Abstract
Synergistic actions between all-trans-retinoic acid (atRA) and interferon γ (IFNγ) on modulation of cellular functions have been reported both in vitro and in vivo. However, the mechanism of atRA-mediated regulation of IFNγ signaling is poorly understood. In this study, we have used the human lung epithelial cell line A549 to examine the effect of atRA on IFNγ-induced expression of IFN regulatory factor-1 (IRF-1), an important transcription factor involved in cell growth and apoptosis, differentiation, and antiviral and antibacterial immune responses. At least 4 h of pretreatment with atRA followed by suboptimal concentrations of IFNγ induced a faster, higher, and more stable expression of IRF-1 than IFNγ alone. Actinomycin D completely blocked the induction of IRF-1 by the combination, suggesting regulation at the transcriptional level. Further, we found that activation of signal transducer and activator of transcription-1 was induced more dramatically by atRA and IFNγ than by IFNγ alone. Expression of IFNγ receptor-1 on the cell surface was also increased upon atRA pretreatment. Experiments using receptor-selective retinoids revealed that ligands for retinoic acid receptor-α (RARα), including atRA, 9-cis-retinoic acid, and Am580, sequentially increased the levels of IFNγ receptor-1, activated signal transducer and activator of transcription-1, and IRF-1 and that an RARα antagonist was able to inhibit the effects of atRA and Am580. In addition, atRA pretreatment affected the transcriptional functions of IFNγ-induced IRF-1, increasing its nuclear localization and DNA binding activity as well as the transcript levels of IRF-1 target genes. These results suggest that atRA, an RARα ligand, regulates IFNγ-induced IRF-1 by affecting multiple components of the IFNγ signaling pathway, from the plasma membrane to the nuclear transcription factors.
Both vitamin A (1,2) and interferons (3) have long been recognized as potent regulators of antibacterial and antiviral immune responses. All-trans-retinoic acid (atRA),2 an active metabolite of vitamin A, acts through retinoic acid receptors (RARs) to transcriptionally activate target genes (4). Interferons (IFNs), on the other hand, signal through interferon signaling pathways and use signal transducer and activator of transcription (STAT) proteins as transcription factors (5). Recently, more attention has been drawn to potential interactions between these two pathways, since synergistic effects of atRA and IFNs have been observed in both animal models (6, 7) and cultured cell lines (8–10). However, the molecular events involved in atRA-dependent regulation of IFNγ signaling or vice versa are poorly understood.
IFN regulatory factor-1 (IRF-1) was discovered in studies of virus-induced IFNα/β gene regulation and IFN-mediated antiviral responses (11). IFNγ is one of the strongest inducers of IRF-1. Upon binding of IFNγ to its receptor, IRF-1 is induced through activation of STAT-1 and binding of activated STAT-1 to the γ-interferon-activated site within the IRF-1 promoter. IRF-1 protein, a transcription factor itself, has a very short half-life and is degraded via the ubiquitin-proteasome pathway (12). IRF-1 is known to regulate cell growth and apoptosis (13,14). The tumor suppressor function of IRF-1 was first demonstrated in oncogenic transformation of primary IRF-1−/− mouse embryonic fibroblasts (15). Recently, it was also shown that IRF-1 reversed the transformed phenotype of tumor cells not only in vitro but also in vivo (16).
RA synergizes with IFNγ to increase the level of IRF-1, which subsequently activates promoters of IRF-1 target genes, such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (17). Caspase-1 (previously termed interleukin (IL)-1β-converting enzyme, or ICE) is another target gene of IRF-1 (18), which plays a critical role in the production of proinflammatory cytokines IL-1α, IL-1β, IL-18, and IFNγ (19). However, the molecular basis of IRF-1 regulation by the combination is not fully understood. atRA by itself activates IRF-1 gene expression in NB4 promyelocytic leukemia cells (20). It induces IRF-1 through a γ-interferon-activated site motif, but not a putative retinoic acid response element, of the IRF-1 promoter (21), indicating a role of STAT-1 between atRA and IRF-1. However, IRF-1 induction by atRA in squamous carcinoma cells is STAT-1-independent (22). In cervical squamous carcinoma SiHa cells, STAT-1 dependence was observed only when high doses of atRA (1 μm and above) were used (23). Collectively, it is still unclear how atRA regulates IRF-1 transcription and whether STAT-1 is involved. RARs, common regulators of atRA functions, may also play important roles in the regulation of IRF-1.
In this study, we hypothesized that atRA increased IFNγ-induced expression and functions of IRF-1 through, at least in part, its regulation of the key molecules of IFNγ signaling pathways. Using a human lung epithelial cell line, A549, we have observed that overnight pretreatment with atRA increases the levels of IFNγ receptor (IFNGR)-1 on the cell surface, thereby enhancing tyrosine phosphorylation (activation) of STAT-1 upon low-dose IFNγ stimulation. Faster, higher, and more stable levels of IRF-1 are then induced by the combination of atRA and IFNγ compared with IFNγ alone. RARα mediates the effect of atRA in increasing cell surface IFNGR-1, activated STAT-1, and IRF-1. atRA pretreatment also potentiates the transcriptional activity of IFNγ-induced IRF-1, increasing its nuclear localization and DNA binding activity. As anticipated, IFNγ-induced caspase-1 and TRAIL are further increased by pretreatment with atRA. Collectively, these results indicate that atRA regulates multiple components of the IFNγ IRF-1 pathway, from the plasma membrane to the nuclear transcription factors.
Experimental Procedures
Reagents, Antibodies, and Cell Culture
atRA (prepared in ethanol), 9-cis-retinoic acid (9cRA), all-trans-retinol, retinyl trimethoxybenzyl ether (RTMBE), actinomycin D, and cycloheximide were obtained from Sigma. Recombinant human IFNγ was obtained from PreproTech Inc. (Rocky Hill, NJ). Receptor-selective retinoids were provided by Michael Klaus (Hoffmann-La Roche, Nutley, NJ). They include Am580 (RARα agonist), Ro19-0645 (RARβ agonist), CD437 (RARγ agonist), Ro25-7386 (RXR panagonist), and Ro41-5253 (RARα antagonist). 4′,6′-Diamidino-2-phenylindole was obtained from Molecular Probes, Inc. (Eugene, OR). IRF-1 polyclonal antibody, IFNGR-1 monoclonal antibody, and consensus IRF-1 gel shift oligonucleotides were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). IRF-1 and STAT-1 monoclonal antibodies were obtained from Transduction Laboratories (Lexington, KY). Polyclonal antibody against phospho-STAT-1 at residue Tyr-701 was obtained from Cell Signaling Technology (Beverly, MA). A549 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in F-12K medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum at 37 °C in a 5% CO2-air incubator. In most experiments, the cells were plated at ∼70% confluence, allowed to attach in complete medium, and adjusted to low serum medium (supplemented with 1% fetal bovine serum) for 2 h before the addition of stimuli.
Preparation of Whole Cell and Nuclear Extracts
A549 cells were lysed in radioimmune precipitation buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS in phosphate-buffered saline (PBS)) containing 10% (v/v) of protease inhibitor mixture (Roche Applied Science) and 1 mM sodium orthovanadate as phosphatase inhibitor (24). Whole cell lysates were obtained by centrifugation at 13,000 × g for 15 min at 4 °C. To obtain nuclear extract, cells were homogenized in a hypotonic buffer (10 mM HEPES, pH 7.9,1.5 mM MgCl2,10 mM KCl,0.2 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 1 mM sodium orthovanadate, 0.5% Nonidet P-40). After centrifugation at 2,500 × g at 4 °C for 5 min, and the supernatant (cytoplasmic fraction) was removed. Pellets were washed once with hypotonic buffer containing no detergent, and hypertonic buffer (final concentrations: 20 mM HEPES, pH 7.9, 10% glycerol, 1.5 mM MgCl2, 400 mM KCl, 0.2 mM EDTA), 0.2 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 1 mM sodium orthovanadate) was added to extract nuclear proteins. After a 30-min incubation on ice, the mixture was centrifuged at 13,000 × g for 30 min. The supernatant was then collected as the nuclear extract (24). Protein concentrations of whole cell and nuclear extracts were determined using a Bio-Rad protein assay.
Western Blot Analysis
Whole cell lysates (25 μg) or nuclear extract (15 μg) was denatured and separated by polyacrylamide gel electrophoresis. After separation, proteins were electrophonically transferred to nitrocellulose membranes, which were then sequentially incubated in primary antibody and horseradish peroxidase-conjugated secondary antibody (24). Detection of the horseradish peroxidase conjugate was done using the ECL system (Pierce). For equal loading controls, the membranes were blotted with an anti-β-actin antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or stained by Ponceau S.
RT-PCR
Total cellular RNA was isolated using a Qiagen RNeasy kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. Total RNA (0.5 μg) was subjected to reverse transcription, and one-tenth of the reaction mixture was used for PCR analysis. A pair of primers was designed to detect differential expression of IRF-1 mRNAs: 5′-GGCTGG GAC ATC AAC AAG GAT G-3′ (forward) and5′-GAG CTG CTG AGT CCA TCA GAG AA-3′ (reverse), amplicon size 330 bp. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an internal control: 5′-TGA AGG TCG GAG TCA ACG GAT TTG GT-3′ (forward) and 5′-CAT GTG GGC CAT GAG GTC CAC CAC-3′ (reverse), amplicon size 980 bp. Primer sequences for IRF-1 target genes were as follows: TRAIL, 5′-TGC GTG CTG ATC GTG ATC TT-3′ (forward) and 5′-CCA ACT AAA AAG GCC CCG AA-3′ (reverse), amplicon size 800 bp; caspase-1, 5′-AGG ACA AAC CGA AGG TGA TC-3′ (forward) and 5′-TGT CCT GGG AAG AGG TAG AA-3′ (reverse), amplicon size 390 bp. During PCR amplification, 0.5 μCi of [α-33P]dATP was added to each reaction as described previously (24). PCR products were separated on a 5% native polyacrylamide gel. The gel was then dried and exposed to Kodak Biomax MS film (Eastman Kodak Co.). Individual bands were cut from the dried gel and counted in 3 ml of ScintiVerse scintillation fluid (Fisher) using a liquid scintillation counter (Beckman Instruments) to quantify relative gene expression levels.
Flow Cytometry Analysis of IFNGR-1
Freshly harvested A549 cells were washed twice in cold wash buffer (0.1% NaN3, 1% fetal bovine serum in PBS, pH 7.2). Anti-IFNGR-1 monoclonal antibody (0.1 μg/reaction) was added at 25 μd/well to a U-bottom 96-well plate. Washed cells (4 × 105 in 25 μl of low serum medium) were then added to each corresponding well, and the plate was incubated for 1 h at room temperature with mixing several times during the incubation. Cells were pelleted and washed once before the addition of 50 μ1 of wash buffer containing PBXL-3L-conjugated anti-mouse secondary antibody (5 μl/reaction; Martek Biosciences, Columbia, MD). The cells were further incubated for 30 min at room temperature in the dark. After they were pelleted again and washed twice in wash buffer, 200 μ1 of wash buffer was added to each well to resuspend and transfer the cells to a flow tube (130 × 10 mm) containing 400 μ1 of fixing solution (1% paraformaldehyde in PBS). Fixed samples were stored at 4 °C in the dark before being analyzed using a Coulter Elite ESP flow cytometer (heliumneon laser) within 3 days.
Immunocytochemistry Analysis
A549 cells were plated in 12-well plates at 50% confluence and treated appropriately. At times of harvesting, cells were washed twice in PBS and immediately fixed with 3.7% formaldehyde (w/v) in PBS for 20 min at room temperature. They were then permeabilized with 0.2% Triton X-100 in PBS for 5 min, and the reactions were quenched with freshly prepared 0.1% sodium borohydride in PBS for 5 min. Afterward, the fixed and permeabilized cell monolayer was sequentially incubated in blocking buffer containing 10% fetal bovine serum, 1% bovine serum albumin, 0.02% NaN3 in PBS, anti-IRF-1 polyclonal antibody in 1% bovine serum albumin in PBS, and fluorescein isothiocyanate-labeled anti-rabbit secondary antibody in 1% bovine serum albumin in PBS in the dark. After washing the monolayer twice in PBS, cells were counterstained with 1.5 μg/m l4′,6′-diamidino-2-phenylindole for 5 min in the dark. Samples were visualized under an Olympus IX70 inverted system fluorescence microscope equipped with appropriate filters (Hitech Instruments; Edgemont, PA). Images were captured using a SPOT camera (Diagnostic Instruments, Sterling Heights, MI).
Electrophoretic Mobility Shift Assay
Nuclear extract was prepared as described earlier, aliquoted, and stored at −80 °C. For each electrophoretic mobility shift assay reaction, 5 μg of nuclear protein was incubated with 30,000 cpm of [γ-32P]ATP-labeled consensus IRF-1 gel shift oligonucleotide (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 30 min on ice. For competition or supershift assay, unlabeled IRF-1 consensus or mutant oligonucleotides (in 10- or 50-fold in excess) or anti-IRF-1 monoclonal antibody (1 μl) was incubated with nuclear extracts for 10 min on ice prior to the addition of radiolabeled oligonucleotides. Reaction mixtures were then separated on a 5% native polyacrylamide gel. After electrophoresis, the gel was dried and subjected to autoradiography (24).
Statistical Analysis
Statistical analysis was performed by using SuperANOVA software (Abacus Concepts, Berkeley, CA) for Student's t test, one-way analysis of variance (ANOVA), two-way ANOVA, and simple regression. Three-dimensional plot and multiple regression were performed by using SigmaPlot software (SPSS Inc., Chicago, IL). All data, unless specified, are shown as the mean ± S.E., and difference was considered statistically significant when the p value was less than 0.05.
Results
atRA Increases IFNγ-Induced IRF-1 Protein Expression
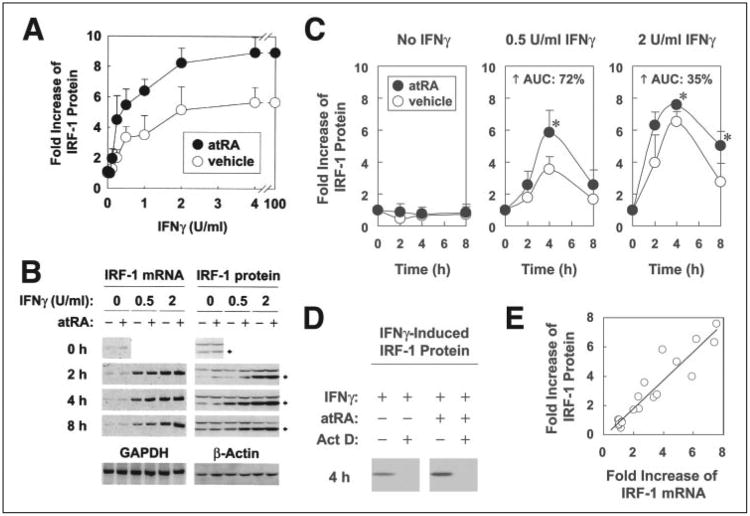
The dose-dependent regulation by IFNγ on IRF-1 protein in the presence or absence of atRA was investigated by pretreating A549 cells with atRA or vehicle overnight, and then with atRA and different concentrations of IFNγ for 4 h (Fig. 1 A). IRF-1 protein was induced dose-dependently by IFNγ. atRA by itself slightly induced IRF-1 transcripts (Fig. 1B), consistent with the reported findings (20); however, a more marked increase was found in the combination between atRA and IFNγ, where atRA was able to synergize with each concentration of IFNγ to further increase IRF-1 expression (Fig. 1A). Maximal induction of IRF-1 was observed around 2–4 units/ml IFNγ, with similar to a ∼5.5-fold increase with IFNγ alone and ∼9-fold with IFNγ plus atRA. Since half-maximal induction was produced by 0.5 units/ml IFNγ, this concentration was chosen as the suboptimal concentration of IFNγ for use in most of the subsequent experiments. These results demonstrate that pretreatment with atRA can augment the effect of low doses of IFNγ in IRF-1 induction.
Figure 1. Overnight pretreatment with atRA followed by suboptimal concentrations of IFNγ induces a faster, higher, and more stable expression of IRF-1.
A, A549 cells were pretreated with either atRA (0.1 μm) or vehicle overnight and then with IFNγ (0,0.03125, 0.0625,0.125, 0.25,0.5, 1,2,4, or 100 units/ml) and atRA for 4 h.Cell lysates were prepared and assayed by Western blot. -Fold increases of IRF-1 protein compared with the untreated cells are shown (n = 3; except for 100 units/ml of IFNγ, n = 1). Two-way ANOVA showed significant differences for atRA (p < 0.002) and IFNγ dose (p < 0.0001). B, A549 cells were pretreated with either atRA (0.1 μm) or vehicle overnight and then with atRA and 0.5 or 2 units/ml of IFNγ for 2, 4, and 8 h. Cell lysates were prepared and assayed for mRNA and protein levels of IRF-1 by RT-PCR and Western blot, respectively. GAPDH (for RT-PCR) and β-actin (for Western blot) were used as negative controls. The Western blot membranes were also stained by Ponceau S to assure equal loading of proteins. C, -fold increases of IRF-1 protein within each IFNγ concentration group (n = 6) are shown; the asterisks indicate significant differences between atRA and vehicle groups (Student's t test); AUC, area under the curve. D, cells were pretreated with either atRA or vehicle overnight and then with actinomycin D (Act D; 5 μg/ml) for 1 h before the addition of IFNγ (0.5 units/ml). IRF-1 pro-teinat4h (time of maximal expression) is shown. E, a correlation test between -fold increases of mRNA and protein levels of IRF-1 was performed by using simple regression (R2 = 0.89).
The kinetics of atRA and IFNγ-regulated IRF-1 were investigated by pretreating A549 cells with atRA or vehicle overnight and then with atRA and IFNγ (0.5 units/ml (suboptimal) or 2 units/ml (maximal)) for 2, 4, and 8 h. Both mRNA (Fig. 1B) and protein (Fig. 1, B and C) results showed that IRF-1 was induced transiently by low concentrations of IFNγ. The expression peaked at 4 h. Treatments with atRA significantly increased IFNγ-induced production of IRF-1, with an overall increase of 72% in the case of 0.5 units/ml and 35% in the case of 2 units/ml IFNγ (Fig. 1C). Specifically, the combination of atRA and IFNγ, compared with IFNγ alone, induced significantly higher levels of IRF-1 protein at 4 h for 0.5 units/ml IFNγ and 4 and 8 h for 2 units/ml IFNγ. Thus, atRA promotes an increase of IFNγ-induced IRF-1 expression that is faster, higher, and more stable than that induced by IFNγ alone.
Induction of IRF-1 by atRA and IFNγ Is Transcriptionally Regulated
Actinomycin D (5 μg/ml) completely blocked the protein expression of IFNγ-induced IRF-1 either with or without atRA (Fig. 1D), indicating that production of nascent transcripts is required for IRF-1 protein induction. This was confirmed by correlation analysis (Fig. 1E) between IRF-1 mRNA and protein (R2 = 0.89). These results suggest that regulation of IRF-1 transcription by atRA and IFNγ is important for their modulation of IRF-1 protein.
Pretreatment with atRA Is Required to Sensitize A549 Cells
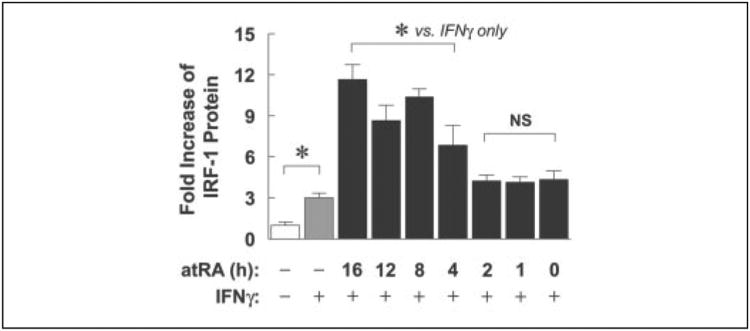
To test whether pretreatment with atRA was required for sensitizing A549 cells to better respond to low concentrations of IFNγ, cells were pretreated with atRA for periods of 0 –16 h, followed by treatments with atRA and IFNγ (0.5 units/ml) for 4 h (Fig. 2). Shorter than 4 h of atRA pretreatment did not produce a significant effect; however, 4 h of atRA pretreatment was able to significantly increase IFNγ-induced IRF-1. The increase was even more marked with longer pretreatments (8–16 h). This demonstrates that at least 4 h of pretreatment with atRA is required in atRA-mediated increase of IFNγ-induced IRF-1 protein. We hypothesize that protein(s) related to IFNγ signaling, such as those important for STAT-1 activation, may be regulated during atRA pretreatment.
Figure 2. At least 4 h of pretreatment with atRA is required to sensitize A549 cells to better respond to low dose IFNγ.
A549 cells were either untreated or treated with 0.5 units/ml IFNγ only or pretreated with atRA (0.1 μm) for 16,12,8,4,2,1, or 0 h before they were treated with IFNγ (0.5 units/ml) and atRA for 4 h. Cell lysates were prepared and assayed by Western blot. -Fold increases of IRF-1 protein compared with untreated are shown (n = 3 or 6). The asterisks above the bars mark significant differences among treatment groups (one-way ANOVA). NS, not significantly different from IFNγ only.
Pretreatment with atRA Increases IFNγ-induced STAT-1 Tyrosine Phosphorylation
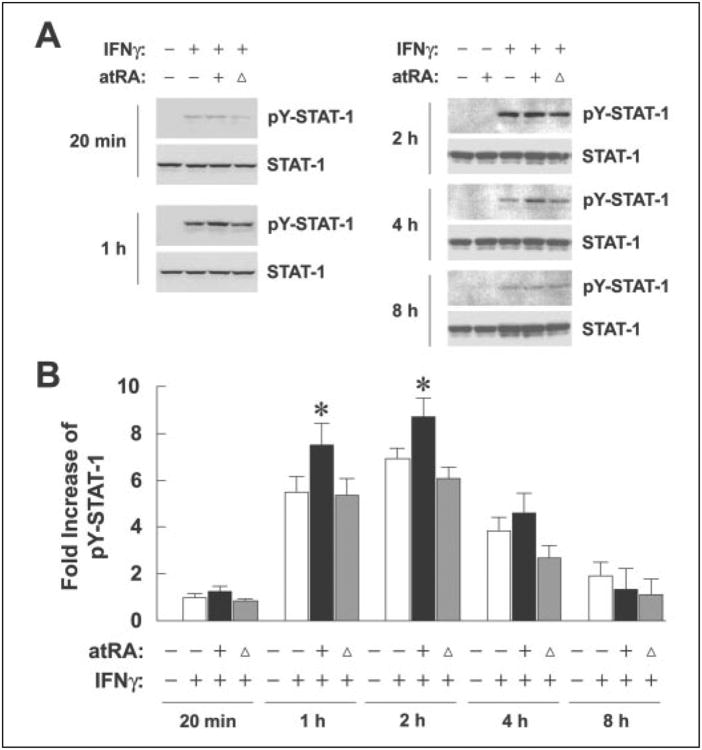
IFNγ signals through binding of the cytokine to IFNγ receptors and generation of a kinase cascade that phosphorylates and activates STAT-1, which in turn transactivates the IRF-1 promoter. STAT-1 phosphorylation at residue Tyr-701 is required for its activation and transcriptional functions. Thus, we measured tyrosine-phosphorylated STAT-1 (pY-STAT-1) to investigate its involvement in atRA-mediated IRF-1 induction (Fig. 3A). pY-STAT-1 protein bands were enumerated by densitometry and plotted in Fig. 3B. As anticipated, neither vehicle nor atRA in the absence of IFNγ could induce pY-STAT-1. IFNγ at 0.5 units/ml, on the other hand, induced STAT-1 phosphorylation. pY-STAT-1 was visible after only 20 min of stimulation with IFNγ and peaked between 1–2 h. Comparisons between any two time points tested showed significant differences except for 20 min and 8 h, indicating rapid and transient increase of pY-STAT-1. More importantly, pretreatment with atRA significantly increased IFNγ-induced pY-STAT-1; however, the effect of a signal dose of atRA at time 0 h was similar to that of IFNγ alone, confirming the requirement of atRA pretreatment. Protein expression of total STAT-1 was not affected by any of the treatments (Fig. 3A).
Figure 3. Pretreatment with atRA increases IFNγ-induced STAT-1 tyrosine phosphorylation.
A, A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with atRA and 0.5 units/ml IFNγfor20min, 1,2,4, and 8 h. An additional treatment group was designed that included overnight pretreatment with the vehicle and co-treatment of atRA with IFNγ, shown in the figure with triangles. Cell lysates were prepared and assayed for protein levels of STAT-1 and pY-STAT-1 by Western blot. B, pY-STAT-1 bands were enumerated using densitometry, and -fold increases of pY-STAT-1 levels compared with the one treated with IFNγ alone are shown (n = 6). pY-STAT-1 was not detectable in the absence of IFNγ. Two-way ANOVA showed atRA and time as significant factors (p < 0.0001). For 1 and 2 h, +atRA was significantly different from both −atRA and co-treatment (*, p < 0.01).
When compared with the expression pattern of IRF-1 mRNA or protein, induction of pY-STAT-1 occurred earlier, suggesting that STAT-1 activation was upstream of IRF-1 expression. Overall, these results suggest that atRA pretreatment increases IFNγ-induced IRF-1 through a STAT-1-dependent pathway.
Expression of IFNGR-1 on the Cell Surface Is Increased upon atRA Pretreatment
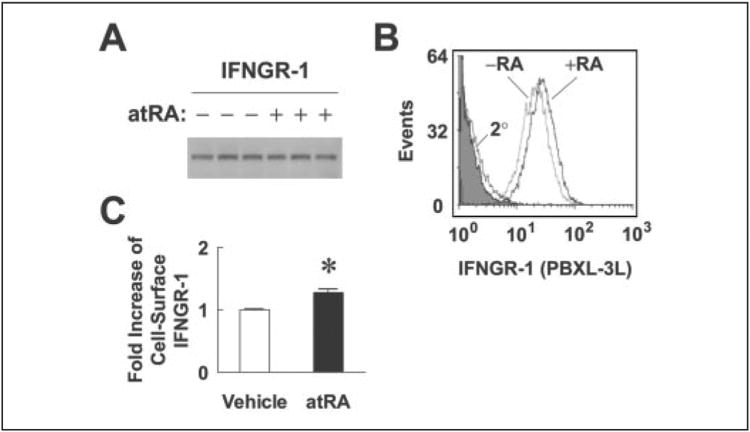
IFNGR-1 is the ligand-binding chain of IFNγ receptor complex. It is also important for Janus kinase-1 binding and STAT-1 recruitment (25). Thus, if total or surface expression of IFNGR-1 is regulated by overnight pretreatment with atRA, an increase of IFNGR-1 expression might explain the observed effects of atRA on increasing IFNγ-induced STAT-1 activation and IRF-1 mRNA and protein expression. To test this hypothesis, A549 cells were treated with vehicle or atRA overnight and harvested at time 0 h for Western blot and flow cytometry analysis (Fig. 4). Although the total protein level of IFNGR-1, assessed by Western blot (Fig. 4A), was not changed by atRA, multiple experiments showed consistently that atRA significantly increased the average intensity of IFNGR-1 on the cell surface (Fig. 4, B and C). The increase was modest, but considering that IFNGR-1 is the start point of IFNγ signaling, a small increase in initial signaling could be amplified by the downstream events. The increase of IFNGR-1 surface expression by atRA is parallel with our earlier observations that both STAT-1 activation and IRF-1 expression induced by IFNγ were further increased by pretreatment with atRA.
Figure 4. Pretreatment with atRA increases IFNGR-1 cell surface expression.
A549 cells were treated with atRA (0.1 μm) or vehicle overnight and harvested for Western blot and flow cytometry analysis. A, Western blot of IFNGR-1 on triplicate samples after overnight treatment with either vehicle or atRA. B, cells after the treatments were stained with IFNGR-1 monoclonal antibody, followed by PBXL-3L-conjugated anti-mouse secondary (2°) antibody. The cells were analyzed by a 633-nm excitation laser flow cytometer. The shaded peak represents an unstained sample. The histogram shown is a representative of six independent experiments. C, -fold increases of the average intensity of PBXL-3L dye (representing the surface expression of IFNGR-1) are shown (n = 6). The asterisk indicates a significant difference between the two treatments (simple t test).
Ligandsfor RARα Sequentially Increased the Levels of IFNGR-1, Activated STAT-1, and IRF-1
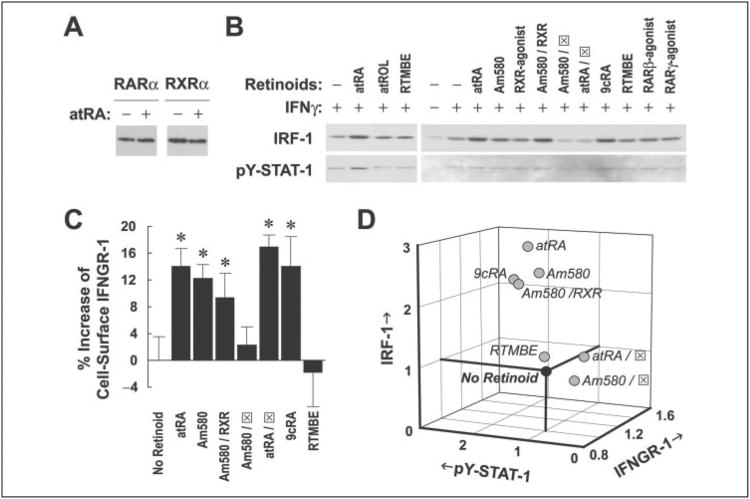
Retinoids regulate transcription of genes through binding to members of the retinoid receptor family. Western blot analysis showed that at least two retinoid receptors, RARα and RXRα, were constitutively expressed in A549 cells (Fig. 5A). To sort out which retinoid receptors were involved in the effect of atRA on IFNγ signaling and IRF-1 expression, we used several receptor-selective retinoids to treat A549 cells overnight and at time 0 h. Cells were harvested after overnight pretreatment (for IFNGR-1 analysis) or at either 1 h 40 min (approximate peaking time of pY-STAT-1), or 4 h (approximate peaking time of IRF-1 protein) after IFNγ stimulation. The results showed that 9cRA, an RARα agonist (Am580), and the combination of Am580 and a pan-RXR agonist significantly increased cell surface IFNGR-1 and IFNγ-induced pY-STAT-1 and IRF-1 to an extent similar to the effect of atRA (Fig. 5, B and C). RARα antagonist blocked the effect of atRA on pY-STAT-1 and IRF-1 and that of Am580 on all three levels, suggesting an important role of RARα in regulating IRF-1 induction. In fact, we found that atRA increased nuclear levels of RARα (2.1-fold) and RXRα (2.4-fold) after overnight pretreatment with atRA, indicating that these retinoid receptors translocate into the nucleus and act to mediate the effect of atRA on IFNγ signaling. RTMBE, a retinoid analog without known binding activity to retinoid receptors, was used as a negative control. All-trans-retinol, a precursor of atRA, did not increase either pY-STAT-1 or IRF-1, suggesting limited conversion of all-trans-retinol to atRA during overnight pretreatment. Protein expression of total STAT-1 was not affected by any of the treatments (data not shown).
Figure 5. Ligands for RARα, including atRA, 9cRA, and Am580, sequentially increase the levels of IFNGR-1, activated STAT-1,and IRF-1.
A, protein expression of RARα and RXRα in A549 cells after overnight treatment with vehicle or atRA (0.1 μm). B, cells were pretreated with different retinoids (0.1 μm; except for RARα antagonist, which was 50 × in excess) overnight and with 0.5 units/ml of IFNγ for 1 h 40 min or 4 h. Cell lysates were prepared and assayed for IRF-1 protein and pY-STAT-1 by Western blot. RTMBE was used as a negative control. RARα antagonist is shown as (x2709). C, cells were treated overnight with different retinoids at the same concentrations described above and then stained for the determination of IFNGR-1 cell surface expression by flow cytometry. Percentage increases of the average intensity of PBXL-3L dye compared with the vehicle are shown (n = 6). Comparisons by one-way ANOVA showed that atRA, Am580, Am580/RXR, atRA/RARα antagonist, and 9cRA are significantly different from either no retinoid or RTMBE (*, p < 0.05). D, -fold increases of the levels of cell surface IFNGR-1, pY-STAT-1, and IRF-1 are plotted on a three-dimensional scale. Multiple regression was performed that suggests significant correlations among IFNGR-1, pY-STAT-1, and IRF-1 (R2 = 0.85).
To determine the correlations among the levels of IFNGR-1, pY-STAT-1, and IRF-1, we used a multiple regression model to analyze these three variables. Retinoid-treated levels of cell surface IFNGR-1 before IFNγ stimulation and those of pY-STAT-1 and IRF-1 at 1 h 40 min and 4 h after IFNγ stimulation, respectively, were normalized to the vehicle-treated levels. A three-dimensional plot (Fig. 5D) showed that positive regulators of all three variables (i.e. atRA, Am580, 9cRA, and the combination of Am580 and RXR-agonist) localized in the upper inner portion of the plot. In contrast, atRA or Am580 combined with RARα antagonist was within the lower outer portion, with the vehicle and RTMBE. Multiple regression analysis showed significant correlations among IFNGR-1, pY-STAT-1, and IRF-1 (R2 = 0.85). This suggests that atRA (together with Am580 and 9cRA) sequentially regulates three components of the IFNγ signaling pathway: cell surface IFNGR-1 and, after IFNγ is given, STAT-1 activation (maximum at 1 h 40 min) and IRF-1 expression (maximum at 4 h).
Pretreatment with atRA Increases Nuclear Localization and DNA Binding Activity of IFNγ-induced Nuclear IRF-1
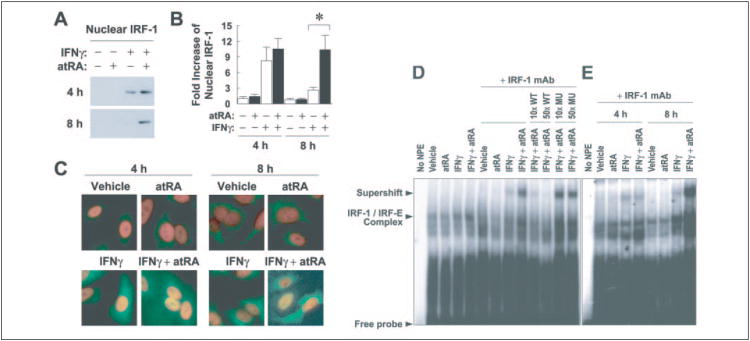
IRF-1 stimulated by 9cRA and IFNγ as part of a coactivation complex activates the promoter of TRAIL (17). However, little is known about the trafficking of IRF-1 between cytoplasm and nucleus. To investigate the effect of IFNγ and atRA on nuclear localization of IRF-1, we performed Western blot of nuclear extracts (Fig. 6, A and B) and immunostaining of formaldehyde-fixed cells (Fig. 6C). IFNγ by itself increased the nuclear level of IRF-1 at 4 h, which declined at 8 h, corresponding to the kinetics of IRF-1 protein expression (Fig. 1C). However, the combination of atRA and IFNγ served to concentrate IRF-1 in the nucleus at both 4 and 8 h. This suggests that atRA not only plays a role in increasing IRF-1 expression but also functions to maintain IRF-1 levels in the nucleus for a longer period of time.
Figure 6. Pretreatment with atRA increases nuclear expression and DNA binding activity of IFNγ-induced IRF-1.
A, A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with IFNγ (0.5 units/ml) and atRA for 4 or 8 h. Nuclear extracts were prepared and analyzed by Western blot. B, -fold increases of the density of nuclear bands are shown (n = 7). The asterisks above the bars mark significant differences between the treatments (simple t test). C, A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with IFNγ (0.5 units/ml) and atRA for 4 or 8 h. They were fixed and stained with IRF-1 polyclonal antibody, followed by a fluorescein isothiocyanate-labeled secondary antibody. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (1.5 μg/ml) before being visualized under a fluorescence microscope. The pictures shown are the overlays of IRF-1/fluorescein isothiocyanate (green) and nuclei/4′,6′-diamidino-2-phenylindole (red) images. D, A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with IFNγ (0.5 units/ml) and atRA for 8 h. Nuclear protein extracts (NPE) were prepared, and 5 μg of NPE were incubated with [γ-32P]ATP-labeled consensus IRF-1 gel shift oligonucleotide with or without unlabeled competitor DNA (wild type (WT) or mutant (MU); 10- or 50-fold molar excess) or were preincubated with IRF-1 monoclonal antibody (mAb) prior to the addition of labeled probe. Protein-DNA complexes were resolved using a 5% polyacrylamide gel. E, A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with IFNγ (0.5 units/ml) and atRA for 4 or 8 h. NPE (5 μg) were prepared, and electrophoretic mobility shift assay was performed.
DNA binding activity of IRF-1 was also tested as an indication of its transcriptional functions. As shown in Fig. 6D, IRF-1 binding to the consensus element (IRF-E) was induced after IFNγ stimulation, and the induction was further increased by atRA pretreatment. The IRF-1 IRF-E complex migrated so close to nonspecific bands that the complex was indistinct; however, supershift experiments clearly identified the complex. atRA pretreatment markedly increased the supershifted band induced by IFNγ. Competition and specificity controls indicated that the complex was specific. Moreover, kinetics experiments showed that atRA markedly increased IFNγ-induced IRF-1 binding to IRF-E at 8 h (Fig. 6E). This is consistent with the earlier demonstration that with atRA pretreatment, IFNγ-induced nuclear IRF-1 stayed at a higher level for a longer period of time (Fig. 6B).
The effect of atRA on IRF-1 binding was abolished by co-pretreatment with RARα antagonist (data not shown), indicating that RARα was involved in mediating the DNA binding activity of IRF-1. Determination of whether this is directly related to the effect of RARα on IRF-1 protein levels requires further investigation.
atRA and IFNγ Synergistically Increase Transcription of IRF-1 Target Genes
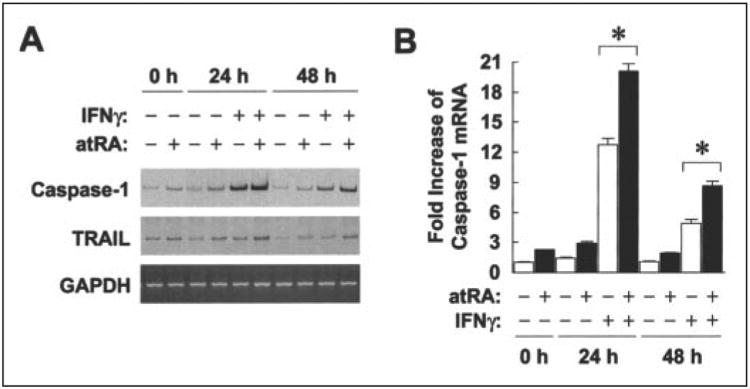
Transcript levels of two IRF-1 target genes, caspase-1 (18) and TRAIL (17), were examined by RT-PCR as indicators of biological effects of atRA and IFNγ-induced IRF-1. Both genes were induced by IFNγ and the combination of IFNγ and atRA (Fig. 7A). Specifically, atRA pretreatment synergized with low dose IFNγ to induce caspase-1 mRNA to more than 20-fold compared with the untreated cells at 24 h, whereas IFNγ alone induced only 12-fold (Fig. 7B). TRAIL mRNA was induced about 3-fold by the combination. These results support the concept that pretreatment with atRA increases the transcriptional functions of IFNγ-induced IRF-1.
Figure 7. atRA and IFNγsynergistically increase transcription of IRF-1 target genes.
A A549 cells were pretreated with atRA (0.1 μm) or vehicle overnight and then with IFNγ (0.5 units/ml) and atRA for 24 or 48 h. Total RNA was isolated, and transcript levels of caspase-1, TRAIL, and GAPDH were measured by RT-PCR. B, radioactivity of caspase-1 bands was counted as described under “Experimental Procedures.” -Fold increases of caspase-1 mRNA are shown (n = 3). The asterisks above the bars mark significant differences between the treatments (simple t test).
Discussion
Vitamin A is important in the maintenance of normal immune functions. This is supported by studies of vitamin A deficiency, where disease risk is increased (26). However, therapeutic effects of vitamin A supplementation vary, depending on the types of illness and the populations studied (27). In addition, vitamin A has failed to affect the prevalence of illness, even in the presence of a large impact on mortality. In contrast, studies on vitamin A and morbidity have consistently showed an important effect of this nutrient on the severity of illness. In one study monitoring 1455 young children weekly for a year (28), vitamin A-supplemented children had less frequent vomiting and anorexia, although the prevalence of diarrhea or acute respiratory infections was not changed. The supplementation also led to fewer clinic attendances, hospital admissions, and death. Thus, vitamin A appears to reduce the severity of infection, possibly by enhancing functions of innate and cell-mediated immune responses and helping to repair virus-damaged epithelia (1).
Our present study was designed to study how atRA, an active metabolite of vitamin A, increases IFNγ-induced response as part of innate immunity. Type I and II IFNs were discovered as proteins that inhibit virus replication (29). They are induced in response to virus infection, secreted, and function to activate a global antiviral state. Type II IFN (IFNγ) is generally expressed at a low level in the early stages of an infection (30); thus, enhancing the functions of albeit small amounts of IFNγ during early infection may potentially improve the overall IFN response and encourage faster recovery.
IFNγ signaling pathway is initiated upon binding of IFNγ to IFNGR-1. The extracellular portion of IFNGR-1 chain contains multiple disulfide bonds that are required for maximal binding of the ligand. The intracellular portion of IFNGR-1 contains a tyrosine residue that becomes phosphorylated upon ligand binding and acts in STAT-1 recruitment (25). STAT-1 is phosphorylated at Tyr-701, which is required for the activation and dimerization of STAT-1 through Src homology 2 domains (31). The activated STAT-1 homodimer binds and activates transcription of target genes through the conserved response element γ-interferon-activated site (32). STAT-1 is also phosphorylated on Ser-727 in order to exert maximal transcriptional activity (33).
In the present study, we found that atRA of a subpharmacologic concentration (0.1 μm) increased the effect of low-dose IFNγ (0.5 units/ml) on IRF-1 induction by about 2-fold, potentially enhancing the antiviral functions of IFNγ. IRF-1, a target of IFNγ signaling, was originally identified as a mouse nuclear factor that specifically bound to the upstream regulatory region of the IFNβ gene. In 1988, Taniguchi and co-workers (11) cloned and characterized the cDNA encoding IRF-1, and a role of IRF-1 in viral infections was then suggested, since it possessed a virus-inducible promoter (34).
In previous studies, Matikainen et al. (20) showed that atRA (1 μm) rapidly up-regulated IRF-1 in promyelocytic leukemia NB4 cells, inducing the mRNA level to nearly 8-fold within 3 h of treatment. There was also a 3-fold increase of IRF-1 protein levels by atRA (1 or 100 μm) in cervical squamous carcinoma SiHa cells (23). In our studies of lung epithelial A549 cells, we observed minimal effect of atRA (0.1 μm) by itself after 16-h (overnight) treatment or longer, probably due to the transient action of lower dose atRA on IRF-1 induction (23). However, atRA at this concentration significantly potentiated the effect of low dose IFNγ on IRF-1 induction (Fig. 1).
atRA pretreatment of a certain length of time (4 h or longer) was required for sensitizing these cells to better respond to IFNγ (Fig. 2). We also used a protein synthesis inhibitor, cycloheximide (10 μg/ml), together with atRA pretreatment, to determine whether the increase of IRF-1 transcription was blocked by inhibiting protein synthesis (data not shown). The result showed that both IFNγ-induced and IFNγ plus RA-induced transcription was blocked by cycloheximide, indicating the need of certain protein(s) in both processes. Such proteins may regulate the migration of IFNGR-1 from the cytoplasm to the cell surface. However, whether or not these proteins are targets of atRA pretreatment requires further investigation. Nevertheless, our study has demonstrated that treatment with atRA increases the levels of cell surface IFNGR-1 (Fig. 4). This is supported by reports that up-regulation of type I IFN receptors by retinoids is associated with increased signaling of IFNα/β (35, 36).
STAT-1 is tyrosine-phosphorylated and activated upon IFNγ stimulation (37). atRA was observed in our model to enhance tyrosine phosphorylation (activation) of STAT-1 induced by low dose IFNγ, without increasing STAT-1 protein levels (Fig. 3). This effect of atRA required pretreatment, suggesting that the increase of STAT-1 activation is due to the increase of cell surface IFNGR-1 during atRA pretreatment. In comparison, the rapid effect of atRA on IRF-1 induction may not involve STAT-1 functions (20, 22), despite a report that atRA induces IRF-1 through a γ-interferon-activated site motif of the IRF-1 promoter (21). However, the present study indicates that STAT-1 activation is required in the regulation of IFNγ-induced IRF-1 by atRA.
The use of receptor-selective retinoids in the present study revealed strong correlations among the levels of IFNGR-1, activated STAT-1, and IRF-1 (Fig. 5), indicating that atRA and RARα together influenced several components of the IFNγ signaling pathway. Our results identified RARa ligands as important regulators of IFNγ-induced IRF-1. atRA has been shown to enhance transactivation of STAT-1 in the presence of RARα in acute promyelocytic leukemia cells (38). However, it was unknown if RARα mediated other components of IFNγ signaling. In the present study, atRA also increased nuclear levels of RARα, which, together with RXRα, may mediate the regulatory effect of atRA on multiple components of the IFNγ signaling pathway. How atRA and RARα function in this context requires further investigation. We hypothesize that during the pretreatment period of 4 h or longer, atRA and RARα regulate the synthesis or posttranslational modification of proteins involved in surface expression of IFNGR-1, tyrosine phosphorylation of STAT-1, and/or transcription of IRF-1.
atRA as a panagonist for all RARs may have increased IFNGR-1 surface expression via receptors other than RARα, since the antagonist specific for RARα did not block the effect of atRA on IFNGR-1. This is supported by the observation that RARβ agonist also increased cell surface IFNGR-1 (data not shown). Hence, additional mechanisms of atRA action might affect IFNGR-1 but not pY-STAT-1 or IRF-1. Nevertheless, our current findings suggest an essential role of RARα in mediating the effect of atRA on IFNγ signaling.
atRA pretreatment also potentiated the transcriptional activity of IFNγ-induced IRF-1, increasing its nuclear localization and DNA binding activity (Fig. 6). Notably, pretreatment with atRA maintained higher IRF-1 levels in the nucleus for a longer period of time, with a nearly 4-fold difference between IFNγ alone and atRA plus IFNγ at 8 h. This is possibly due to an unknown role of atRA in facilitating nuclear translocation of IRF-1 and/or preventing shuttling of the protein out of the nucleus. Reports are scarce on this subject; however, a retinoid-medi-ated increase of nuclear localization of calcyclin-binding protein was observed during neuronal differentiation (39). Another report has shown that nucleocytoplasmic translocation of RARβ is facilitated by RXR in a ligand-dependent manner (40). Our results are consistent with these findings, where atRA facilitated or maintained the nuclear localization of RARα, RXRα, and IRF-1 and may thereby prolong the trans-activation of IRF-1.
IFNγ-induced caspase-1, an enzyme important for the production of IFNγ during an innate immune response, was further increased by pretreatment with atRA (Fig. 7). Caspase-1 processes the precursors of IL-1β (41) and IL-18 (19) to their biologically active forms. IL-1β and IL-18, alone or in synergy with IL-12, increases lipopolysaccharide-induced IFNγ production by natural killer cells, evidenced by marked reduction of lipopolysaccharide-induced serum IFNγ titers in caspase-1−/− mice (19) and IL-12-deficient mice (42). It is very likely that IFNγ (low dose)-induced caspase-1, which can be enhanced by atRA pretreatment, feeds forward IFNγ signaling during early stages of an infection.
TRAIL, another target gene of IRF-1, induces apoptosis selectively in cancer cells, sparing normal cells that are generally TRAIL-insensitive (43). IFNγ-9cRA cotreatment synergistically induces TRAIL mRNA levels due to sustained occupancy of IRF-1 on the TRAIL promoter (17), supporting the use of combination therapies between IFNs and retinoids in cancer treatments. The induction of TRAIL could also be important in the cellular response to infection but has received little attention in this context. Our data indicate that the retinoid status of the cell can affect the induction of TRAIL by low doses of IFNγ (Fig. 7), suggesting another mechanism by which retinoids may improve the response of the host to infectious disease.
In summary, the present study has detailed the regulating roles of atRA in IFNγ signaling and IRF-1 induction. Using RARα as a mediator, atRA pretreatment sensitizes A549 cells to better respond to low dose IFNγ, increasing cell surface IFNGR-1, STAT-1 activation, levels of IRF-1 mRNA and protein, and transcriptional functions of IRF-1 (Fig. 8). Could these results in a model system of the cellular response to low dose IFNγ be relevant to human studies of vitamin A status and infectious disease? In epidemiological studies conducted in populations where vitamin A deficiency remains a public health problem (see World Health Organization review (27)), supplementation with vitamin A has been shown to reduce the severity of infectious diseases (28) and to significantly reduce all-cause mortality rates in children and pregnant women (44, 45). It is possible that vitamin A, through conversion to physiological levels of atRA, results in the generation of an “active atRA state,” which serves to improve the integrity of epithelial tissue barriers as a first line of host defense and to enhance the initial response to viral and bacterial infections through potentiation of the response to low levels of IFNγ, such as are likely to be present in the early stages of an infection. A heightened host response to IFNγ, produced when the “active atRA state” is adequate, may provide an important advantage in quickly responding to viral and bacterial agents and could be part of the mechanism whereby vitamin A has been shown to be effective in reducing morbidity and mortality in human populations.
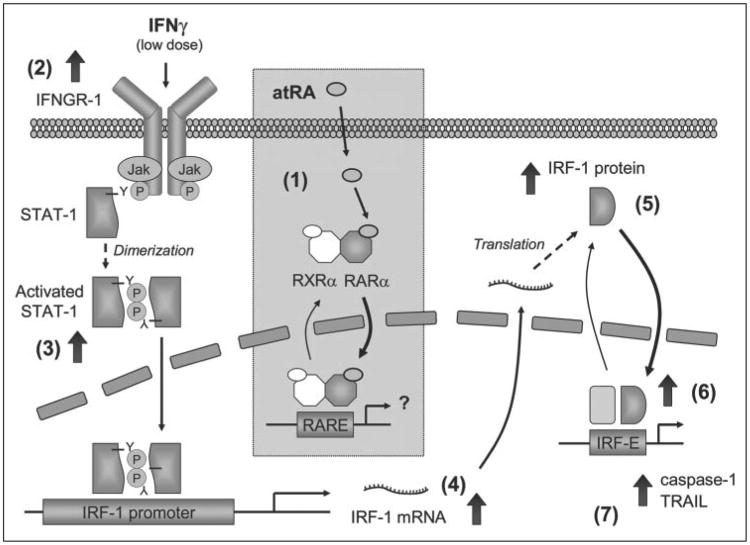
Figure 8. Working model of the regulation of IFNγ-induced IRF-1 by pretreatment with atRA.
In A549 cells, atRA pretreatment of 4 h or longer facilitates the shuttling of RARα and its dimerization partner, RXRα, from the cytoplasm to the nucleus in a ligand-dependent manner (1). This enhances the transactivation activity of RARα/RXRα heterodimer, leading to yet to be characterized changes that prepare the cells for increased response to low dose IFNγ. Via the actions of RARα, atRA increases the levels of IFNGR-1 on the cell surface (2), enhancing tyrosine phosphorylation (activation) of STAT-1 upon IFNγ stimulation (3). IRF-1, a target gene of STAT-1, is then induced (4) and translated (5); its nuclear localization and DNA binding activity (6), and target gene (caspase-1 and TRAIL) transcription (7), are also increased.
Acknowledgments
We are grateful to Elaine Kunze (Center for Quantitative Cell Analysis, The Pennsylvania State University) for technical assistance in flow cytometry and fluorescence microscopy.
Footnotes
The abbreviations used are: atRA, all-trans-retinoic acid; IFN, interferon; IRF, interferon regulatory factor; STAT, signal transducers and activators of transcription; IFNGR-1, IFNγ receptor-1; RARα, retinoic acid receptor-α; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; IL, interleukin; PBS, phosphate-buffered saline; RT, reverse transcription; pY-STAT-1, tyrosine-phosphorylated STAT-1; 9cRA, 9-cis-retinoic acid; RTMBE, retinyl trimethoxybenzyl ether; ANOVA, analysis of variance; GAPDH, glycer-aldehyde-3-phosphate dehydrogenase.
This work was supported by National Institutes of Health Grants DK41479 and CA 90214.
References
- 1.Ross AC, Stephensen CB. FASEB J. 1996;10:979–985. [PubMed] [Google Scholar]
- 2.Semba RD. J Nutr. 1999;129:783–791. doi: 10.1093/jn/129.4.783. [DOI] [PubMed] [Google Scholar]
- 3.Samuel CE. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petkovich M, Brand NJ, Krust A, Chambon P. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 5.Darnell JE, Jr, Kerr IM, Stark GR. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.DeCicco KL, Youngdahl JD, Ross AC. Immunology. 2001;104:341–348. doi: 10.1046/j.1365-2567.2001.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeCicco KL, Zolfaghari R, Li N, Ross AC. J Infect Dis. 2000;182(1):29–36. doi: 10.1086/315908. [DOI] [PubMed] [Google Scholar]
- 8.Austenaa LM, Ross AC. J Leukocyte Biol. 2001;70:121–129. [PubMed] [Google Scholar]
- 9.Garattini E, Mologni L, Ponzanelli I, Terao M. Leuk Lymphoma. 1998;30:467–475. doi: 10.3109/10428199809057559. [DOI] [PubMed] [Google Scholar]
- 10.Chelbi-Alix MK, Pelicano L. Leukemia. 1999;13:1167–1174. doi: 10.1038/sj.leu.2401469. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa K, Yokosawa H. Eur J Biochem. 2000;267:1680–1686. doi: 10.1046/j.1432-1327.2000.01163.x. [DOI] [PubMed] [Google Scholar]
- 13.Harada H, Taniguchi T, Tanaka N. Biochimie(Paris) 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 14.Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. J Interferon Cytokine Res. 2002;22:39–47. doi: 10.1089/107999002753452647. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lam-phier MS, Aizawa S, Mak TW, Taniguchi T. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 16.Kroger A, Dallugge A, Kirchhoff S, Hauser H. Oncogene. 2003;22:1045–1056. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- 17.Clarke N, Jimenez-Lara AM, Voltz E, Gronemeyer H. EMBO J. 2004;23:3051–3060. doi: 10.1038/sj.emboj.7600302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwase S, Furukawa Y, Kikuchi J, Saito S, Nakamura M, Nakayama R, Horiguchi-Yamada J, Yamada H. FEBS Lett. 1999;450:263–267. doi: 10.1016/s0014-5793(99)00515-3. [DOI] [PubMed] [Google Scholar]
- 19.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, Kamen R, Tracey D, Allen H. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 20.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I. Blood. 1996;88:114–123. [PubMed] [Google Scholar]
- 21.Pelicano L, Li F, Schindler C, Chelbi-Alix MK. Oncogene. 1997;15:2349–2359. doi: 10.1038/sj.onc.1201410. [DOI] [PubMed] [Google Scholar]
- 22.Percario ZA, Giandomenico V, Fiorucci G, Chiantore MV, Vannucchi S, Hiscott J, Affabris E, Romeo G. Cell Growth & Differ. 1999;10:263–270. [PubMed] [Google Scholar]
- 23.Arany I, Whitehead WE, Grattendick KJ, Ember IA, Tyring SK. Clin Diagn Lab Immunol. 2002;9:1102–1106. doi: 10.1128/CDLI.9.5.1102-1106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q, Ma Y, Ross AC. Immunology. 2002;107:199–208. doi: 10.1046/j.1365-2567.2002.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestka S, Kotenko SV, Muthukumaran G, Izotova LS, Cook JR, Garotta G. Cytokine Growth Factor Rev. 1997;8:189–206. doi: 10.1016/s1359-6101(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 26.Sommer A, Katz J, Tarwotjo I. Am J Clin Nutr. 1984;40:1090–1095. doi: 10.1093/ajcn/40.5.1090. [DOI] [PubMed] [Google Scholar]
- 27.The Vitamin A and Pneumonia Working Group. Bulletin of the World Health Organization. 1995;73:609–619. [PMC free article] [PubMed] [Google Scholar]
- 28.Kirkwood BR, Arthor P, Ross DA, Morris SS, Gyapong JO, Tomkins AM. Lancet. 1993;342:7–12. [Google Scholar]
- 29.Sen GC. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 30.van den Broek MF, Muller U, Huang S, Zinkernagel RM, Aguet M. Immunol Rev. 1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 31.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 32.Khan KD, Shuai K, Lindwall G, Maher SE, Darnell JE, Jr, Bothwell AL. Proc Natl Acad Sci U S A. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen Z, Zhong Z, Darnell JE., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 34.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, Taniguchi T. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 35.Obora A, Shiratori Y, Okuno M, Adachi S, Takano Y, Matsushima-Nishiwaki R, Yasuda I, Yamada Y, Akita K, Sano T, Shimada J, Kojima S, Okano Y, Friedman SL, Moriwaki H. Hepatology. 2002;36:1115–1124. doi: 10.1053/jhep.2002.36369. [DOI] [PubMed] [Google Scholar]
- 36.Hamamoto S, Fukuda R, Ishimura N, Rumi MA, Kazumori H, Uchida Y, Kadowaki Y, Ishihara S, Kinoshita Y. J Lab Clin Med. 2003;141:58–66. doi: 10.1067/mlc.2003.8. [DOI] [PubMed] [Google Scholar]
- 37.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 38.Gianni M, Terao M, Fortino I, LiCalzi M, Viggiano V, Barbui T, Rambaldi A, Garattini E. Blood. 1997;89:1001–1012. [PubMed] [Google Scholar]
- 39.Wu J, Tan X, Peng X, Yuan J, Qiang B. J Biochem Mol Biol. 2003;36:354–358. doi: 10.5483/bmbrep.2003.36.4.354. [DOI] [PubMed] [Google Scholar]
- 40.Maruvada P, Baumann CT, Hager GL, Yen PM. J Biol Chem. 2003;278:12425–12432. doi: 10.1074/jbc.M202752200. [DOI] [PubMed] [Google Scholar]
- 41.Howard AD, Kostura MJ, Thornberry N, Ding GJ, Limjuco G, Weidner J, Salley JP, Hogquist KA, Chaplin DD, Mumford RA, Schmidt JA, Tocci MJ. J Immunol. 1991;147:2964–2969. [PubMed] [Google Scholar]
- 42.Magram J, Connaughton SE, Warrier RR, Carvajal DM, Wu CY, Ferrante J, Stewart C, Sarmiento U, Faherty DA, Gately MK. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 43.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 44.Beaton GH, Martorell R, Aronson KJ, Edmonston B, McCabe G, Ross AC, Harvey B. United Nations Administrative Committee on Coordination/Sub-Committee on Nutrition (ACC/SCN) State-of-the-art Series. Nutrition Policy Discussion Paper No.13. ACC/SCN; Geneva, Switzerland: 1993. [Google Scholar]
- 45.Villamor E, Fawzi WW. J Infect Dis. 2000;182(1):122–133. doi: 10.1086/315921. [DOI] [PubMed] [Google Scholar]