Summary
X11-like (X11L) is neuronal adaptor protein that interacts with the amyloid β-protein precursor (APP) and regulates its metabolism. The phosphotyrosine interaction/binding (PI/PTB) domain of X11L interacts with the cytoplasmic region of APP695. We found that X11L-APP interaction is enhanced in osmotically stressed cells and X11L modification is required for the enhancement. Amino acids 221-250 (X11L221-250) are required for the enhanced association with APP in osmotically stressed cells; this motif is 118 amino acids closer to the amino-terminal end of the protein than the PI/PTB domain (amino acids 368-555). We identified two phosphorylatable seryl residues, Ser236 and Ser238, in X11L221-250 and alanyl substitution of either seryl residue diminished the enhanced association with APP. In brain Ser238 was found to be phosphorylated and phosphorylation of X11L was required for the interaction of X11L and APP. Both seryl residues in X11L221-250 are conserved in neuronal X11, but not in X11L2, a non-neuronal X11 family member that did not exhibit enhanced APP association in osmotically stressed cells. These findings indicate that the region of X11L that regulates association with APP is located outside of, and amino-terminal to, the PI/PTB domain. Modification of this regulatory region may alter the conformation of the PI/PTB domain to modulate APP binding.
Keywords: Alzheimer’s disease, X11, X11L, APP, cell stress, protein phosphorylation
Introduction
X11-like (X11L) is a member of the X11 adaptor protein family, comprised of the X11/X11α/MINT1, X11L/X11β/MINT2, and X11L2/X11γ/MINT3 genes (reviewed in [Miller et al. 2006]). X11L and X11 are expressed predominantly in neurons, while X11L2 is expressed ubiquitously [Ho et al. 2003]. The X11 family (X11s) associate with various other proteins, including synaptic proteins such as the calcium/calmodulin-dependent serine kinase-related protein (CASK) and motor proteins such as KIF17 [Borg et al. 1999; Setou et al. 2000]. The interaction of X11s with synaptic proteins, as in the formation of the X11-CASK-Veli complex, may regulate presynaptic release probability in neurons [Ho et al. 2003]. Additionally, the association of Veli with the N-methyl-D-aspartate (NMDA) receptor 2B subunit (NR2B) and of X11 with KIF17 regulates transport of a neurotransmitter receptor [Setou et al, 2000]. Therefore, one of the functions of the X11 proteins is to regulate the formation and maintenance of synaptic structure.
The X11s also bind the cytoplasmic regions of APP and Alcadein (Alc). This interaction suppresses APP metabolism, including the generation of amyloid β-protein (Aβ) [Borg et al, 1998; Tomita et al, 1999; Tanahashi & Tabira, 1999], which is produced by consecutive cleavages of APP and is believed to be a primary cause of Alzheimer’s disease (AD) pathogenesis (reviewed in [Selkoe, 2001; Gandy 2005]). Formation of APP-X11L-Alc strengthens the suppression of cleavage of both APP and Alc [Araki et al, 2003 & 2004]. Therefore, another function of the X11 proteins is to regulate APP metabolism in the brain.
Indeed, transgenic Tg2576 mice with the APP Swedish mutation which also over-express X11 or X11L showed decreased levels of cerebral Aβ and a reduction in Aβ plaques in the cortex and hippocampus [Lee et al, 2003 & 2004]. Moreover, X11L gene knockout mice showed a significant increase in the amyloidogenic carboxyl-terminal fragment (CTFβ) of APP, generated by β-site cleavage, together with increased levels of Aβ in the brain [Sano et al, 2006]. X11 proteins are thought to regulate the translocation of APP into lipid rafts, a detergent resistant membrane fraction, where β-site cleaving enzyme (BACE) is enriched [Saito et al, 2008]. These observations clearly indicate that X11L is a key regulator of amyloidogenic metabolism of APP in the brain in vivo.
The X11 proteins are composed of a relatively divergent amino-terminal region, a central phosphotyrosine interaction/binding (PI/PTB) domain, and two carboxyl-terminal PDZ domains. The PI/PTB domain is responsible for binding the 681GYENPTY687 motif in the cytoplasmic region of APP (numbering for the APP695 isoform) (reviewed in [Suzuki et al, 2006; Suzuki & Nakaya, 2008]). However, the molecular mechanisms that regulate this interaction remain unclear. Because dysregulation of the mechanism governing the X11L-APP interaction in the human brain may increase the production of Aβ and generate pathogenic alterations, it is important to understand the regulation of X11L-APP binding. Herein, we demonstrate that X11L contains a regulatory region, X11L221-250, in its amino-terminal region, separate from the APP-binding PI/PTB domain. Phosphorylation of amino acid residues Ser236 and Ser238 in the regulatory region are critical for increasing the association of X11L and APP, and are conserved in X11, a neuronal X11 family protein, but not in the non-neuronal X11L2. Furthermore, phosphorylated X11L derived from mouse brain exhibits enhanced binding to APP when compared to the dephosphorylated form. These findings contribute to our understanding of the mechanism of regulation of X11L-APP binding.
Experimental Procedures
Plasmids and Peptides
The constructs pcDNA3-APP695, pcDNA3-FLAG-APP695, pcDNA3-FLAG-APP695T668A, pcDNA3.1-FLAG-X11L, and pcDNA3-HA-X11L were described previously [Ando et al, 1999; Tomita et al, 1999; Araki et al, 2003; Taru & Suzuki, 2004]. ThecDNAs encoding the amino-terminal (pcDNA3.1-FLAG-X11L-N+PI, amino acids 1-555) and the carboxyl-terminal (pcDNA3.1- FLAG-X11L-PI+C, amino acids 368-749) regions adjacent to the PI/PTB domain of X11L are the same as described [Tomita et al, 1999; Taru & Suzuki, 2004]. The cDNA construct pcDNA3.1-FLAG-X11L F520V (Phe to Val mutation at amino acid 520) was generated by PCR. The cDNA construct pcDNA3-FLAG-X11L Δ221-250 (deletion of amino acids 221-250 of X11L) was generated by PCR using pcDNA3-FLAG-X11L as the template and the generated fragment was ligated into pcDNA3-FLAG-X11L at the KpnI/EcoRI site [Taru & Suzuki, 2004]. The pcDNA3.1-HA-X11 and pcDNA3.1-HA-X11L2 constructs were generated by replacing the FLAG-tag of pcDNA3.1-FLAG-X11 and pcDNA3.1-FLAG-X11L2 with an HA-tag. The pcDNA3-HA-X11L ST/A1, pcDNA3-HA-X11L ST/A2, pcDNA3-HA-X11L ST/A3, and pcDNA3-HA-X11L ST/A4 constructs were generated by PCR using pcDNA3-HA-X11L as the template and the generated fragments were ligated into the AccIII/EcoRI sites. The pcDNA3.1-FLAG-X11L S236A, pcDNA3.1-FLAG-X11L S238A, pcDNA3.1-FLAG-X11L S236A/S238A, and pcDNA3.1-FLAG-X11L S236D/S238D constructs were generated by PCR using pcDNA3-HAX11L as the template and the generated fragments were ligated into pcDNA3.1-FLAG-X11L at the AccIII/EcoRI sites. pGEX-4T-1-APPcyt and pGEX-4T-1-X11L were used to produce the GST fusion proteins of the APP cytoplasmic region (APPcyt, APP652-695) and of X11L, respectively, as described previously [Tomita et al, 1999]. pGEX-4T-1-APPcytAAAA (NPTY to AAAA mutation at amino acids 684-687 of APP695) was generated by PCR.
Antibodies
Monoclonal anti-FLAG (M2, Sigma), anti-HA (12CA5, Roche Diagnostics), anti-GST (Upstate Biotechnology), and polyclonal anti-APP C-terminus (APP/C, Sigma #8717 product) antibodies were purchased. Mouse monoclonal anti-X11L 4A10 antibody was raised against a synthetic peptide that is composed of Cys plus the region of human X11L735-749, C+AMFRLLTGQETPLYI. This antibody recognizes X11L specifically (data not shown). Phosphorylation state-specific polyclonal antibodies to X11L Ser236 (UT158) and Ser238 (UT162) were raised against chemically phosphorylated synthetic peptides of human X11L223-241, C+IKMpSLSMTS (pSer236 peptide) or C+IKMpSLpSMTS (pSer236/pSer238 peptide). The anti-pSer236 UT158 antibody was affinity-purified using antigen coupled resin. The UT162 serum was pre-absorbed with pSer236 peptide-coupled resin and the flow-through serum was used as an antibody specific for pSer238. The specificity of both antibodies is shown in Supplementary Fig. S1.
Preparation of Proteins
Production and purification of recombinant GST fusion proteins were performed as described [Tomita et al, 1999; Taru et al, 2002]. Briefly, GST fusion proteins were generated in E. coli BL21 transformed with pGEX-4T-1 cDNA constructs and purified with glutathione-Sepharose 4B (GE Healthcare Bioscience).
Cell Culture, Expression of Proteins and Induction of osmotically stressed cells
Human embryonic kidney 293 (HEK293) cells were cultured in DMEM supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS). To express proteins, ~2 × 105 cells were transiently transfected with 0.5–1 μg of each plasmid using LipofectAMINE 2000 (Invitrogen) according to the manufacturer’s protocol and cultured for 24 h in DMEM containing 10% (v/v) FBS. Osmotic stress was induced by treatment of 0.5 M sorbitol for 45 min in medium prior to harvesting the cells [Taru & Suzuki, 2004]. The cells were lysed in CHAPS lysis buffer (phosphate-buffered saline containing 10 mM CHAPS, 5 μg/ml chymostatin, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, 1 mM Na3VO4, and 1 mM NaF) and centrifuged at12,000 × g for 10 min at 4 °C. The resulting supernatants were used for binding assays.
λPPase treatment
For λPPase treatment, brains of adult mice (2–3 months old) were homogenized in eight volumes of CHAPS lysis buffer without protein phosphatase inhibitors and centrifuged at 100,000 × g for 60 min at 4 °C. The supernatant was subjected to dephosphorylation with lambda protein phosphatase (λPPase) (Sigma) for 2 h in a Sigma-supplied buffer. For control, 1 mM NaF, 1 mM Na3VO4, and 1 μM microcystin-LR were added to the buffer instead of λPPase.
Binding assays
In the coimmunoprecipitation assay, cell lysates were incubated with anti-FLAG M2 at 4 °C for 2–16 h. Immunocomplexes were recovered using protein G-Sepharose beads (GE Healthcare Biosciences). In the GST pull-down assay, cell or mouse brain lysates were incubated with glutathione-Sepharose beads coupled to purified GST protein at 4 °C for 2 h, respectively. The lysates and the recovered proteins were analyzed by immunoblottingwith the indicated antibodies
Results
Enhanced association of X11L with APP in osmotically stressed cells
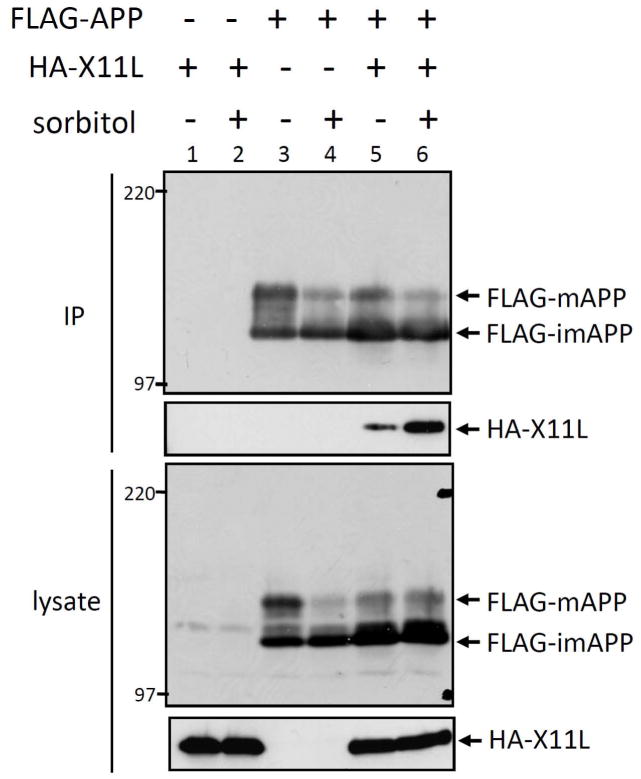
HEK293 cells transiently expressing FLAG-APP (lanes 3–6) and HA-X11L (lanes 1, 2, 5 and 6) were treated with (+) or without (−) 0.5 M sorbitol for 45 min, and APP was recovered by immunoprecipitation with anti-FLAG M2 antibody. The immunoprecipitates (IP) and cell lysates were analyzed by immunoblotting with anti-APP/c and anti-HA antibodies (Fig. 1). HA-X11L coimmunoprecipitated with FLAG-APP (lane 5), and interestingly, greater amounts of HA-X11L were recovered from sorbitol-treated cells (lane 6). This result suggests that the X11L-APP interaction was facilitated in osmotically stressed cells. We hypothesized that the increased association of X11L with APP is due to a change in conformation or modifications of APP, X11L, or both, in the stressed cells.
Figure 1. Enhanced association of X11L with APP in osmotically stressed cells.
HEK293 cells (~2 × 105) were transiently cotransfected with 0.3 μg of pcDNA3-FLAG-APP695 (+; lanes 3–6) and pcDNA3-HA-X11L (+; lanes 1, 2, 5, 6). To standardize the plasmid concentrations, pcDNA3 vector (−) was added to yield 0.6 μg of plasmid in total. The cells were treated with (+; lanes 2, 4, 6) or without (−; lanes 1, 3, 5) 0.5 M sorbitol for 45 min and cell lysates were immunoprecipitated with anti-FLAG M2 antibody. The immunoprecipitates (IP) and lysates were analyzed by immunoblotting with anti-APP (APP/C) and anti-HA (12CA5) antibodies. FLAG-APP indicates mature FLAG-APP (FLAG-mAPP) and immature FLAG-APP (FLAG-imAPP) forms. Molecular weight standards (in kDa) are indicated at the left.
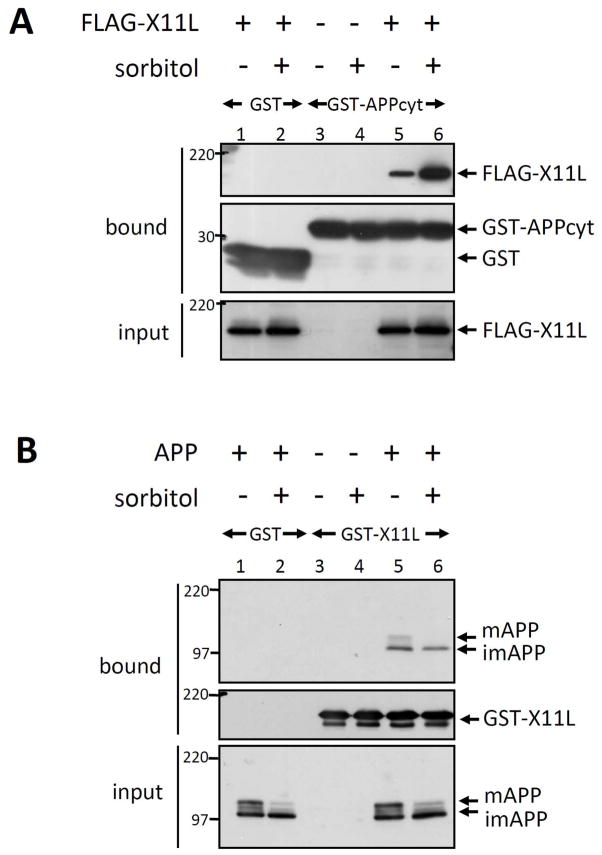
To examine which protein was responsible for the enhanced association in response to hyperosmotic conditions, we performed GST pull-down assays in vitro. HEK293 cells transiently expressing FLAG-X11L (lanes 1, 2, 5, 6) were treated with (+) or without (−) sorbitol. Cell lysates were incubated with the purified GST-APP cytoplasmic domain (GST-APPcyt) or with GST alone (GST) (Fig. 2A). Bound protein (bound) and cell lysate (input) were probed with anti-FLAG M2 and anti-GST antibodies. Binding of FLAG-X11L from sorbitol-treated cells to APPcyt was increased relative to FLAG-X11L isolated from untreated cells (compare lane 5 with 6). In contrast, APP derived from sorbitol-treated cells did not exhibit enhanced binding to purified GST-X11L (compare lane 5 with 6 in Fig. 2B). These results indicate that changes in X11L, but not in APP, were responsible for the enhanced X11L-APP association, although phosphorylation of APP at Thr668 was induced and maturation (O-glycosylation) of APP was slightly suppressed in osmotically stressed cells (see lane 6 of the “input” panel in Fig. 2B) [Nakaya & Suzuki, 2006; Nakaya, Kawai & Suzuki, 2008]. We confirmed that the phosphorylation of APP at Thr668 did not influence the interaction with X11L in sorbitol-treated cells using an APPT668A mutant in which Ala was substituted for Thr668 (Supplementary Fig. S2). HEK293 cells transiently expressing FLAG-APPT668A and HA-X11L were treated with or without sorbitol and APPT668A was recovered by immunoprecipitation with anti-FLAG antibody. Sorbitol increased the amount of HA-X11L coimmunoprecipitated with FLAG-APPT668A to the same extent as with wild-type APP (compare lanes 11 and 12 to lanes 7 and 8 in second row of Supplementary Fig. S2). Thus APP phosphorylation at Thr668 is not largely responsible for the enhanced association with X11L-APP.
Figure 2. Modification of X11L enhances the association with APP in osmotically stressed cells.
(A) Enhanced association of APP with X11L expressed in osmotically stressed cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (+; lanes 1, 2, 5, 6) or pcDNA3 (−; lanes 3, 4), and treated with (+; lanes 2, 4, 6) or without (−; lanes 1, 3, 5) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt or GST alone, and lysates (input) and proteins bound to GST-APPcyt (bound) were analyzed by immunoblotting with anti-FLAG M2 and anti-GST antibodies. Molecular weight standards (in kDa) are indicated at the left.
(B) Association of X11L with APP is not affected by APP expressed in osmotically stressed cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-APP695 (+; lanes 1, 2, 5, 6) or pcDNA3 (−; lanes 3, 4), and treated with (+; lanes 2, 4, 6) or without (−; lanes 1, 3, 5) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-X11L (lanes 3–6) or GST alone (lanes 1, 2), and lysate (input) and proteins bound to GST-X11L (bound) were analyzed by immunoblotting with anti-APP (APP/C) and anti-GST antibodies. APP indicates mature APP (mAPP) and immature APP (imAPP) forms. Molecular weight standards (in kDa) are indicated at the left.
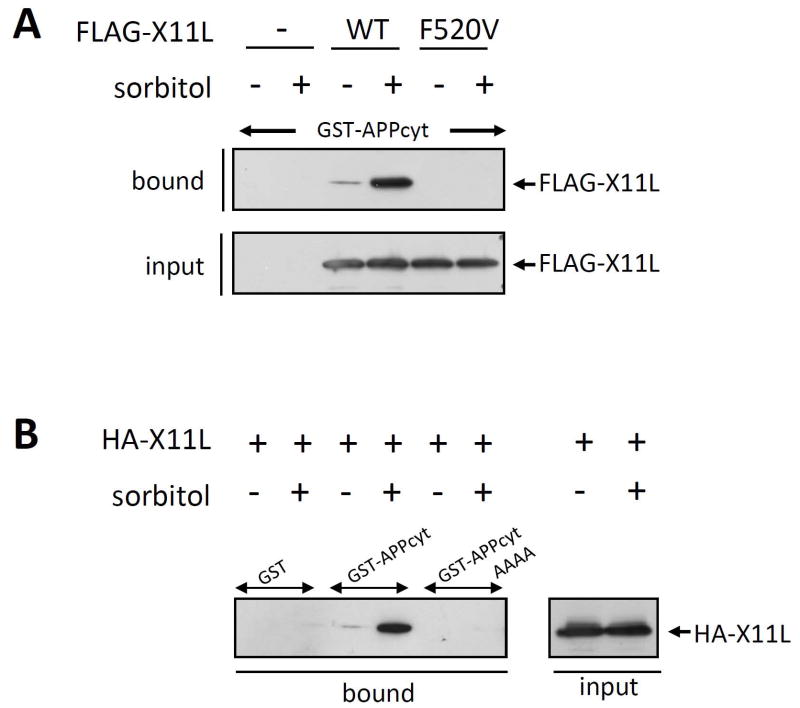
The PI/PTB domain of X11s associates with the cytoplasmic region of APP (Borg et al, 1998; Tomita et al, 1999; Tanahashi et al, 1999). Phe486 (now Phe608, given the corrected, complete amino acid sequence of X11) in the X11 PI/PTB domain is an amino acid critical for interaction with APP [Zhang et al, 1997]. Phe520 in the X11L PI/PTB domain corresponds to Phe608 of X11. FLAG-tagged X11L carrying a Val substitution for Phe520 (F520V) or wild-type X11L (WT) were expressed in HEK293 cells. Cells were treated with (+) or without (−) sorbitol treatment, and the cell lysates were analyzed for X11L-APP binding using pull-down assays. FLAG-X11LF520V was unable to associate with GST-APPcyt (Fig. 3A), indicating that X11L association with APP is mediated by the PI/PTB domain of X11L under both normal and osmotically stressed conditions. We confirmed this finding using pull-down assays with GST-APPcyt carrying the AAAA mutation, in which amino acids in the APP 684NPTY687 X11L-binding core element were altered to 684AAAA687 (Fig. 3B). HEK293 cells transiently expressing HA-tagged X11L were treated with (+) or without (−) sorbitol, and the cell lysates were incubated with GST alone, GST-APPcyt, or GST-APPcytAAAA. HA-X11L derived from cells treated with (+) or without (−) sorbitol could not bind to GST-APPcytAAAA, while HA-X11L bound to GST-APPcyt, as in Fig. 2. Taken together, the increased binding of X11L to APP in stressed cells was likely due to enhancement of a “basal” or “physiological” level of binding observed in unstressed cells, mediated by binding of the PI/PTB domain of X11L to the NPTY element of the APP cytoplasmic region.
Figure 3. Interaction of the NPTY element of APP with the PI/PTB domain of X11L recovered from osmotically stressed cells.
(A) Interaction of APPcyt with X11L and X11L PI/PTB domain mutants derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (WT), pcDNA3.1-FLAG-X11L F520V (F520V), or pcDNA3 alone (−), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and FLAG-X11L bound to GST-APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-FLAG M2 antibody.
(B) Interaction of APPcyt carrying a mutation in the NPTY element with X11L derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (+), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST alone, GST-APPcyt, or GST-APPcytAAAA in which the NPTY sequence was altered to AAAA. HA-X11L bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody.
Requirement for the amino-terminal region of X11L in its enhanced interaction with APP in stressed cells
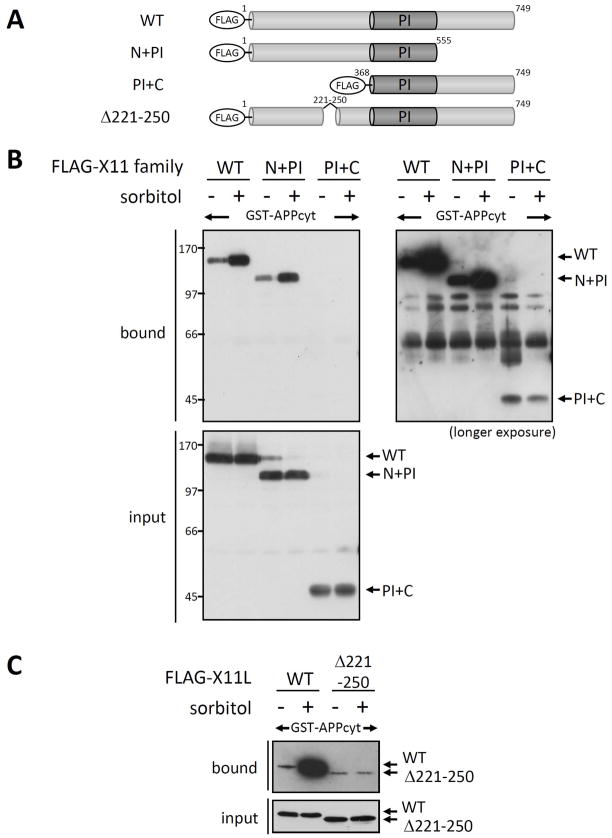
To determine the region of X11L responsible for enhanced association with APP in stressed cells, we generated constructs of X11L lacking the regions on the amino- or carboxyl-terminal side of the central PI/PTB domain, and assayed for APP binding (Fig. 4). The FLAG-tagged amino-terminal domain attached to the PI/PTB domain (N+PI, X11L1-555), the PI/PTB domain attached to the carboxyl-terminal domain (PI+C, X11L368-749), and full-length X11L (WT) were transiently expressed in HEK293 cells with or without 0.5 M sorbitol treatment for 45 min. Cell lysates were then examined for binding to GST-APPcyt using pull-down assays (Fig. 4A and B). Bound proteins and cell lysates (input) were detected by immunoblotting with anti-FLAG M2 antibody. Enhanced binding to GST-APPcyt by the full-length X11L (WT) and the N+PI construct, but not the PI+C construct, was observed. PI+C showed very weak APP binding, as described previously [Tomita et al, 1999], and association of PI+C with APPcyt was not enhanced by cellular stress (see longer exposure in Fig. 4B). These results suggest that the amino-terminal region of X11L (X11L1-368) contains a region responsible for the enhanced binding to APP. Next, we further delineated the region responsible using several amino-terminally deleted X11L proteins (unpublished observations). X11L containing a deletion of amino acids 221-250 (Δ221-250) almost completely lacked the enhanced APPcyt binding in response to sorbitol treatment (Fig. 4A and C), suggesting that X11L221-250 contains the element responsible for the enhanced X11L-APP association in osmotically stressed cells.
Figure 4. Identification of the region of X11L responsible for enhanced binding to APP in stressed cells.
(A) Schematic structure of X11L deletion mutants. The FLAG-tagged X11L proteins used in this study are shown. PI indicates the PI/PTB domain which interacts with APP. Numbers indicate the amino acid positions.
(B) Association of APPcyt with X11L lacking the N- or C-terminal region derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (WT), pcDNA3.1-FLAG-X11L N+PI (N+PI), or pcDNA3.1-FLAG-X11L PI+C (PI+C) and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and FLAG-X11L proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-FLAG M2 antibody. A longer film exposure for bound proteins is also shown (right panel). Numbers indicate molecular weight standards (kDa).
(C) Association of APPcyt with X11L containing a deletion of amino acids 221-250 derived from sorbitol treated cells. HEK293 cells (~2 × 105 cells) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (WT) or pcDNA3-FLAG-X11L Δ221-250 (Δ221-250), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and FLAG-X11L proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-FLAG M2 antibody.
Reversible modification of X11L is involved in its enhanced association with APP in stressed cells
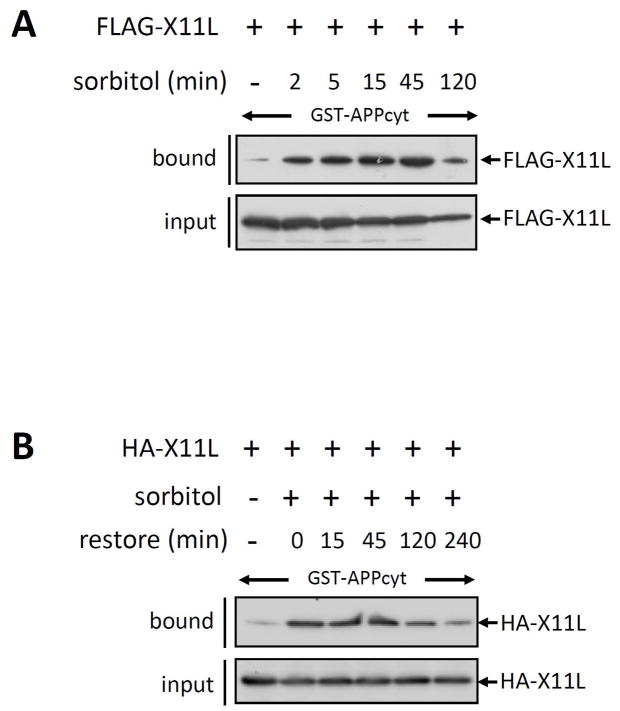
To investigate whether the enhanced X11L-APP association is reversible, we performed X11L pull-down assay with GST-APPcyt using X11L derived from sorbitol treated cells, which were further cultured in medium from which sorbitol was withdrawn (Fig. 5). Maximal binding of FLAG-X11L to GST-APPcyt was detected with FLAG-X11L derived from cells treated with 0.5 M sorbitol for 45 min (Fig. 4A). Therefore, cells expressing HA-X11L were treated with (+) or without (−) 0.5 M sorbitol for 45 min and then cultured for 0, 15, 45, 120 or 240 min in medium lacking sorbitol (Fig. 4B). The enhanced association (0 h) of HA-X11L with GST-APPcyt was maintained for 45 min after sorbitol withdrawal, but decreased thereafter. These observations suggested that a reversible posttranslational modification of X11L, such as phosphorylation, may occur.
Figure 5. Reversibility of the enhanced association of X11L with APP by withdrawal of osmotic stress.
(A) Time course of enhanced association of X11L with APP in cells treated with 0.5 M sorbitol. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (+), and treated with (2, 5, 15, 45 and 120 min) 0.5 M sorbitol or left untreated (−). The cell lysates were incubated with GST-APPcyt, and lysates (input) and FLAG-X11L bound to GST-APPcyt (bound) were analyzed by immunoblotting with anti-FLAG M2 antibody.
(B) Effect of sorbitol withdrawal on the enhanced association of X11L with APP. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (+), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cells were then further cultured for 0, 15, 45, 120 and 240 min in the absence of sorbitol (restore). The cell lysates were incubated with GST-APPcyt, and HA-X11L bound to GST-APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody.
Our previous studies have indicated that X11L is phosphorylated in primary cultured neurons (unpublished observation). In cultured cells, we found that X11L was phosphorylated at multiple sites, with the majority of the sites being located within the amino-terminal region, but not in the PI/PTB or carboxyl-terminal PDZ domains (Supplementary Fig. S3). In addition, we analyzed the effect of the osmotic stress on X11L phosphorylation and found that phosphorylation of X11L was altered in sorbitol-treated cells (Supplementary Fig. S3). This result suggests that phosphorylation of X11L may contribute to the enhanced association with APP in stressed cells.
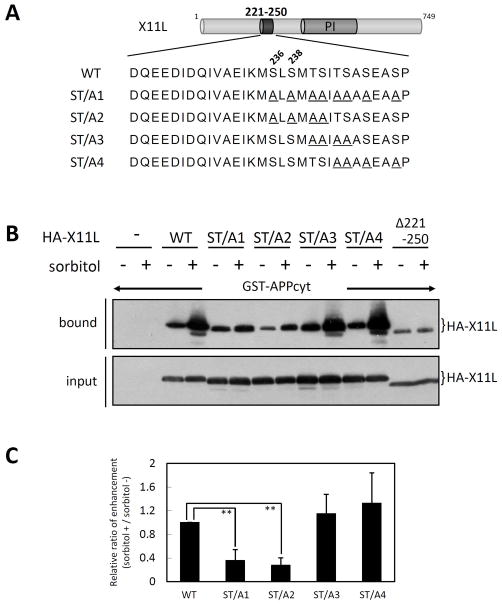
There are eight seryl and threonyl residues in the carboxyl-half of X11L221-250 which might be phosphorylated. HA-tagged X11L constructs carrying Ala substitutions (Fig. 6A) were transiently expressed in HEK293 cells. Cells were treated with (+) or without (−) sorbitol, and pull-down assays with cell lysates and GST-APPcyt were performed (Fig. 6B). The ST/A1 (all eight seryl and threonyl residues replaced with Ala) and ST/A2 (amino-terminal four seryl and threonyl residues replaced with Ala) mutants of X11L bound to APPcyt, but the enhancement of APP binding in stressed cells was diminished, similar to X11LΔ221-250. In contrast, the ST/A3 (central four threonyl and seryl residues replaced with Ala) and ST/A4 (carboxyl-terminal four threonyl and seryl residues replaced with Ala) mutants of X11L retained their ability to bind APP and showed enhanced APP-binding in response to sorbitol treatment. The relative ratio of enhanced binding of the X11L mutants to APP is shown in Fig. 6C. These assays suggested that the first two seryl residues, Ser236 and Ser238, in the carboxyl-half of X11L221-250 play a role in mediating the enhanced binding to APP in osmotically stressed cells.
Figure 6. Dissection of the role of the X11L221-250 region in enhanced association with APPcyt.
(A) Sequences of amino acid mutations in the X11L221-250 region. Alanyl residues substituted for seryl and threonyl residues are underlined. WT indicates wild-type X11L. PI indicates the PI/PTB domain.
(B) Association of APPcyt with X11L mutants derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (WT), pcDNA3-HA-X11L ST/A1 (ST/A1), pcDNA3-HA-X11L ST/A2 (ST/A2), pcDNA3-HA-X11L ST/A3 (ST/A3), pcDNA3-HA-X11L ST/A4 (ST/A4), or pcDNA3-HA-X11L Δ221-250 (Δ221-250), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and HA-X11L proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody.
(C) Quantification of X11L proteins bound to APPcyt. X11L proteins bound to APPcyt were quantified using the VersaDoc imaging system (Bio-Rad). The data are presented as the ratio (sorbitol (+)/sorbitol (−)) relative to the level of wild type X11L-binding (WT, 1.0). Results represent means ± s.d. for n=5 experiments. **, p<0.001 by Student’s t-test.
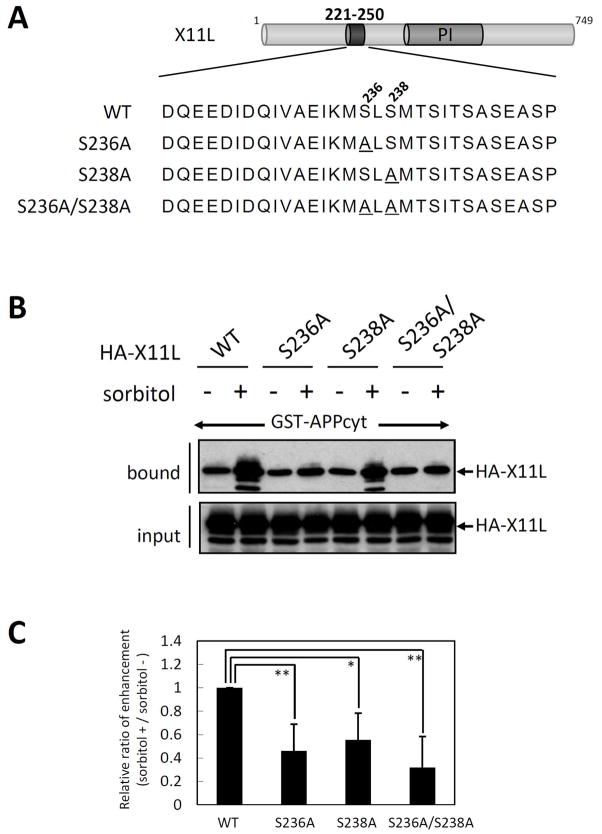
To determine whether each seryl residue, or both together, contributed to the enhanced binding to APP in stressed cells, HA-X11L mutants carrying Ala substitutions for Ser236, Ser238, or both were expressed in HEK293 cells. Cells were treated with (+) or without (−) sorbitol, and the cell lysates were examined for binding to GST-APPcyt (Fig. 7). X11LS236A (S236A) and X11LS238A (S238A) both exhibited a decrease in the enhanced binding to APP when compared to wild-type X11L (WT). X11LS236A/S238A(S236A/S238A) also exhibited a significantly reduced binding enhancement in response to sorbitol treatment. These results show that both Ser236 and Ser238 are required to enhance the association of X11L with APP in stressed cells.
Figure 7. Role of Ser236 and Ser238 in X11L221-250 in the enhanced association with APPcyt.
(A) Sequences of amino acid substitution mutants at Ser236 and/or Ser238. Alanyl residues substituted for Ser236 (S236A) and Ser238 (S238A) are underlined. WT indicates the wild-type X11L. PI indicates the PI/PTB domain.
(B) Association of APPcyt with X11L mutants derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (WT), pcDNA3-HA-X11L S236A (S236A), pcDNA3-HA-X11L S238A (S238A), or pcDNA3-HA-X11L S236A/S238A (S236A/S238A), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and HA-X11L proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody.
(C) Quantification of X11L proteins bound to APPcyt. X11L proteins bound to APPcyt were quantified with the VersaDoc imaging system (Bio-Rad). The data are presented as the ratio (sorbitol (+)/sorbitol (−)) relative to the level of wild type X11L-binding (WT, 1.0). Results represent means ± s.d. for n=4 experiments. *, p<0.05, **, p<0.01 by Student’s t-test.
We expected that amino acid substitution of Asp for both Ser236 and Ser238 of X11L (S236D/S238D) might enhance the association with APP since Asp or Glu substitution for Ser or Thr residues in a protein sometimes can mimic the phosphorylated state of the protein by increasing negative charge [Tomita et al., 2005]. Wild-type HA-X11L or an HA-X11L mutant carrying Asp substitutions at both Ser236 and Ser238 (X11LS236D/S238D) were expressed in HEK293 cells and cells were treated with or without sorbitol. Cell lysates were then subjected to binding to GST-APPcyt (Supplementary Fig. S4). Like X11LS236A/S238A, there was no enhanced binding of X11LS236D/S238D to GST-APPcyt following treatment with sorbitol. The results suggest that Asp substitution for Ser236 and Ser238 did not mimic the effect of phosphorylation on the regulation of the association of X11L with APP.
We next analyzed whether Ser236 and/or Ser238 can be phosphorylated in sorbitol-treated cells. We developed phosphorylation-state specific antibodies against peptides containing the phosphorylated Ser236 and/or Ser238 sequences. The UT158 antibody recognizes pSer236 predominantly, whereas the UT162 antibody recognizes pSer238 (Supplementary Figure S1A). The phosphorylation state-specificity of these antibodies was demonstrated by treatment of lysates with λ-phosphatase, which eliminated the signal from phosphorylated X11L (Supplementary Figure S1B).
HEK293 cells transiently expressing FLAG-X11L carrying the S236A, S238A, or Δ221-250 mutations, as well as FLAG-X11L wild-type (WT), were treated with (+) or without (−) sorbitol and cell lysates were analyzed by immunoblotting with UT158 and UT162 (Fig. 8). X11LWT was phosphorylated at both Ser236 and Ser238. In response to sorbitol treatement, increased phosphorylation at Ser236 (upper row), but not Ser238 (middle row), was observed in cells expressing X11LWT. Interestingly, Ser236 phosphorylation was very weak in the X11LS238A mutant, suggesting that the mutation at or phosphorylation state of Ser238 may influence the phosphorylation of Ser236. Taken together, both Ser236 and Ser238 were phosphorylated in cells, and the increased phosphorylation at Ser236 may enhance the association of X11L with APP under stress conditions.
Figure 8. Phosphorylation of Ser236 and Ser238 in normal and osmotically stressed cells.
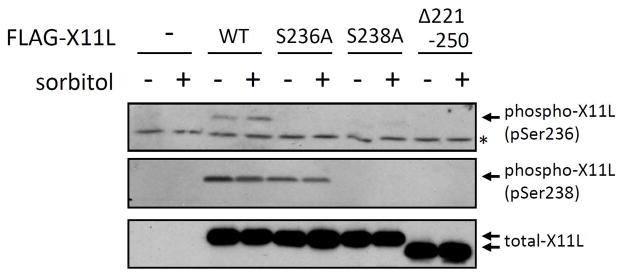
HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3.1-FLAG-X11L (WT), pcDNA3.1-FLAG-X11L S236A (S236A), pcDNA3.1-FLAG-X11 S238A (S238A), pcDNA3.1-FLAG-X11 S236A/S238A (S236A/S238A), or pcDNA3-FLAG-X11Δ220-250, and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were analyzed by immunoblotting with anti-pSer236 UT158 (pSer236), anti-pSer238 UT162 (pSer238), and anti-FLAG M2 (total-X11L) antibodies. *, non-specific band recognized by anti-pSer236 UT158.
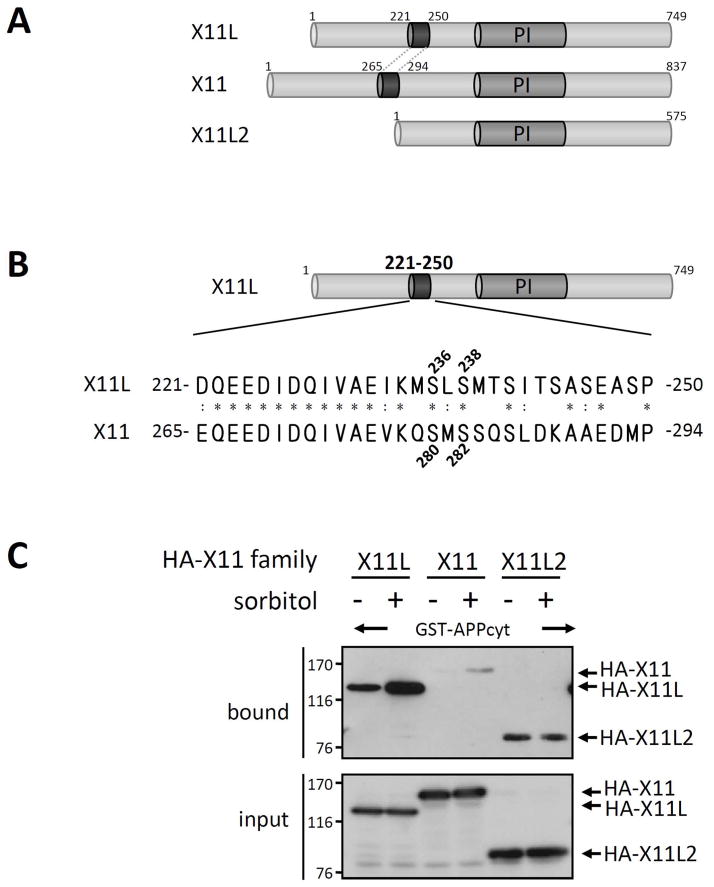
Neuron-specific X11 and X11L, but not the ubiquitously expressed X11L2, displayed enhanced association with APP
The X11 family consists of neuronally expressed X11 and X11L, and the ubiquitously expressed X11L2 (Fig. 9A). The amino acid sequence of X11L221-250, including Ser236 and Ser238, are highly conserved in X11265-294, but not in X11L2 (Fig. 9B). We examined whether X11 and X11L2 exhibit an enhanced association with APP in response to sorbitol treatment of cells using pull-down assays (Fig. 9C). HEK293 cells expressing HA-tagged X11L, X11, or X11L2 were treated with (+) or without (−) sorbitol. The cell lysates were incubated with GST-APPcyt, and the bound proteins and cell lysate (input) were probed with anti-HA. Although X11 binding to APP was weak in unstressed cells (−), HA-X11 binding to GST-APPcyt was enhanced when HA-X11 derived from stressed cells was used. In contrast to X11 and X11L, X11L2 binding to APPcyt was not enhanced by sorbitol treatment. These results indicate that the enhanced association of X11s with APP depends on Ser236 and Ser238 in X11L and Ser280 and Ser282 in X11, and suggest that the neuronal X11 and X11L utilize a common mechanism to regulate their interactions with APP.
Figure 9. Interaction of APPcyt with X11L, X11, and X11L2 derived from sorbitol treated cells.
(A) Schematic structures of X11L, X11 and X11L2. Human X11 family proteins are shown. PI indicates the PI/PTB domain, which associates with APP. Numbers indicate amino acid positions.
(B) Amino acid sequence in the highly conserved regulatory region of X11L221-250 and X11265-294. Positions of Ser236 and Ser238 in X11L221-250 and Ser280 and Ser282 in X11265-294 are shown. The protein sequences are presented using the single-letter amino acid code. Amino acid residues of X11 identical (*) or similar (:) to X11L are indicated.
(C) Association of APPcyt with X11, X11L and X11L2 derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (X11L), pcDNA3.1-HA-X11 (X11), or pcDNA3.1-HA-X11L2 (X11L2), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and HA-X11 family proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody. Numbers indicate molecular weight standards (kDa).
X11L phosphorylated in brain shows the enhanced binding to APP
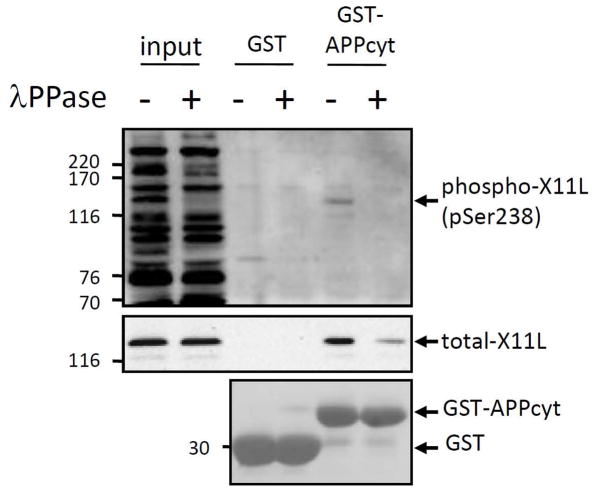
We next examined whether X11L is phosphorylated physiologically in brain by immunoblotting with the phosphorylation state-specific antibodies to pSer236 and pSer238. We detected X11L phosphorylated at Ser238 with mouse brain lysate (Fig. 10), while the phosphorylation at Ser236 was not detectable (data not shown). To test whether the phosphorylation of X11L effected its binding to APP, we performed GST pull-down assay using mouse brain lysates containing endogenously phosphorylated X11L. Lysate prepared from adult mice brains was treated with (+) or without (−) λ protein phosphatase (λPPase) and incubated with the purified GST-APPcyto or GST alone (Fig. 10). Bound X11L was probed with anti-X11L (middle panel) and anti-pSer238 (top panel) antibodies. Dephosphorylated of X11L decreased binding to APPcyt, indicating that phosphorylation of X11L regulates APP-binding in the brain with Ser238 in X11L221-250 being one of the likely sites involved.
Figure 10. Suppressed binding to APP by dephosphorylation of X11L derived from mouse brain.
Brains from adult male mice (C57/BL6, 2–3 months old) were lysed and treated with (+) or without (−) λPPase. The lysates (50 μg protein; input) were incubated with GST-APPcyt or GST alone, and bound proteins were eluted and analyzed by immunoblotting with anti-phosphoSer238 (UT162, top panel, phospho-X11L) and anti-X11L (4A10, middle panel, total-X11L) antibodies. The amounts of GST-APPcyt and GST alone were estimated by staining of membrane with ponceau S (bottom panel). Molecular weight standards (in kDa) are indicated at the left.
Discussion
APP interacts with several adaptor proteins in the X11, JIP, and FE65 families through its cytoplasmic region, and these interactions play critical roles in the intracellular trafficking and metabolism of APP and may be implicated in AD pathogenesis [reviewed in Suzuki et al, 2006; Suzuki & Nakaya, 2008, Borg et al, 1998; Tomita et al, 1999; Tanahashi & Tabira, 1999; Taru et al, 2002; MeLoughlin et al, 1999; Fiore et al, 1995; Araki et al, 2007].
The cytoplasmic region of APP contains at least two functional motifs, 667VTPEER672 and 681GYENPTY687 [reviewed in Suzuki et al, 2006; Suzuki & Nakaya, 2008]. The 667VTPEER672 motif forms a type I β-turn and amino-terminal helix-capping box structure, which contribute to stabilization of the overall structure of the cytoplasmic region of APP [Ramelot, Gentile & Nicholson, 2000]. Phosphorylation at Thr668 in this motif induces a significant conformational change in the cytoplasmic domain of APP [Ando et al, 2001; Ramelot & Nicholson, 2001] and regulates its association with FE65, a neural adaptor protein that may be a signal transducer [Ando et al, 2001; Nakaya & Suzuki, 2006; Nakaya, Kawai & Suzuki, 2008]. The 681GYENPTY687 motif contains the NPXY core element, a typical internalization signal in membrane proteins [Chen, Goldstein & Brown, 1990; Norstedt et al, 1993], to which several adaptor proteins such as the X11, JIP, and FE65 families bind [Borg et al, 1998; Tomita et al, 1999; Tanahashi & Tabira, 1999, Taru et al, 2002; MeLoughlin et al, 1999; Fiore et al, 1995].
X11 and X11L are APP-binding proteins which can suppress amyloidogenic metabolism in the brain in vivo [Lee et al, 2003 & 2004; Sano et al, 2006; Saito et al, 2008], but the mechanism which regulates X11L binding to APP remains unclear. In the present study, we identified an enhanced association of X11L and X11 with APP in osmotically stressed cells. In sorbitol treated cells: (i) enhanced association of X11L with APP is regulated by modification of X11L alone; (ii) the regulatory region is located in the region amino-terminal to the PI/PTB domain, which binds APP; (iii) Ser236 and Ser238 within the highly conserved X11L221-250 region are the amino acids mediating enhanced association with APP; (iv) both seryl residues are phosphorylatable and phosphorylation of either seryl residue may regulate the enhanced association of X11L with APP. Furthermore, in brain; (v) X11L is phosphorylated at least at Ser238; (vi) endogenously phosphorylated X11L binds to APP stronger than dephosphorylated X11L. These data provide the first evidence for that modification of X11L is involved in regulation of its binding to APP.
Our previous results implied that the presence of the amino-terminal region of X11L somehow affected binding to APP through the PI/PTB domain [Tomita et al, 1999]. Thus, the changes in phosphorylation state in that region may alter the conformation of the PI/PTB domain and lead to enhanced association with APP. The regulatory region of X11L221-250, including Ser236 and Ser238, is conserved in X11265-294, but not in X11L2. X11 and X11L respond to osmotic stress by enhanced APP binding, whereas X11L2 does not. This result suggests that this regulatory region plays a significant role in the function of the neuronal X11 and X11L, but not the ubiquitously expressed X11L2.
Although we proposed here that the regulatory region of X11L is distinct from the APP binding domain, and that the interaction between APP and X11L is modified by phosphorylation within the regulatory region, there are several issues that remain for future studies. At this point, we have largely studied this regulatory mechanism in a model cell system, in which the changes were induced by an osmotic stress stimulus. It will be important to characterize the phosphorylation of X11L in neurons, especially the temporal and spatial regulation of Ser236 and/or Ser238 phosphorylation. Such data would indicate how the interaction between APP and X11L is dynamically regulated in vivo. In the current study, we detected X11L phosphorylated at Ser238 in brain, while Ser236 was not detected with the available phospho-antibody. This observation suggests that phosphorylation at Ser238 alone may be sufficient to influence the the interaction of APP and X11L in brain. It will also be important to identify the kinase(s) involved in the phosphorylation of Ser236 and Ser238. In preliminary studies, other osmolytes such as mannitol or NaCl showed an effect identical to sorbitol, while genotoxic stresses such as H2O2 showed a weaker effect on binding of X11L to APP (data not shown). Although genotoxic stresses activate c-Jun NH2-terminal kinase (JNK) cascade [Ip & Davis, 1998], protein kinases in JNK cascade did appear to participate in the phosphorylation of Ser236 and/or Ser238 (unpublished observation).
X11 and X11L are APP-binding proteins which can suppress amyloidogenic metabolism in the brain in vivo [Lee et al, 2003 & 2004; Sano et al, 2006; Saito et al, 2008]. Our observations raise the possibility that insufficient association of APP with X11 and/or X11L caused by aberrant phosphorylation of Ser236 and/or Ser238 in brain may increase amyloidogenic metabolism of APP and trigger AD pathologies.
Supplementary Material
Acknowledgments
This study was supported by Grants-in-Aid for Scientific Research 207900049 (T.N), 20012001 (T.S.), 207900049 (T.S), and by The Naito Foundation (T.S.). M.S is a recipient of research fellowships from the Japan Society for the Promotion of Science for young scientists. ACN was supported by NIH (AG 09464).
Abbreviations
- AD
Alzheimer’s disease
- APP
amyloid β-protein precursor
- CHAPS
3-[(3-cholamidopropyl)dimethylammonil]-1-propanesulfonic acid
- PBS
phosphate buffered saline
- X11L
X11-like
- HA
hemaglutinin
References
- Ando K, Oishi M, Takeda S, Iijima K, Isohara T, Nairn AC, Kirino Y, Greengard P, Suzuki T. Role of phosphorylation of Alzheimer’s amyloid precursor protein during neuronal differentiation. J Neurosci. 1999;19:4421–4427. doi: 10.1523/JNEUROSCI.19-11-04421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of β-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M104059200. [DOI] [PubMed] [Google Scholar]
- Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid β-protein precursor metabolism. J Biol Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- Araki Y, Miyagi N, Kato N, Yoshida T, Wada S, Nishimura M, Komano H, Yamamoto T, De Strooper B, Yamamoto K, Suzuki T. Coordinated metabolism of Alcadein and amyloid β-protein precursor regulates FE65-dependent gene transactivation. J Biol Chem. 2004;279:24343–24354. doi: 10.1074/jbc.M401925200. [DOI] [PubMed] [Google Scholar]
- Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, Suzuki T. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg JP, Yang Y, De Taddeo-Borg M, Margolis B, Turner RS. The X11α protein slows cellular amyloid precursor protein processing and reduces Aβ40 and Aβ42 secretion. J Biol Chem. 1998;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- Borg JP, Lopez-Figueroa MO, de Taddeo-Borg M, Kroon DE, Turner RS, Watson SJ, Margolis B. Molecular analysis of the X11-mLin-2/cask complex in brain. J Neurosci. 1999;19:1307–1316. doi: 10.1523/JNEUROSCI.19-04-01307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- Gandy S. The role of cerebral amyloid β accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Morishita W, Hammer RE, Malenka RC, Sudhof TC. A role for Mints in transmitter release: Mint 1 knockout mice exhibit impaired GABAergic synaptic transmission. Proc Natl Acad Sci U S A. 2003;100:1409–1414. doi: 10.1073/pnas.252774899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Davis RJ. Signal transduction by the c-JUN N-terminal kinase (JNK)-from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lau KF, Perkinton MS, Standen CL, Shemilt SJ, Mercken L, Cooper JD, McLoughlin DM, Miller CC. The neuronal adaptor protein X11α reduces Aβ levels in the brains of Alzheimer’s APPswe Tg2576 transgenic mice. J Biol Chem. 2003;278:47025–47029. doi: 10.1074/jbc.M300503200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lau KF, Perkinton MS, Standen CL, Rogelj B, Falinska A, McLoughlin DM, Miller CC. The neuronal adaptor protein X11β reduces amyloid β-protein levels and amyloid plaque formation in the brains of transgenic mice. J Biol Chem. 2004;279:49099–49104. doi: 10.1074/jbc.M405602200. [DOI] [PubMed] [Google Scholar]
- McLoughlin DM, Irving NG, Brownlees J, Brion JP, Leroy K, Miller CC. Mint2/X11-like colocalizes with the Alzheimer’s disease amyloid precursor protein and is associated with neuritic plaques in Alzheimer’s disease. Eur J Neurosci. 1999;11:1988–1994. doi: 10.1046/j.1460-9568.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Miller CC, McLoughlin DM, Lau KF, Tennant ME, Rogelj B. The X11 proteins, Aβ production and Alzheimer’s disease. Trends Neurosci. 2006;29:280–285. doi: 10.1016/j.tins.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Suzuki T. Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes Cells. 2006;11:633–645. doi: 10.1111/j.1365-2443.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Kawai T, Suzuki T. Regulation of FE65 nuclear translocation and function by amyloid β-protein precursor in osmotically stressed cells. J Biol Chem. 2008;283:19119–19131. doi: 10.1074/jbc.M801827200. [DOI] [PubMed] [Google Scholar]
- Norstedt C, Caporaso GL, Thyberg J, Gandy E, Greengard P. Identification of the Alzheimer β/A4 amyloid precursor protein in clathrin-coated vesicles purified from PC12 cells. J Biol Chem. 1993;268:608–612. [PubMed] [Google Scholar]
- Ramelot TA, Gentile LN, Nicholson LK. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Nicholson LK. Phosphorylation-induced structural changes in the amyloid precursor protein cytoplasmic tail detected by NMR. J Mol Biol. 2001;307:871–884. doi: 10.1006/jmbi.2001.4535. [DOI] [PubMed] [Google Scholar]
- Saito Y, Sano Y, Vassar R, Gandy S, Nakaya T, Yamamoto T, Suzuki T. X11 proteins regulate the translocation of amyloid β-protein precursor (APP) into detergent-resistant membrane and suppress the amyloidogenic cleavage of APP by β-site cleaving enzyme in brain. J Biol Chem. 2008;283:35763–35771. doi: 10.1074/jbc.M801353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y, Syuzo-Takabatake A, Nakaya T, Saito Y, Tomita S, Itohara S, Suzuki T. Enhanced amyloidogenic metabolism of the amyloid β-protein precursor in the X11L-deficient mouse brain. J Biol Chem. 2006;281:37853–37860. doi: 10.1074/jbc.M609312200. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Araki Y, Yamamoto T, Nakaya T. Trafficking of Alzheimer’s disease-related membrane proteins and its participation in disease pathogenesis. J Biochem. 2006;139:949–955. doi: 10.1093/jb/mvj121. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nakaya T. Regulation of APP by phosphorylation and protein interactions. J Biol Chem. 2008;283:29633–29637. doi: 10.1074/jbc.R800003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi H, Tabira T. X11L2, a new member of the X11 protein family, interacts with Alzheimer’s β-amyloid precursor protein. Biochem Biophys Res Commun. 1999;255:663–667. doi: 10.1006/bbrc.1999.0265. [DOI] [PubMed] [Google Scholar]
- Taru H, Suzuki T. Facilitation of stress-induced phosphorylation of β-amyloid precursor protein family members by X11-like/Mint2 protein. J Biol Chem. 2004;279:21628–21636. doi: 10.1074/jbc.M312007200. [DOI] [PubMed] [Google Scholar]
- Taru H, Iijima K, Hase M, Kirino Y, Yagi Y, Suzuki T. Interaction of Alzheimer’s β-amyloid precursor family proteins with scaffold proteins of the JNK signaling cascade. J Biol Chem. 2002;277:20070–20078. doi: 10.1074/jbc.M108372200. [DOI] [PubMed] [Google Scholar]
- Tomita S, Ozaki T, Taru H, Oguchi S, Takeda S, Yagi Y, Sakiyama S, Kirino Y, Suzuki T. Interaction of a neuron-specific protein containing PDZ domains with Alzheimer’s amyloid precursor protein. J Biol Chem. 1999;274:2243–2254. doi: 10.1074/jbc.274.4.2243. [DOI] [PubMed] [Google Scholar]
- Tomita S, Stein V, Stocker TJ, Nicoll RA, Bredt DS. Bidirectional synaptic plasticity regulated by phosphorylation of stargazing-like TARPs. Neuron. 2005;45:269–277. doi: 10.1016/j.neuron.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lee CH, Mandiyan V, Borg JP, Margolis B, Schlessinger J, Kuriyan J. Sequence-specific recognition of the internalization motif of the Alzheimer’s amyloid precursor protein by the X11 PTB domain. EMBO J. 1997;16:6141–6150. doi: 10.1093/emboj/16.20.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.