staining and eliminating potential loss of captured cells. The CTC observation can be easily automated and individual cells can be analyzed using HMI to explore a large number of molecular markers for better delineation of the status of the captured cells.
CONCLUSION
Thus, we have developed versatile immunomagnetic nanocarriers for labeling of rare cancer cells in the whole blood. The combination of nanocarriers and a magnetic microfluidic chip allows highly efficient capture, enumeration and molecular characterization of CTCs. This platform provides flexibility in capturing phenotypically different CTCs by using nanocarriers that are targeted to different molecular tumor biomarkers; this can significantly improve the effectiveness of CTC assays. Furthermore, the use of small targeted nanoparticles allows a straightforward extension to multiplexed approaches where capture of cancer cells is carried out by a mixture of nanoparticles with different target specificities. Our platform allows a flexible approach to improve capture of CTC and to delineate some of the many reasons used to explain poor capture to date. This approach provides the basis for the development of a low cost simple CTC assay.
MATERIALS AND METHODS
Synthesis and characterization of gold shell/magnetic core nanoparticles
The gold-coated iron oxide nanoparticles were prepared according to a previously published protocol with a number of modifications.24 Briefly, 1 mM iron(III) acetylacetonate was mixed in 10 ml phenyl ether, followed by addition of 3 mM oleic acid, 2 mM oleylamine, and 3 mM 1,2-hexadecanediol. The mixture was stirred vigorously, heated to 260 °C, and refluxed for 1 h yielding a suspension of highly uniform 5.1 nm Fe3O4 nanoparticles. Five ml of the Fe3O4 nanoparticle suspension was cooled to room temperature followed by addition of 1.1 mM gold acetate, 0.75 mM oleic acid, 3 mM oleylamine, 3 mM 1,2-hexadecanediol, and 15 ml phenyl ether under vigorous stirring. The reaction mixture was heated to 180 °C and was refluxed for 1 h. Then, a dark purple precipitate was formed after addition of ethanol and centrifugation. The recovered gold-coated iron oxide nanoparticles were dispersed in hexane.
To render gold-coated iron oxide nanoparticles hydrophilic, the transfer of the nanoparticles from organic to aqueous phase was modified as previously described.25 Nanoparticles in hexane at ca. 0.6 mg nanoparticle/ml and equal volume of 5 mM α-cyclodextrin (α-CD) were mixed and stirred overnight at room temperature. Then, the nanoparticles were recovered by centrifugation, and the supernatant was discarded. The precipitate was resuspended in 0.2 mM sodium citrate using 10 min sonication.
Transmission election microscopy (TEM) images were obtained using a FEI TECNAI G2 F20 X-TWIN TEM at 80 keV. The samples were prepared by depositing 10 μl of a nanoparticle suspension onto a carbon-coated copper TEM grid for observation. UV-V is spectra were collected with a BioTek Synergy HT micro-titer plate spectrometer.
Conjugation of antibodies to nanoparticles
Anti-Epithelial Cell Adhesion Molecule (EpCAM), anti-Epidermal Growth Factor Receptor 2 (HER2), anti-Epidermal Growth Factor Receptor 1 (EGFR), anti-Mucin 1 (MUC1), and anti-Cytokeratin (CK) antibodies (Sigma Aldrich Co., St. Louis, MO) were attached to the gold shell/magnetic core nanoparticles via a heterofunctional PEG linker with hydrazide and dithiol moieties - dithiolaromatic PEG6-CONHNH2 (Nanoscience Instruments Inc., Phoenix, AZ). Antibody solution (100 μl, 1 mg/ml) in 4 mM HEPES was incubated in the dark with 10 μl 100 mM NaIO4 for 30 min at room temperature, followed by quenching the reaction with 500 μl phosphate buffered saline (PBS). Then, 2 μl of 46.5 mM linker solution was added to the antibody solution and shaken gently for 1 h. The excess linker was removed by filtration in a 10,000 MWCO centrifuge filter (Millipore Inc., Bedford, MA) at 2,000g, 8 °C for 16 min. The retained antibodies were resuspended in PBS to a concentration of 1 mg/ml. The modified antibodies were mixed with gold shell/magnetic core nanoparticles in 4 mM HEPES for 1 h at room temperature. Then, 10−5 M 10 kD PEG-thiol (SensoPath Technologies, Inc., Bozeman, MT) was added to passivate the remaining nanoparticles surface. The functionalized nanoparticles were recovered by centrifugation at 2,000g for 5 min and were resuspended in 2 % w/v 10 kD PEG in PBS.
Labeling Specificity Assays
COLO 205, SK-BR-3, and A-431 cells (ATCC, Manassas, VA) were used as cancer cell models to demonstrate molecular specific cellular imaging. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, NY) supplemented with 5 % fetal bovine serum (FBS, Hyclone, Logan, UT), and harvested at ~90% confluence with trypsin. Cell suspensions containing ~3×105 cells were resuspended in complete media. Then, approximately 6×1012 immunotargeted nanocarriers conjugated with either anti-HER2, anti-EGFR, or anti-EpCAM antibodies were added to a cell suspension for 2 h at room temperature under mild mixing. To target the intracellular cytokeratins cells were fixed with 4% formaldehyde for 10 min followed by permeabilized with 1% Triton X-100 (Sigma Aldrich Co., St. Louis, MO) for 15 min before incubating with immunotargeted nanocarriers against anti-CK. After incubation with nanocarriers, cells were washed in phosphate buffered saline and were spun down to remove any unbound nanoparticles followed by imaging using 20×, 0.5-NA darkfield objective under Leica DM6000 upright microscope.
Microchannel design
To screen blood samples we used polydimethylsiloxane (PDMS)-based microchannel combined with magnetic field gradient generated by arrayed magnets with alternate polarities.35 The microchannel with the height of 500 μm was fabricated through soft lithography using PDMS (Sylgard 184, Dow Corning, Midland, MI, 10:1 prepolymer to curing agent), with subsequent steps of bonding the PDMS channel with a glass coverslip (24 × 40 × 0.15 mm, Fisher Scientific Co., Pittsburgh, PA).8 Dimensions of the microchannel are shown in Figure 5a. The inlet of the microchannel was connected to a reservoir for sample loading and the outlet was connected to a syringe pump (Harvard Apparatus, South Natick, MA) to control flow rates.
Screening procedures and analysis of whole blood samples
Blood samples from a healthy donor were spiked with a known number of cancer cells to determine sensitivity of the immunomagnetic nanocarrier platform. Whole blood samples were collected with CellSave tubes (Veridex LLC, Raritan, NJ). Three cell lines - COLO 205, SK-BR-3, and A-431 - with known phenotypes were harvested, centrifuged, and resuspended in phosphate buffered saline. Ten μl cell suspension at a concentration of approximately 20,000 cells per ml was added to a conical CellSave tube containing 2.5 ml of whole blood. The same amount of cell suspension was distributed on three glass slides to calculate the mean of cells spiked into the blood sample. Then, a suspension of functionalized nanocarriers (100 μl, 50 nM) conjugated with either anti-EpCAM, anti-HER2, or anti-EGFR antibodies was added to the blood samples spiked with COLO 205, SK-BR-3, and A-431 cells, respectively. In addition, a combination of anti-EGFR and anti-EpCAM nanoparticles was used in capture experiments with A-431 cells and a combination of anti-HER2 and anti-EpCAM nanoparticles was applied for detection of SK-BR-3 cells. Each nanocarrier in the combinations was administered with equal volume (100 μl) and concentration (50 nM). BT-20 (ATCC, Manassas, VA) breast cancer cell line was used as a model of cells with a low EpCAM expression; this cell line was labeled with both anti-EpCAM nanocarriers and a combination of anti-EpCAM and anti-MUC1 nanocarriers.
The labeling with anti-CK nanoparticles targeting the intracellular cytokeratin biomarkers required a cell fixation/permeabilization step before addition of the nanoparticles. The whole blood samples containing cancer cells were incubated with 4% formaldehyde for 10 min at room temperature. Then, 1% Triton X-100 was added to the solution for 15 min followed by two washing steps in phosphate buffered saline.
The whole blood samples with spiked cancer cells and immunotageted nanocarriers were incubated for 2 h under gentle shaking. Then, cancer cells were separated from the whole blood in the microfluidic chip described above that was operating at a continuous flow rate of 2.5 ml per hour. After the separation step, the microchannel was flushed with 3 to 4 ml of phosphate buffered saline to wash blood cells. Subsequently, 1 ml of ice-cold acetone was administered to the microchannel to fix captured cancer cells. The slides were stained using nuclear dye 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories Inc., Peterborough, UK), anti-cytokeratin pan-FITC (Sigma Aldrich Co., St. Louis, MO), and the anti-CD45 antibodies labeled with Alexa Fluor 568 (Invitrogen, Carlsbad, CA). Captured cancer cells were defined as DAPI+/CK+/CD45−, and white blood cells were classified as DAPI+/CK−/CD45+. The capture yield was calculated by dividing the number of cells found in the sample by the mean number of spiked cells.
To determine dependence of capture efficiency on the number of cancer cells in whole blood, we conducted a series of experiments with a number of spiked COLO 205 cells ranging from 5 to 500 in 2.5 ml of whole blood. Anti-EpCAM nanocarriers were used for cell capture and the experiments were carried out as described above.
Acknowledgments
This work was supported by the National Institute of Health (NIH) National Cancer Institute (NCI) grants 1R01CA139070 and R01CA103830.
References
- 1.Meng S, Tripathy D, Shete S, Ashfaq R, Saboorian H, Haley B, Frenkel E, Euhus D, Leitch M, Osborne C, et al. uPAR and HER-2 Gene Status in Individual Breast Cancer Cells from Blood and Tissues. Proc Natl Acad Sci US A. 2006;103:17361–17365. doi: 10.1073/pnas.0608113103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, Chia D, Wong DT. Salivary Transcriptomic Biomarkers for Detection of Resectable Pancreatic Cancer. Gastroenterology. 2010;138:949–957.e7. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, et al. A First-Generation Multiplex Biomarker Analysis of Urine for the Early Detection of Prostate Cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, et al. Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 6.Coumans FA, Doggen CJ, Attard G, de Bono JS, Terstappen LW. All Circulating EpCAM+CK+CD45- Objects Predict Overall Survival in Castration-Resistant Prostate Cancer. Ann Oncol. 2010;21:1851–1857. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 7.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the Cellsearch System. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X. Microchip-Based Immunomagnetic Detection of Circulating Tumor Cells. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem Cell and Epithelial-Mesenchymal Transition Markers Are Frequently Overexpressed in Circulating Tumor Cells of Metastatic Breast Cancer Patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-Epithelial Cell Adhesion Molecule Antibodies and the Detection of Circulating Normal-Like Breast Tumor Cells. J Natl Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostert B, Kraan J, Bolt-de Vries J, van der Spoel P, Sieuwerts AM, Schutte M, Timmermans AM, Foekens R, Martens JW, Gratama JW, et al. Detection of Circulating Tumor Cells in Breast Cancer May Improve through Enrichment with Anti-CD146. Breast Cancer Res Treat. 2011;127:33–41. doi: 10.1007/s10549-010-0879-y. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Lee E, Kim do K, Jang NK, Jeong YY, Jon S. Antibiofouling Polymer-Coated Superparamagnetic Iron Oxide Nanoparticles as Potential Magnetic Resonance Contrast Agents for in vivo Cancer Imaging. J Am Chem Soc. 2006;128:7383–7389. doi: 10.1021/ja061529k. [DOI] [PubMed] [Google Scholar]
- 14.Farokhzad OC, Langer R. Impact of Nanotechnology on Drug Delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 15.Barcikowski S, Compagnini G. Advanced Nanoparticle Generation and Excitation by Lasers in Liquids. Phys Chem Chem Phys. 2013;15:3022–3026. doi: 10.1039/c2cp90132c. [DOI] [PubMed] [Google Scholar]
- 16.Bigall NC, Parak WJ, Dorfs D. Fluorescent, Magnetic and Plasmonic-Hybrid Multifunctional Colloidal Nano Objects. Nano Today. 2012;7:282–296. [Google Scholar]
- 17.Larson TA, Bankson J, Aaron J, Sokolov K. Hybrid Plasmonic Magnetic Nanoparticles as Molecular Specific Agents for MRI/Optical Imaging and Photothermal Therapy of Cancer Cells. Nanotechnology. 2007;18:325101. [Google Scholar]
- 18.Song HM, Wei Q, Ong QK, Wei A. Plasmon-Resonant Nanoparticles and Nanostars with Magnetic Cores: Synthesis and Magnetomotive Imaging. ACS Nano. 2010;4:5163–5173. doi: 10.1021/nn101202h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Q, Song HM, Leonov AP, Hale JA, Oh D, Ong QK, Ritchie K, Wei A. Gyromagnetic Imaging: Dynamic Optical Contrast Using Gold Nanostars with Magnetic Cores. J Am Chem Soc. 2009;131:9728–9734. doi: 10.1021/ja901562j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Aaron J, Sokolov KV. Directional Conjugation of Antibodies to Nanoparticles for Synthesis of Multiplexed Optical Contrast Agents with Both Delivery and Targeting Moieties. Nat Protoc. 2008;3:314–320. doi: 10.1038/nprot.2008.1. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Harrison N, Richards-Kortum R, Sokolov K. Plasmonic Nanosensors for Imaging Intracellular Biomarkers in Live Cells. Nano Lett. 2007;7:1338–1343. doi: 10.1021/nl070365i. [DOI] [PubMed] [Google Scholar]
- 22.Huang K, Ma H, Liu J, Huo S, Kumar A, Wei T, Zhang X, Jin S, Gan Y, Wang PC, et al. Size-Dependent Localization and Penetration of Ultrasmall Gold Nanoparticles in Cancer Cells, Multicellular Spheroids, and Tumors in vivo. ACS Nano. 2012;6:4483–4493. doi: 10.1021/nn301282m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with Plasmonic Nanosensors. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Luo J, Fan Q, Suzuki M, Suzuki IS, Engelhard MH, Lin Y, Kim N, Wang JQ, Zhong CJ. Monodispersed Core-Shell Fe3O4@Au Nanoparticles. J Phys Chem B. 2005;109:21593–21601. doi: 10.1021/jp0543429. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wong JF, Teng X, Lin XZ, Yang H. “Pulling” Nanoparticles into Water: Phase Transfer of Oleic Acid Stabilized Monodisperse Nanoparticles into Aqueous Solutions of α-Cyclodextrin. Nano Lett. 2003;3:1555–1559. [Google Scholar]
- 26.Devriese LA, Voest EE, Beijnen JH, Schellens JH. Circulating Tumor Cells as Pharmacodynamic Biomarker in Early Clinical Oncological Trials. Cancer Treat Rev. 2011;37:579–589. doi: 10.1016/j.ctrv.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Hermanson GT. Bioconjugate Techniques. 2. Academic Press; Waltham, Massachusetts: 2008. pp. 761–764. [Google Scholar]
- 28.Bobbitt JM. Periodate Oxidation of Carbohydrates. Adv Carbohydr Chem. 1956;11:1–41. doi: 10.1016/s0096-5332(08)60115-0. [DOI] [PubMed] [Google Scholar]
- 29.Rae JM, Scheys JO, Clark KM, Chadwick RB, Kiefer MC, Lippman ME. EGFR and EGFRvIII Expression in Primary Breast Cancer and Cell Lines. Breast Cancer Res Treat. 2004;87:87–95. doi: 10.1023/B:BREA.0000041585.26734.f9. [DOI] [PubMed] [Google Scholar]
- 30.Aaron J, Nitin N, Travis K, Kumar S, Collier T, Park SY, Jose-Yacaman M, Coghlan L, Follen M, Richards-Kortum R, et al. Plasmon Resonance Coupling of Metal Nanoparticles for Molecular Imaging of Carcinogenesis in vivo. J Biomed Opt. 2007;12:034007. doi: 10.1117/1.2737351. [DOI] [PubMed] [Google Scholar]
- 31.Aaron J, Travis K, Harrison N, Sokolov K. Dynamic Imaging of Molecular Assemblies in Live Cells Based on Nanoparticle Plasmon Resonance Coupling. Nano Lett. 2009;9:3612–3618. doi: 10.1021/nl9018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Boriskina SV, Wang H, Reinhard BM. Illuminating Epidermal Growth Factor Receptor Densities on Filopodia through Plasmon Coupling. ACS Nano. 2011;5:6619–6628. doi: 10.1021/nn202055b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Yu X, Boriskina SV, Reinhard BM. Quantification of Differential ErbB1 and ErbB2 Cell Surface Expression and Spatial Nanoclustering through Plasmon Coupling. Nano Lett. 2012;12:3231–3237. doi: 10.1021/nl3012227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crow MJ, Seekell K, Ostrander JH, Wax A. Monitoring of Receptor Dimerization Using Plasmonic Coupling of Gold Nanoparticles. ACS Nano. 2011;5:8532–8540. doi: 10.1021/nn201451c. [DOI] [PubMed] [Google Scholar]
- 35.Hoshino K, Chen P, Huang YY, Zhang X. Computational Analysis of Microfluidic Immunomagnetic Rare Cell Separation from a Particulate Blood Flow. Anal Chem. 2012;84:4292–4299. doi: 10.1021/ac2032386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, et al. Molecular Biomarker Analyses Using Circulating Tumor Cells. PloS one. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]