Abstract
Background
Epstein-Barr virus (EBV)-mediated lymphomagenesis in the setting of HIV infection has been widely accepted. However, little is known about how EBV impacts prognosis. We investigated the hypothesis that EBV infection is associated with expression of specific B-cell oncogenic markers in HIV-related diffuse large B-cell lymphoma (DLBCL), and examined the prognostic utility of detecting EBV infection.
Study Design
HIV-related DLBCL cases diagnosed between 1996–2007 within Kaiser Permanente California were identified. Immunohistochemistry staining was used to analyze the expression of selected markers that are cell cycle regulators, B-cell activators, and anti-apoptotic proteins among others. EBV infection was determined by in situ hybridization of EBV RNA. Correlations between EBV and marker expression were examined using Spearman’s correlation coefficient. The prognostic utility of EBV status was examined in multivariable Cox model adjusting for international prognostic index (IPI). Receiver-operating characteristics (ROC) analysis was used to evaluate improvement in model discrimination.
Results
Seventy HIV-related DLBCL cases were included (31% EBV+). EBV+ tumor was associated with increased expression of BLIMP1 and CD30, and reduced expression of BCL6 and LMO2. EBV+ tumor was independently associated with elevated 2-year overall mortality [hazard ratio=3.3 (95% CI: 1.6–6.6)]. Area under the ROC curve demonstrated improved model discrimination when incorporating tumor EBV status with IPI in the prediction model [0.65 vs. 0.74 (IPI only)].
Conclusion
Our results suggest that EBV infection was associated with expression of several tumor markers that are involved in the NF-κB pathway, and that detecting tumor EBV status may have prognostic utility in HIV-related DLBCL.
Keywords: EBV, Lymphoma, HIV, Diffuse large B-cell lymphoma, Prognosis
Introduction
HIV-infected persons remain at significantly elevated risk for developing non-Hodgkin lymphoma (NHL) in the era of combined antiretroviral therapy (cART)(1). Compared to NHL in persons without HIV infection, HIV-related NHL often presents at an advanced disease stage, frequently with extranodal involvement, and has an aggressive clinical course(2). Epstein-Barr virus (EBV) has been implicated in the development of many non-Hodgkin lymphomas (NHL) subtypes in HIV-infected individuals(3), including the most common subtype of diffuse large B-cell lymphoma (DLBCL). EBV is associated with HIV-related DLBCL in 30–60% of cases(4, 5), compared with only 10% in the general population(6, 7).
EBV is a ubiquitous γ-herpesvirus that infects most individuals early in life(8). In healthy adults, the infection is controlled by the body’s anti-viral T-cell response(9). However, EBV maintains a latent lifetime infection in B lymphocytes. Lack of functional immunoregulation is the key risk factor for EBV-mediated lymphomagenesis(10, 11). Studies have shown that altered EBV antibody patterns and detectable serum viral levels precede the onset of NHL(12–15) and the loss of EBV specific T-cell immunity is seen to precede the development of EBV-positive HIV-related DLBCL(16, 17). In vitro, EBV causes B cells to transform into lymphoblastoid cell lines in the absence of T cell immune responses to this virus(10).
While EBV-mediated lymphomagenesis in the setting of HIV infection has been widely accepted, little is known about how EBV impacts prognosis. It is thought that EBV contributes to the B cell cancer pathogenesis by expressing EBV-encoded transforming proteins (e.g., LMP1) as well as enhancing genetic instability through mutation, translocation and dysregulated expression of proto-oncogenes(9, 18). EBV-induced genetic instability, in turn, may also predispose to poorer prognosis of the lymphoma. In the general population, it has been reported that EBV-associated tumor was associated with shorter survival in DLBCL patients(6, 7). However, the prognostic role of EBV in HIV-infected patients with DLBCL has not been extensively examined. In this study, we examined the association between tumor EBV infection and the expression of a number of B-cell oncogenic/prognostic markers, as well as the prognostic utility of detecting tumor EBV infection in our cohort of HIV-infected patients with DLBCL.
Methods
Study Design, Population and Setting
We conducted an observational cohort study of incident HIV-related DLBCL cases diagnosed between 1996 and 2007 in the Kaiser Permanente (KP) Southern and Northern California Health Plans. These health plans are large integrated health care delivery systems providing comprehensive medical services to more than six million health plan members, representing roughly 30% of insured Californians in the most populated areas. DLBCL cases were ascertained from KP’s Surveillance, Epidemiology, and End Results-affiliated cancer registries. Cancer case ascertainment is considered highly valid since reporting of cancers is mandated under state law. The KP cancer registries include data on histopathology, cancer stage, tumor size, extension, extranodal involvement and initial course of treatment. DLBCL diagnoses were identified by International Classification of Disease (ICD)-Oncology version 3 histology code 9678–9680, 9684, 9675.
HIV infection status was identified through record linkage with KP’s HIV registries, which include all known cases of HIV infection dating back to the early 1980’s for Kaiser Permanente Northern California and 2000 for Kaiser Permanente Southern California. HIV-infected individuals are initially identified for inclusion in the registries by a positive HIV antibody test, detectable HIV viral ribonucleic acid (RNA), prescription for an HIV antiretroviral, HIV/AIDS-related diagnosis, or other evidence of HIV infection from electronic sources. Confirmation of cases is done by medical chart review and comparisons of case lists with KP HIV clinics. HIV-infected patients diagnosed with all stages of DLBCL, of both genders, and aged over 18 years were eligible for the study. The appropriate KP institutional review boards approved this study and provided waivers of informed consent.
Pathology Review and Tissue Microarray Construction
The study pathologist (Said J and Zha H) reviewed all pathology reports associated with the DLBCL diagnosis to select accessions appropriate for laboratory analysis. Archived tumor specimens were retrieved and hematoxylin and eosin stained (H&E) slides were reviewed to confirm the DLBCL diagnosis as well as to identify representative tumor blocks for tissue microarray (TMA) construction (at the UCLA Core Microarray Facility). Tumor blocks at risk for exhaustion were excluded from TMA construction. Using an H&E slide from the representative block, the most tumor-rich areas were circled. The H&E slide was matched up with the paraffin tumor block to determine the areas of the block to be included in the TMA. Whenever possible three 0.6-mm cores from different areas of the donor block were obtained from each case and inserted in a grid pattern into a recipient paraffin block using a tissue arrayer (Beecher Instruments, Silver Spring, MD). Sections of 5 μm were then cut from each TMA and dried for 16 hours at 56°C before being dewaxed in xylene and rehydrated through a graded ethanol series and washed with phosphate-buffered saline.
EBV Status and Tumor Marker Expression
EBV infection was determined by in situ hybridization of EBV encoded RNA and was considered positive if ≥75% of the DLBCL cells had detectable EBV. Immunohistochemistry staining was performed on TMA cores to analyze the expression of selected B-cell oncogenic markers in the following categories: (1) cell cycle promoters, including cyclin D2, cyclin E, cMYC, p27, SKP2; (2) B-cell activators/differentiation, including BCL6, FOXP1, PKC-beta 2, CD21 and CD10; (3) apoptotic regulators, including BCL2, p53, survivin, BAX, GAL3, and BLIMP1; and (4) others, including MUM1, Ki-67, CD44, CD30, CD43, LMO2, and MMP9. Expression of CD10, MUM1 and BCL6 were used to determine the germinal center (GC) phenotype using the Hans’ algorithm(19). In addition to the 25 markers listed above, immunohistochemical detection of EBV latent membrane protein-1 (LMP1) was also performed. Percent of DLBCL cells with visible marker staining, including that for EBV, was scored on a scale from 0–4 (0: 0–9%, 1: 10–24%, 2: 25–49%, 3: 50–74% and 4: ≥75%). Scoring was performed manually by a study pathologist for all markers except for Ki-67, which was scored on a computerized automated platform.
Immunohistochemistry Staining
Sections from paraffin-embedded blocks were cut at 4 μm and paraffin removed with xylene and rehydrated through graded ethanols. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 10 min. Heat-induced antigen retrieval and proteolytic induced epitope retrieval were used. Following this pretreatment the slides were incubated with primary antibodies for the markers of interest. The signal was detected using the Dakocytomation Envision⊕ System labeled polymer horseradish perixoidae (HRP) anti-mouse or anti- rabbit (DakoCytomation); or MACH 2 Rabbit/Mouse HRP Polymer (Biocare Medical). For Gal 3 and Blimp1, the sections were incubated with secondary rabbit and rat immunoglobulin for 30 min at 1:200 dilution (DakoCytomation) followed by a 30 min incubation with Dakocytomation Envision⊕ System labeled Polymer HRP anti-rabbit. Novolink Polymer Detection System (Leica) was used for LMO2. For MMP9, CSA II System/HRP, Mouse (DakoCytoation) combined with CSA II Rabbit Link (DaKoCytomation) was used. All staining was performed manually. Detailed information on antibody source, pre-treatment, dilution and incubation for all markers is presented in Table 1. For quality control, normal tonsillar lymphoid tissue was used as positive controls. Negative controls for each case consisted of substituting the primary antibody with isotype specific non-cross reacting antibody matching the primary antibody. Laboratory staff who performed the staining procedures was blinded to the outcome status of each subject.
Table 1.
Antibody source, pre-treatment, dilution and incubation duration for tumor markers
| Primary Antibody | Clone | Species | Manufacturer | Catalog# | Pretreatment | Dilution | Diluent | Incubation Duration | Secondary Antibody | Detection Method |
|---|---|---|---|---|---|---|---|---|---|---|
| FOXP-1 | RABBIT | Sigma Aldrich, St. Louis, MO | HPA003876-100UL | VS6 *1 | 1/50 | BSA | OVN | MACH-2 Rabbit | ||
| SKP2 | 2C8D9 | MOUSE | Invitrogen, Grand Island, NY | 18-7334 | VSE *2 | 1/50 | BSA | 90 min | MACH-2 MOUSE | |
| CD44 | IM7 | MOUSE | EBIOSCIENCE, San Diego, CA | 14-0441 | VS6 *1 | 1/200 | BSA | 45′ | ENV MOUSE | |
| P53 | 1801 | MOUSE | ONCOGENE, Gibbstown, NJ | OP09-100ug | VS6 | 1/200 | BSA | 45′ | MACH-2 MOUSE | |
| CD10 | SS2/36 | MOUSE | DAKO, Carpinteria, CA | M0727 | PCE **2 | 1/100 | BSA | 45′ | ENV MOUSE | |
| BCL2 | 124 | MOUSE | DAKO, Carpinteria, CA | M0887 | VSE *2 | 1/300 | BSA | 45′ | ENV MOUSE | |
| Ki67 | MIB1 | MOUSE | DAKO, Carpinteria, CA | M7240 | VS6*1 | 1/100 | CaCl | 45′ | MACH-2 MOUSE | |
| CD30 | BERH2 | MOUSE | DAKO, Carpinteria, CA | M0751 | VS6*1 | 1/40 | BSA | 45′ | ENV MOUSE | |
| CD43 | DF-T1 | MOUSE | DAKO, Carpinteria, CA | M0786 | VS6*1 | 1/100 | BSA | 45′ | ENV MOUSE | |
| CD21 | IF8 | MOUSE | DAKO, Carpinteria, CA | M0784 | PRO K*** | 1/20 | BSA | 45′ | ENV MOUSE | |
| Cyclin D2 | DCS-3.1 + DCS-5.2 | RABBIT | Abcam, Cambridge, MA | ab3087 | VSE *2 | 1/200 | BSA | 2HR | ENV RABBIT | |
| Cyclin E | RABBIT | Abcam, Cambridge, MA | ab52189 | VSE *2 | 1/50 | BSA | OVN | ENV RABBIT | ||
| cMYC | Y69 | RABBIT | Epitomics, Burlingame, CA | 1472-1 | VSE *2 for 40 min | 1/50 | BSA | OVN | ENV RABBIT | |
| p27/Kip1 | MOUSE | BD Transduction Laboratory, San Diego, CA | 610241 | VS6*1 | 1/200 | BSA | 2hr | ENV MOUSE | ||
| Survivin | RABBIT | Novus Biological, Littleton, CO | NB500-201 | VS6 *1 | 1/50 | BSA | 45′ | ENV RABBIT | ||
| BAX | B9 | MOUSE | Santa Cruz, Santa Cruz, CA | sc-7480 | PC10**3 | 1/20 | BSA | OVN | ENV MOUSE | |
| Galectin 3 | M3/38 | RAT | Biolegend, San Diego, CA | 125402 | VS6 *1 | 1/50 | BSA | OVN | Rb anti rat | ENV RABBIT |
| BLIMP1 | 6D3 | RAT | Santa Cruz, Santa Cruz, CA | sc-47732 | VSE *2 | 1/50 | BSA | 45′ | Rb anti rat | ENV RABBIT |
| BCL6 | PG-B6P | MOUSE | DAKO, Carpinteria, CA | M7211 | VSE *2 | 1/30 | BSA | 45′ | ENV MOUSE | |
| PKC-beta 2 | Y125 | RABBIT | Abcam, Cambridge, MA | ab32026 | VSE *1 | 1/200 | BSA | OVN | ENV RABBIT | |
| MUM1 | MUM1p | MOUSE | DAKO, Carpinteria, CA | M7259 | PCE **2 | 1/100 | BSA | 45′ | ENV MOUSE | |
| LMO2 | SP51 | RABBIT | Spring Bioscience, Pleasanton, CA | M3510 | VSE *2 | 1/500 | BSA | 45′ | Novolink | |
| MMP9 | RABBIT | DAKO, Carpinteria, CA | A0150 | PC10 **3 | 1/100 | BSA | 60 min | CSA Rabbit link | CSA |
Vegetable steamer, 95°C for 25 min.
Pressure cooker, pressure cooker at 115°C for 2–3min.
Proteinase K, at 37°C for 10 min.
0.01M sodium citrate buffer, pH = 6.00.
0.001M EDTA, pH = 8.0.
0.05M Tris pH= 10.00.
Scoring of Tumor Marker Expression
All sections were visualized with the diaminobenzidine reaction and counterstained with hematoxylin. For computerized evaluation of Ki-67 staining, slides were analyzed using the Ariol SL-50 automated slide scanner (Applied Imaging, San Jose, CA). Thresholds for each image were applied using the Ariol analytical software based on multiple parameters: RGB algorithm, shape and size. All analyses were performed with the MultiStain script. Threshold classifiers were customized for each stain. Accuracy of thresholding was verified by a licensed pathologist prior to analysis. Study pathologist who performed the scoring of marker expression was blinded to the outcome status of each subject.
DLBCL Subtyping
DLBCL variant subtyping was performed independently by the two study pathologists by reviewing pathology reports, H&E slides and stained tumor marker expression data. Minor classification discrepancies on two cases were resolved in review by the two pathologists applying criteria for classification according the World Health Organization 2008 classification of tumors of the heamatopoietic and lymphoid tissues. Both pathologists were blinded to the outcome status of study subjects.
Ascertainment of Patient Survival
Information on 2-year mortality among the DLBCL patients was ascertained through record linkage with a combination of electronic health records, including KP’s membership and utilization files, California’s state death file, and Social Security records. Two-year mortality was chosen as the outcome since most deaths (85% in our study) occurred within 2 years after DLBCL diagnosis. Cause of death was electronically obtained from the primary cause of death filed in the death certificate. We evaluated the consistency of cause of death data by comparing results between the medical chart review by the study oncologist (Abrams DI) with the electronic cause of death ascertained from death certificates. Among 19 deaths evaluated, 79% had the same cause of death from each approach, suggesting reasonable consistency. Therefore, we decided to use the electronic cause of death as the primary source since this information was available for all 34 deaths observed. By contrast, chart note on cause of death was not always available for all deaths since death could have occurred outside the health plan facilities. The following ICD-9 and ICD-10 diagnosis codes were used to define lymphoma-specific deaths (based on primary causes): ICD-9 diagnosis codes 042.2, 200.8, 202.8; and ICD-10 diagnosis code B212, B217, C834, C835, C851, C859. All patients had complete two years of follow-up for assessing mortality outcome (i.e., there was no loss-to-follow up for these outcomes).
Data Collection for Other Covariates
Covariates evaluated as potential prognostic factors included demographics (age, sex, race/ethnicity), CD4 cell count, prior AIDS diagnosis, use of cART, duration of known HIV infection, HIV transmission risk group, and DLBCL characteristics including stage, subtype, extranodal involvement, elevated serum lactose dehydrogenase (LDH) level, Eastern Cooperative Oncology Group (ECOG) performance status, B symptoms and chemotherapy. Data on demographics and HIV disease factors were ascertained from the HIV registries. Data on ECOG performance status, B symptoms and chemotherapy were obtained from standardized medical chart review. Measurements of serum LDH and CD4 cell counts were obtained from the KP laboratory databases. Antiretroviral medications were ascertained from the KP pharmacy databases. cART was defined as a regimen of three or more antiretrovirals(20). DLBCL characteristics were obtained from KP’s cancer registries (i.e., stage, grade, extranodal involvement, and presence of B symptoms) and by pathology review (e.g., DLBCL subtype). The International Prognostic Index (IPI), an established prognostic score for NHL in the general population, which has also been validated in HIV-related NHL(21, 22) was then calculated based on age, stage, extranodal involvement, elevation in serum LDH level, and ECOG performance status. Because information was not complete for some covariates, the multiple imputation method proposed by Rubin(23) was used to handle the missing data.
Statistical Analysis
Those with an adequate tumor block for TMA construction and a readable result for EBV staining constituted the sub-cohort for the analysis. We compared the demographics, HIV disease factors, DLBCL characteristics and co-morbidity history between those who had an adequate tumor specimen vs. those who did not, using t-test for continuous variables and chi-square test or Fisher’s exact test for categorical variables. Next, among cases with adequate tumor specimen, we compared demographics and DLBCL characteristics, including GC phenotype, between those with EBV+ and EBV− tumors. The association between EBV status and tumor marker expression was examined using Pearson’s correlation coefficients, treating the expression score of each marker as a continuous variable (from 0 to 4). Due to the small sample size in the analytical subcohort, p-value <0.10 was used as the cut-off for statistical significance in this study. Bonferroni’s method was used to adjust for multiple comparisons. The mean and standard deviation of expression level of each of the tumor markers of interest among EBV+ vs. EBV− tumors were then calculated. As an exploratory exercise, among EBV+ tumors, mean tumor marker expression levels were also calculated by LMP1 expression status without formal statistical testing.
Kaplan-Meier survival curves for EBV+ and EBV− tumors were generated. The crude association between DLBCL EBV status, demographics, clinical prognostic factors and 2-year overall mortality as well as lymphoma-specific mortality was examined using bivariate Cox regression. The predictive utility of tumor EBV status on 2-year mortality was examined in multivariable Cox model, adjusting for IPI. In an alternative model, we adjusted for all demographics (i.e., age, gender, ethnicity) and previously established prognostic factors (i.e., DLBCL subtype, clinical stage, ECOG performance status, extranodal involvement, and elevated LDH level at diagnosis), as well as any other factors that showed a crude association at p<0.10 level with the mortality outcome (i.e., prior AIDS diagnosis and CD4 cell count at DLBCL diagnosis). Given the small sample size, we used the propensity score approach to adjust for these factors. The propensity score function for EBV infection status was modeled using logistic regression. To evaluate the prognostic utility of tumor EBV status accounting for the DLBCL treatment, we repeated the analyses restricting to those who received chemotherapy. We also conducted stratified analysis for the most common DLBCL subtype: centroblastic DLBCL.
To assess the improvement in the model discrimination in distinguishing those who experienced a mortality outcome vs. those who did not, we constructed the receiver-operating characteristics (ROC) curve(24) for two prediction models: (1) IPI alone; and (2) IPI + tumor EBV status. The area under the ROC curve (AUC) was then calculated, and compared between the two models using chi-square test. All analyses in this study were performed with SAS Version 9.1; Cary, North Carolina, USA. The PROG MI procedure in SAS was used to analyze the datasets with multiple imputation for missing data.
Results
A total of 194 incident HIV-related DLBCL cases were identified between 1996 and 2007. Of these, 70 cases had adequate tissue for analysis and were included in the study. The remaining 124 cases were excluded for the following reasons: 1) lack of an appropriate accession for TMA (i.e., with only core biopsy, fluid, bone marrow smear or a small tissue block, n=99); 2) missing tumor specimen (n=9); 3) risk of exhaustion of tissue (n=6); and 4) unsuccessful staining of EBV (n=10). We found no important difference, either qualitatively or statistically, in the demographic or clinical characteristics between those who were included in the tumor marker analysis vs. those who were not. A total of 34 deaths were found during the two-year follow up; 20 of these were lymphoma-specific deaths.
Twenty-two (31%) of the 70 DLBCL were EBV+. Table 2 presents the characteristics of the 70 patients by DLBCL EBV infection status. Patients with EBV+ DLBCL were more likely to be immunoblastic (23% vs. 17% for EBV+ and EBV−) and plasmablastic subtype (18% vs. 4% for EBV+ and EBV−) (p=0.095), had lower mean CD4 cell count at diagnosis (128/mm3 vs. 248/mm3, p=0.007), and a shorter mean duration of HIV infection prior to DLBCL diagnosis (3.1 year vs. 6.2 year, p=0.06). B symptoms (36% vs. 23%, p=0.35) and prior cART use (73% vs. 60%, p=0.32) were more common among EBV+ cases, although these associations were not statistically significant. Those with EBV+ DLBCL and those with EBV− DLBCL did not differ by lymphoma stage, extranodal involvement, serum LDH abnormality, ECOG performance status or HIV transmission risk group.
Table 2.
Demographic and clinical characteristics of HIV+ diffuse large B-cell lymphoma cases by EBV status.
| EBV-negative (N=48) | EBV-positive (N=22) | p-value | |
|---|---|---|---|
| Age (years), mean (sd) | 47.9 (9.3) | 49.6 (9.5) | 0.49 |
| Male gender, n (%) | 45 (93.8%) | 19 (86.4%) | 0.37 |
| Race/Ethnicity, n (%) | |||
| White | 28 (58.3%) | 13 (59.1%) | 0.20 |
| Black | 9 (18.8%) | 2 (9.1%) | |
| Hispanic | 11 (22.9%) | 5 (22.7%) | |
| Asian/Pacific islander | 0 (0%) | 2 (9.1%) | |
| Stage, n (%) | |||
| I (Localized) | 11 (22.9%) | 5 (22.7%) | 0.89 |
| II (Regional) | 8 (16.7%) | 4 (18.2%) | |
| III (Distant) | 25 (52.1%) | 10 (45.5%) | |
| Extranodal involvement, n (%) | |||
| Single site involvement | 12 (25.0%) | 6 (27.3%) | |
| ≥2 sites on the same side of the diaphragm | 9 (18.8%) | 4 (18.2%) | |
| Both sides of the diaphragm | 4 (8.3%) | 4 (18.2%) | 0.36 |
| Disseminated | 23 (47.9%) | 7 (31.8%) | |
| Subtype, n (%) | |||
| Centroblastic | 38 (79.2%) | 13 (59.1%) | 0.10 |
| Immunoblastic | 8 (16.7%) | 5 (22.7%) | |
| Plasmablastic | 2 (4.2%) | 4 (18.2%) | |
| Germinal center phenotype, n (%) | 22 (47.8%) | 5 (22.7%) | 0.06 |
| Detection of LMP1, n(%) | NA | 8 (36.4%) | NA |
| Elevated serum lactose dehydrogenase, n (%) | 25 (69.4%) | 14 (73.7%) | 0.74 |
| ECOG performance status ≥2, n (%) | 9 (20%) | 4 (20%) | 1.00 |
| Presence of B symptoms n (%) | 11 (22.9%) | 8 (36.4%) | 0.35 |
| International prognostic index, n (%) | |||
| 0–1 (low risk) | 11 (32.3%) | 7 (41.2%) | |
| 2 (low-intermediate risk) | 7 (20.6%) | 4(23.5%) | 0.91 |
| 3 (high-intermediate risk) | 11 (32.3%) | 4 (23.5%) | |
| 4–5 (high risk) | 5 (14.7%) | 2 (11.8%) | |
| Chemotherapy, n (%) | 42 (87.5%) | 16 (72.7%) | 0.17 |
| CHOP | 20 (47.6%) | 8 (50.0%) | |
| RCHOP | 15 (35.7%) | 4 (25.0%) | 0.36 |
| Other | 2 (4.8%) | 3 (18.8%) | |
| HIV risk group, n (%) | |||
| Heterosexual | 9 (18.8%) | 6 (27.3%) | 0.83 |
| Intravenous drug user | 1 (2.1%) | 0 (0%) | |
| Men who have sex with men | 22 (45.8%) | 9 (40.9%) | |
| Other/Unknown | 16 (33.3%) | 7 (31.8%) | |
| Known HIV duration (years), mean (sd) | 6.2 (6.2) | 3.1 (4.0) | 0.06 |
| Prior AIDS diagnosis, n (%) | 19 (39.6%) | 12 (54.6%) | 0.24 |
| Prior use of cART, n (%) | 29 (60.4%) | 16 (72.7%) | 0.32 |
| CD4 at diagnosis (cells/mm3), mean (sd) | 247.9 (169.0) | 128.4 (132.3) | 0.01 |
| CD4 lowest KP recorded (cells/mm3), mean (sd) | 85.7 (72.2) | 43.8 (42.5) | 0.02 |
Percentage may not add up to 100% due to missing values.
DLBCL EBV infection status and tumor marker expression
There was a suggestion that BLIMP1, CD30 and MUM1 were more commonly expressed in EBV+, and that BCL6, LMO2 and BAX were more commonly expressed in EBV-DLBCL (Table 3). However, only the association with BCL-6, BLIMP1, LMO2 and CD30 reached statistical significance using p<0.10 with adjustment for multiple comparisons. Of the EBV+ DLBCL, 36% had positive LMP1 expression. Expression level of CD30 appears to differ materially by LMP1 expression status (Table 4).
Table 3.
Mean tumor marker expression score for BCL6, BLIMP1, LMO2 and CD30 among others by tumor EBV infection status.
| DLBCL EBV infection status | BCL6 | BLIMP1 | LMO2 | CD30 |
|---|---|---|---|---|
|
| ||||
| Tumor marker scorea, mean (sd) | ||||
| EBV− (n=48) | 1.78 (1.34) | 0.09 (0.36) | 2.33 (1.55) | 0.62 (1.27) |
| EBV+ (n=22) | 0.55 (1.10) | 0.77 (1.27) | 1.09 (1.02) | 1.76 (1.73) |
| p-valueb | <0.001 | 0.003 | 0.003 | <0.001 |
|
| ||||
| BCL2 | BAX | Cyclin D2 | Cyclin E | |
|
|
||||
| EBV− | 1.83 (1.34) | 2.89 (1.37) | 0.02 (0.15) | 1.24 (1.37) |
| EBV+ | 1.33 (1.24) | 2.05 (1.53) | 0.05 (0.21) | 1.64 (1.33) |
| p-valueb | 0.231 | 0.016 | 0.598 | 0.239 |
|
| ||||
| Ki-67 | FOXP1 | P53 | Survivin | |
|
|
||||
| EBV− | 2.10 (0.79) | 1.25 (1.25) | 0.54 (1.15) | 2.91 (1.43) |
| EBV+ | 2.29 (1.64) | 0.91 (1.27) | 0.77 (1.02) | 2.71 (1.45) |
| p-valueb | 0.534 | 0.326 | 0.110 | 0.342 |
|
| ||||
| P27 | MMP9 | GAL3 | SKP2 | |
|
|
||||
| EBV− | 0.52 (0.80) | 1.82 (1.66) | 1.58 (1.44) | 0.44 (1.03) |
| EBV+ | 0.64 (0.73) | 2.24 (1.70) | 1.33 (1.43) | 0.41 (0.91) |
| p-valueb | 0.353 | 0.495 | 0.345 | 0.632 |
|
| ||||
| PKC-beta 2 | MUM1 | cMYC | CD43 | |
|
|
||||
| EBV− | 2.52 (1.79) | 1.52 (1.50) | 1.46 (1.50) | 0.89 (1.52) |
| EBV+ | 3.00 (1.45) | 2.50 (1.41) | 1.73 (1.49) | 1.52 (1.89) |
| p-valueb | 0.218 | 0.012 | 0.598 | 0.278 |
|
| ||||
| CD10 | CD44 | CD21 | ||
|
|
||||
| EBV− | 1.08 (1.40) | 2.81 (1.28) | 1.12 (0.15) | |
| EBV+ | 1.00 (1.35) | 2.71 (1.19) | 0 | |
| p-valueb | 0.945 | 0.602 | 0.493 | |
Tumor marker score 0=0–9%, 1=10–24%, 2=25–49%, 3=50–74% and 4= ≥75% of DLBCL cells stained positive for the marker.
p-value derived from Pearson’s correlation coefficient statistics. Significance level for p-value <0.10 after adjustment for multiple comparison is 0.004 [0.10/23 (number of markers examined)=0.004].
Table 4.
Tumor marker expression level by LMP1 status among EBV+ DLBCL cases.
| EBV+ DLBCL LMP1 expression status |
BCL6 | BLIMP1 | LMO2 | CD30 |
|---|---|---|---|---|
|
| ||||
| Tumor marker scorea, mean (sd) | ||||
| LMP− (n=14) | 0.79 (1.31) | 1.08 (1.44) | 0.93 (1.00) | 0.77 (1.17) |
| LMP+ (n=8) | 0.13 (0.35) | 0.25 (0.71) | 1.38 (1.06) | 3.38 (1.19) |
|
| ||||
| BCL2 | BAX | Cyclin D2 | Cyclin E | |
|
|
||||
| LMP− | 0.92 (1.04) | 1.64 (1.65) | 0.07 (0.27) | 1.71 (1.33) |
| LMP+ | 2.00 (1.31) | 2.86 (0.90) | 0 (0) | 1.50 (1.41) |
|
| ||||
| Ki-67 | FOXP1 | P53 | Survivin | |
|
|
||||
| LMP− | 2.38 (0.65) | 1.07 (1.49) | 0.79 (1.19) | 2.21 (1.48) |
| LMP+ | 2.13 (0.64) | 0.63 (0.74) | 0.75 (0.71) | 3.71 (0.76) |
|
| ||||
| P27 | MMP9 | GAL3 | SKP2 | |
|
|
||||
| LMP− | 0.79 (0.80) | 1.92 (1.75) | 1.08 (1.19) | 0.50 (1.09) |
| LMP+ | 0.38 (0.52) | 2.75 (1.58) | 1.75 (1.75) | 0.25 (0.46) |
|
| ||||
| PKC-beta 2 | MUM1 | cMYC | CD43 | |
|
|
||||
| LMP− | 2.43 (1.55) | 2.79 (1.42) | 2.00 (1.52) | 1.69 (1.97) |
| LMP+ | 4.00 (0) | 2.00 (1.31) | 1.25 (1.39) | 1.25 (1.83) |
|
| ||||
| CD10 | CD44 | CD21 | ||
|
|
||||
| LMP− | 1.29 (1.59) | 2.62 (1.04) | 0 (0) | |
| LMP+ | 0.50 (0.53) | 2.88 (1.46) | 0 (0) | |
Tumor marker score 0=0–9%, 1=10–24%, 2=25–49%, 3=50–74% and 4= ≥75% of DLBCL cells stained positive for the marker.
DLBCL EBV infection status and 2-year mortality
Figure 1 shows the Kaplan-Meier curve for overall survival by DLBCL EBV infection status. In the crude survival analysis, EBV+ DLBCL was associated with a 3-fold increase in overall mortality hazard within 2 years of diagnosis [hazard ratio (HR) = 2.9 95% confidence interval (1.4–5.6), Table 5]. A slightly stronger association was observed for lymphoma-specific mortality [crude HR=3.9 (1.6–9.4)]. In the analysis adjusting for IPI, EBV infection was still associated with a 3-fold increase in overall mortality hazard [HR = 3.3 (1.6–6.6), Table 6], and a 4-fold increase in hazard for lymphoma-specific mortality [HR= 4.6 (1.8–11.4)]. In the alternative model adjusting for propensity score as well as in the analysis restricted to those who received chemotherapy or analysis restricted to centroblastic DLBCL subtype, tumor EBV status remained predictive of mortality outcomes (Table 6).
Figure 1.
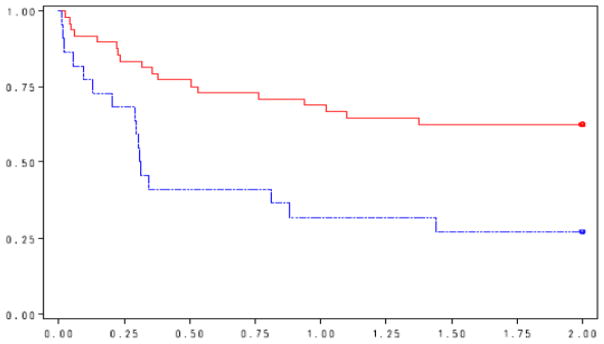
Kaplan-Meier survival curve for two-year overall survival by EBV status.
X-axis: 2-year overall survival
Y-axis: Survival function
Red solid line: EBS-negative tumor
Blue dash line: EBV-positive tumor
Table 5.
Univariate association between covariates of interest and mortality outcomes.
| Overall mortality | Lymphoma specific-mortality | |||
|---|---|---|---|---|
|
| ||||
| Hazard ratio (95% confidence interval) | p-value | Hazard ratio (95% confidence interval) | p-value | |
| EBV+ status | 2.9 (1.4–5.6) | <0.01 | 3.9 (1.6–9.4) | <0.01 |
| Age (per year increase) | 1.0 (1.0–1.0) | 0.73 | 1.0 (1.0–1.1) | 0.81 |
| Female gender | 2.6 (0.9–7.4) | 0.08 | 3.3 (1.0–11.3) | 0.06 |
| Hispanic ethnicity | 1.7 (0.8–3.6) | 0.15 | 1.4 (0.8–3.8) | 0.54 |
| Clinical stage | ||||
| Localized | Ref | - | Ref | - |
| Regional | 1.3 (0.5–3.8) | 0.59 | 4.0 (0.8–19.7) | 0.09 |
| Distant | 1.3 (0.6–3.1) | 0.55 | 2.7 (0.6–12.0) | 0.21 |
| Extranodal involvement: | ||||
| Disseminated | 1.7 (0.7–4.0) | 0.25 | 3.9 (0.9–17.6) | 0.07 |
| DLBCL subtype | ||||
| Centroblastic | Ref | - | - | |
| Immunoblastic | 1.4 (0.6–3.2) | 0.48 | 1.5 (0.5–4.9) | 0.46 |
| Plasmablastic | 4.2 (1.6–11.3) | <0.01 | 8.5 (2.9–25.4) | <0.01 |
| Germinal center phenotype | 0.6 (0.3–1.2) | 0.16 | 1.0 (0.4–2.4) | 0.95 |
| Elevated serum lactose dehydrogenase | 4.2 (1.3–14.1) | 0.02 | 2.7 (0.8–9.4) | 0.12 |
| ECOG performance status ≥2 | 2.5 (1.2–5.5) | 0.02 | 5.1 (2.0–13.0) | <0.01 |
| Presence of B symptoms | 1.3 (0.6–2.7) | 0.55 | 1.9 (0.8–4.9) | 0.17 |
| International Prognostic Index | ||||
| 0–1 (low risk) | Ref | - | Ref | - |
| 2 (low-intermediate risk) | 0.9 (0.2–3.6) | 0.88 | 0.9 (0.2–4.9) | 0.90 |
| 3 (high-intermediate risk) | 2.6 (1.0–7.3) | 0.06 | 2.3 (0.7–8.2) | 0.19 |
| 4–5 (high risk) | 3.5 (1.1–11.5) | 0.04 | 4.1 (1.0–16.4) | 0.05 |
| Prior AIDS diagnosis | 2.9 (1.5–5.8) | <0.01 | 3.8 (1.5–9.5) | <0.01 |
| CD4 cell count ≥200/mm3 | 0.4 (0.2–0.9) | 0.02 | 0.4 (0.1–1.1) | 0.08 |
Table 6.
Adjusted hazard ratios (HR) for EBV status on survival outcomes.
| All DLBCL | Overall mortality | Lymphoma specific-mortality | ||
|---|---|---|---|---|
|
| ||||
| HR (95% confidence interval) | p-value | HR (95% confidence interval) | p-value | |
| Multivariable Model 1: Adjusting for IPIa | 3.3 (1.6–6.6) | <0.01 | 4.6 (1.8–11.4) | <0.01 |
|
| ||||
| Multivariable Model 2: Adjusting for propensity scoreb | 2.1 (1.0–4.5) | 0.06 | 2.8 (1.1–7.7) | 0.04 |
|
| ||||
|
Centroblastic DLBCL
|
||||
| Multivariable Model 1: Adjusting for IPIa | 1.9 (0.9–3.8) | 0.08 | 2.8 (1.2–6.8) | 0.02 |
|
| ||||
| Multivariable Model 2: Adjusting for propensity scoreb | 2.9 (1.1–7.4) | 0.03 | 4.3 (1.1–16.8) | 0.04 |
|
| ||||
| Patients on standard chemotherapy | ||||
|
| ||||
| Multivariable Model 1: Adjusting for IPIa | 3.3 (1.4–7.8) | <0.01 | 3.3 (1.1–10.0) | 0.03 |
|
| ||||
| Multivariable Model 2: Adjusting for propensity scoreb | 2.9 (1.1–7.4) | 0.03 | 3.1 (0.9–10.0) | 0.06 |
IPI: International Prognostic Index.
Model adjusted for propensity score. Variables used to create propensity score include: age (<=60 vs. >60 years), gender, ethnicity (Hispanic vs. non-Hispanic), DLBCL subtype (centroblastic, immunoblastic, plasmablastic), stage (III, IV, vs. others), extranodal involvement (disseminated vs.other), elevated serum lactose dehydrogenase level at DLBCL diagnosis, ECOG performance status (≥2 vs. <2), prior AIDS diagnosis (yes vs. no) and CD4 cell count at diagnosis (< 200 vs. ≥200/mm3).
Area under the ROC comparing IPI vs. IPI+ EBV
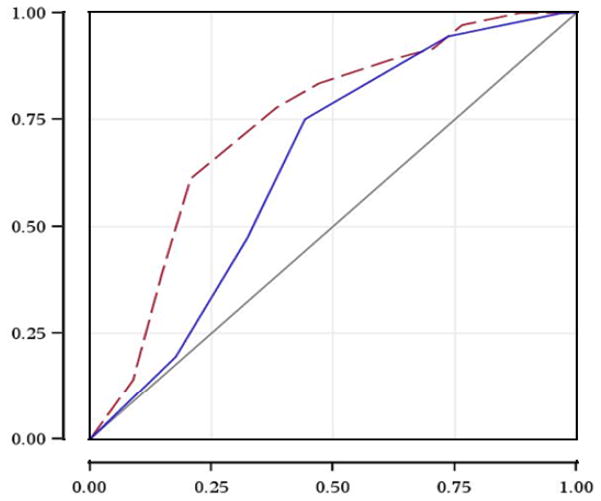
Figure 2 shows the ROC curve for 2-year overall mortality for IPI alone, and for model incorporating both IPI and tumor EBV infection status. The area under the ROC curve (AUC) was 0.65 for IPI alone, and 0.74 when combining IPI and tumor EBV infection status. This increase in AUC was marginally significant (p=0.12), suggesting improved model discrimination when tumor EBV infection status was considered along with IPI for HIV-related DLBCL prognosis.
Figure 2.
ROC curve for 2-year overall mortality: IPI and IPI + EBV status
X-axis: 1-Specificity
Y-axis: Sensitivity
Solid line: International Prognostic Index only (area under the ROC curve: 0.65)
Dash line: International Prognostic Index + tumor EBV infection status (area under the ROC curve: 0.74)
Discussion
We found that 31% of our DLBCL cases were positive for EBV infection. This is consistent with previously reported prevalence of EBV+ DLBCL tumors in the cART era(5). We also found that EBV+ tumor was associated with expression of several of the tumor markers examined, including a positive association with expression of BLIMP1 and CD30, and negative association with BCL6 and LMO2. BLIMP1 is a transcription factor that regulates the differentiation of mature B-cells into antibody-secreting plasma cells(25). BLIMP1 acts in an autoregulatory feedback loop that controls p53 activity through repression of p53 transcription(26). The activity of BLIMP1 hence inhibits apoptosis, and deletion of BLIMP1 in lymphocytes induces apoptosis(26). The positive association between EBV infection and BLIMP1 expression suggested that it may play a role in EBV-induced lymphoproliferation. CD30 is a transmembrane protein that is part of the tumor necrosis factor (TNF) receptor family. When stimulated by CD30 ligand, CD30 interacts with TNF receptor associated factors (TRAF2 and TRAF5), mediating signal transduction that leads to the activation of the NF-κB pathway(27), which has been linked to cellular activation and carcinogenesis. This finding is consistent with an EBV-associated carcinogenic mechanism operating through the NF-κB pathway. EBV LMP1 expression is known to mimic the activity of ligated CD40, another molecule that is a member of the TNF receptor family, which in turn stimulates the NF-κB and stress activated kinase pathways. In our study sample, EBV+ DLBCL, with or without LMP1 expression, expressed CD30. However, CD30 expression was more common in LMP1+ tumors (88% vs. 23% in the EBV+/LMP1−), despite lack of statistical significance.
BCL6 and LMO2, on the other hand, are suspected favorable prognostic factors. BCL6 is a transcription repressor that is commonly translocated in lymphomas. BCL6 represses B-cell receptor signals(28) and plays a central role in inducing the germinal center phenotype in both B and T cells(29). Lack of BCL6 function thus enhances proliferation and inhibits differentiation(28). To this end, BLIMP1 is a target protein repressed by BCL6(28, 30). LMO2 is a transcription factor that critically regulates erythropoiesis, angiogenesis, and embryogenesis(31–34). LMO2 is associated with the GC phenotype, and has been reported as a favorable prognostic factor in DLBCL by previous studies(35–37). The inverse relationship between EBV infection and expression of BCL6 and LMO2 suggested that these two transcription factors may be further repressed in EBV induced lymphomagenesis when compared to other lymphomagenic mechanisms that do not involve EBV.
As noted previously, EBV is thought to contribute to the development of B cell cancers by infecting cells and expressing EBV-encoded transforming proteins which in turn enhances genetic instability through mutation, translocation and aberrant expression of proto-oncogenes(18). LMP1, a viral gene product of EBV, is known to constitutively activate the NF-κB, Jun N-terminal kinase and p38 kinase pathways(38)as well as protect cells from p53 induced apoptosis(9). LMP1 may also contributes to the immortalization of B cells by increasing the expression of anti-apoptotic proteins BCL2 and A20 as well as cell cycle regulator p27(9). Vrzalikova et al reported down-regulation of BLIMP1 by EBV infection, specifically, LMP1, in lymphoblastoid cell lines established from GC B cells(39). This seemly contrasting finding may be due to the fact in our study, most EBV+ tumors are the non-GC type. As a result, the effects of EBV seen in GC cells therefore may not be present in post-GC cells. In our exploratory exercise, no consistent pattern of elevation for markers linked to cancer development was observed in LMP1-positive tumors, although the small sample size of LMP1-positive tumors precludes an informative analysis in this study.
EBV also may up-regulate the receptor CD21, thereby protecting cells from self-destruction(40).While our results provided some support with patient level data for these previously proposed carcinogenic mechanisms of EBV, we did not find association between tumor EBV infection status and expression of p53, BCL2, p27 or CD21. It is possible that these tumor markers were important for all lymphomagenic pathways, regardless of involvement of EBV.
We also found that detecting tumor EBV infection may have independent prognostic utility for survival among patients with HIV-related DLBCL beyond clinical prognostic factors, including IPI and CD4 cell count at diagnosis(41). This contrasts with the findings of Chadburn et al(42), who reported that EBV status was not associated with overall or event-free survival among 78 patients with HIV-related DLBCL. They also did not find any association between EBV status and expression of FOXP1 and BLIMP1. However, patients in the study were enrolled in a clinical trial investigating the efficacy of rituximab in HIV-infected DLBCL patients, which may have limited generalizability to HIV-related DLBCL patients at large. Two other studies in non HIV-related DLBCL patients also reported tumor EBV infection status to be an adverse prognostic factor(6, 7). The utility of EBV status as a prognostic marker in DLBCL should be confirmed in larger studies.
There are several potential limitations of this study. First, a large proportion of patients were excluded from the tumor marker analysis due to lack of an adequate tumor tissue for TMA construction. However, no important differences in demographic and clinical characteristics were found between those with vs. without adequate tumor specimen, suggesting this was not a significant source of bias. Also, our sample size precluded other potentially informative analyses, such as comparing expressions of LMP1 and other selected tumor markers or clinical characteristics with sufficient statistical power, which should be examined in future study to further inform the mechanism of the prognostic effect for EBV. Furthermore, we did not measure other EBV latent proteins nor define the various latent stages of the EBV infection. Despite these limitations, our study is based on a well-defined, representative cohort of HIV-related DLBCL, with comprehensive clinical information and measurement of a large number of tumor markers. To our knowledge, this study is also among the few that have examined the prognostic role of EBV status in HIV-related DLBCL.
In conclusion, we found that EBV infection status in DLBCL is associated with expression of several tumor markers that are involved in the NF-κB pathway. These factors were likely mediated by EBV and contribute to the EBV-related lymphomagenesis through activation of this pathway, as well as inhibition of apoptosis. Our results also suggest that tumor EBV status may have independent prognostic utility beyond conventional clinical prognostic scores such as the IPI, and may be used for risk stratification of patients diagnosed with HIV-related DLBCL.
Statement of Translational Relevance.
In the era of combination antiretroviral therapy, HIV-related diffuse large B-cell lymphoma (DLBCL) is no longer invariably fatal and is heterogeneous in clinical outcomes. Despite the availability of potentially effective regimens for the treatment of DLBCL, more than 50% of patients continue to succumb to the disease. Therefore, understanding factors underlying HIV-related DLBCL aggressiveness and heterogeneity is critical to risk-stratified patient management and novel therapeutic development. In this study, we found that tumor EBV infection status was independently associated with increased mortality in HIV+ DLBCL patients. This finding, if confirmed, may help to improve clinical patient risk stratification, and to identify potential new molecular therapeutic targets (e.g., EBV-targeting therapy) for resistant tumors.
Acknowledgments
Financial Support:
This work is supported by National Cancer Institute grant R01CA134234-01 Prognostic Markers for HIV-Positive Diffuse Large B-Cell Lymphoma.
Footnotes
Conflicts of interest:
Chun Chao received research funding from Merck, Amgen and Pfizer for unrelated studies. Michael J Silverberg received research funding from Merck and Pfizer for unrelated studies. Reina Haque received research funding from Novartis for unrelated studies. Other authors do not have conflict of interest to disclose.
References
- 1.Silverberg MJ, Chao C, Leyden WA, Xu L, Tang B, Horberg MA, et al. HIV infection and the risk of cancers with and without a known infectious cause. AIDS. 2009;23:2337–45. doi: 10.1097/QAD.0b013e3283319184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher SG, Fisher RI. The epidemiology of non-Hodgkin’s lymphoma. Oncogene. 2004;23:6524–34. doi: 10.1038/sj.onc.1207843. [DOI] [PubMed] [Google Scholar]
- 3.Angeletti PC, Zhang L, Wood C. The viral etiology of AIDS-associated malignancies. Adv Pharmacol. 2008;56:509–57. doi: 10.1016/S1054-3589(07)56016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heslop HE. Biology and treatment of epstein-barr virus-associated non-hodgkin lymphomas. Hematology Am Soc HematolEduc Program. 2005:260–6. doi: 10.1182/asheducation-2005.1.260. [DOI] [PubMed] [Google Scholar]
- 5.Aboulafia DM, Pantanowitz L, Dezube BJ. AIDS-related non-Hodgkin lymphoma: still a problem in the era of HAART. AIDS Read. 2004;14:605–17. [PubMed] [Google Scholar]
- 6.Park S, Lee J, Ko YH, Han A, Jun HJ, Lee SC, et al. The impact of the Epstein-Barr Virus status on clinical outcome in diffuse large B cell lymphoma. Blood. 2007;110:972–8. doi: 10.1182/blood-2007-01-067769. [DOI] [PubMed] [Google Scholar]
- 7.Saez AI, Saez AJ, Artiga MJ, Perez-Rosado A, Camacho FI, Diez A, et al. Building an outcome predictor model for diffuse large B-cell lymphoma. Am J Pathol. 2004;164:613–22. doi: 10.1016/S0002-9440(10)63150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rickinson A, Kieff E. Epstein-Barr virus. In: Fields B, Knipe D, Howley P, editors. Fields virology. Philadelphia: Lippincott-Raven; 1996. p. 2397. [Google Scholar]
- 9.Cahir McFarland ED, Izumi KM, Mosialos G. Epstein-barr virus transformation: involvement of latent membrane protein 1-mediated activation of NF-kappaB. Oncogene. 1999;18:6959–64. doi: 10.1038/sj.onc.1203217. [DOI] [PubMed] [Google Scholar]
- 10.Pattle SB, Farrell PJ. The role of Epstein-Barr virus in cancer. Expert Opin Biol Ther. 2006;6:1193–205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 11.Epeldegui M, Vendrame E, Martinez-Maza O. HIV-associated immune dysfunction and viral infection: role in the pathogenesis of AIDS-related lymphoma. Immunol Res. 2010;48:72–83. doi: 10.1007/s12026-010-8168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaidano G, Capello D, Carbone A. The molecular basis of acquired immunodeficiency syndrome-related lymphomagenesis. Semin Oncol. 2000;27:431–41. [PubMed] [Google Scholar]
- 13.Mueller N, Mohar A, Evans A, Harris NL, Comstock GW, Jellum E, et al. Epstein-Barr virus antibody patterns preceding the diagnosis of non-Hodgkin’s lymphoma. Int J Cancer. 1991;49:387–93. doi: 10.1002/ijc.2910490313. [DOI] [PubMed] [Google Scholar]
- 14.Mueller NE, Mohar A, Evans A. Viruses other than HIV and non-Hodgkin’s lymphoma. Cancer Res. 1992;52(19 Suppl):5479s–81s. [PubMed] [Google Scholar]
- 15.Staal SP, Ambinder R, Beschorner WE, Hayward GS, Mann R. A survey of Epstein-Barr virus DNA in lymphoid tissue. Frequent detection in Hodgkin’s disease. Am J Clin Pathol. 1989;91:1–5. doi: 10.1093/ajcp/91.1.1. [DOI] [PubMed] [Google Scholar]
- 16.van Baarle D, Hovenkamp E, Callan MF, Wolthers KC, Kostense S, Tan LC, et al. Dysfunctional Epstein-Barr virus (EBV)-specific CD8(+) T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2001;98:146–55. doi: 10.1182/blood.v98.1.146. [DOI] [PubMed] [Google Scholar]
- 17.Epeldegui M, Widney DP, Martinez-Maza O. Pathogenesis of AIDS lymphoma: role of oncogenic viruses and B cell activation-associated molecular lesions. Curr Opin Oncol. 2006;18:444–8. doi: 10.1097/01.cco.0000239882.23839.e5. [DOI] [PubMed] [Google Scholar]
- 18.Epeldegui M, Hung YP, McQuay A, Ambinder RF, Martinez-Maza O. Infection of human B cells with Epstein-Barr virus results in the expression of somatic hypermutation-inducing molecules and in the accrual of oncogene mutations. Mol Immunol. 2007;44:934–42. doi: 10.1016/j.molimm.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 20.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. October 2008 revision. Available at: http://aidsinfo.nih.gov/ContentFiles/AboutHIVTreatmentGuidelines_FS_en.pdf.
- 21.Rossi G, Donisi A, Casari S, Re A, Cadeo G, Carosi G. The International Prognostic Index can be used as a guide to treatment decisions regarding patients with human immunodeficiency virus-related systemic non-Hodgkin lymphoma. Cancer. 1999;86:2391–7. [PubMed] [Google Scholar]
- 22.Mounier N, Spina M, Gabarre J, Raphael M, Rizzardini G, Golfier JB, et al. AIDS-related non-Hodgkin lymphoma: final analysis of 485 patients treated with risk-adapted intensive chemotherapy. Blood. 2006;107:3832–40. doi: 10.1182/blood-2005-09-3600. [DOI] [PubMed] [Google Scholar]
- 23.Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York: John Wiley & Sons, Inc; 1987. [Google Scholar]
- 24.Soreide K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J Clin Pathol. 2009;62:1–5. doi: 10.1136/jcp.2008.061010. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, Jiang J, Lim CA, Wu Q, Ng HH, Chin KC. BLIMP1 regulates cell growth through repression of p53 transcription. Proc Natl Acad Sci U S A. 2007;104:1841–6. doi: 10.1073/pnas.0605562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aizawa S, Nakano H, Ishida T, Horie R, Nagai M, Ito K, et al. Tumor necrosis factor receptor-associated factor (TRAF) 5 and TRAF2 are involved in CD30-mediated NFkappaB activation. J Biol Chem. 1997;272:2042–5. doi: 10.1074/jbc.272.4.2042. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 29.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nature Immunology. 2010;11:114–20. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunology. 2002;169:1922–9. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 31.Nam CH, Rabbitts TH. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Mol Ther. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y, Pannell R, Forster A, Rabbitts TH. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc Natl Acad Sci U S A. 2000;97:320–4. doi: 10.1073/pnas.97.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A. 1998;95:3890–5. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jais JP, Haioun C, Molina TJ, Rickman DS, de Reynies A, Berger F, et al. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia. 2008;22:1917–24. doi: 10.1038/leu.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, et al. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26:447–54. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 37.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–42. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cahir-McFarland EKE. NF-κB inihibition in EBV-transformed lymphoblastoid cell lines. Recent Results in Cancer Res. 2002;159:44–8. doi: 10.1007/978-3-642-56352-2_6. [DOI] [PubMed] [Google Scholar]
- 39.Vrzalikova K, Vockerodt M, Leonard S, Bell A, Wei W, Schrader A, et al. Down-regulation of BLIMP1alpha by the EBV oncogene, LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: implications for the pathogenesis of EBV-associated B-cell lymphomas. Blood. 2011;117:5907–17. doi: 10.1182/blood-2010-09-307710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tannock IF, Hill RP, Bristow RG, Harrington L. In: The Basic Science of Oncology. Tannock IF, Hill RP, Bristow RG, Harrington L, editors. New Baskerville: McGraw Hill Co, Inc; 2005. [Google Scholar]
- 41.Chao C, Xu L, Abrams D, Leyden W, Horberg M, Towner W, et al. Survival of non-Hodgkin lymphoma patients with and without HIV infection in the era of combined antiretroviral therapy. AIDS. 2010;24:1765–70. doi: 10.1097/QAD.0b013e32833a0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chadburn A, Chiu A, Lee JY, Chen X, Hyjek E, Banham AH, et al. Immunophenotypic analysis of AIDS-related diffuse large B-cell lymphoma and clinical implications in patients from AIDS Malignancies Consortium clinical trials 010 and 034. J Clin Oncol. 2009;27:5039–48. doi: 10.1200/JCO.2008.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]