Abstract
Increasing seed oil content is one of the most important goals in breeding of rapeseed (B. napus L.). To dissect the genetic basis of oil content in B. napus, a large and new double haploid (DH) population containing 348 lines was obtained from a cross between ‘KenC-8’ and ‘N53-2’, two varieties with >10% difference in seed oil content, and this population was named the KN DH population. A genetic linkage map consisting of 403 markers was constructed, which covered a total length of 1783.9 cM with an average marker interval of 4.4 cM. The KN DH population was phenotyped in eight natural environments and subjected to quantitative trait loci (QTL) analysis for oil content. A total of 63 identified QTLs explaining 2.64–17.88% of the phenotypic variation were identified, and these QTLs were further integrated into 24 consensus QTLs located on 11 chromosomes using meta-analysis. A high-density consensus map with 1335 marker loci was constructed by combining the KN DH map with seven other published maps based on the common markers. Of the 24 consensus QTLs in the KN DH population, 14 were new QTLs including five new QTLs in A genome and nine in C genome. The analysis revealed that a larger population with significant differences in oil content gave a higher power detecting new QTLs for oil content, and the construction of the consensus map provided a new clue for comparing the QTLs detected in different populations. These findings enriched our knowledge of QTLs for oil content and should be a potential in marker-assisted breeding of B. napus.
Introduction
Rapeseed (B. napus, AACC, 2n = 38) is the world’s second most important oilseed crop after soybean. Rapeseed oil represents about 13.0–16.0% of the world vegetable oil production, and is also considered as a substitute for producing feedstock oils for biodiesel [1]. With the increasing demand for vegetable oil and canola-oil-based biodiesel usage, oil content has become a key factor for increasing oil production. At present a 1.0% increase in seed oil content is equivalent to an increase of 2.3–2.5% in seed yield in B. napus [2]. Seed oil content is a typical quantitative trait, controlled by a large number of genes and also highly influenced by environment [3], [4]. Better understanding of the genetic determinants of this trait is very important in breeding B. napus for oil content. QTL mapping is an effective approach to dissect the genetic mechanisms of complex quantitative traits [5], and many QTLs for oil content or fatty acid composition have been detected in various crops, such as maize [6]–[8], soybean [9]–[11], sunflower [12], [13], peanut [14], [15] and B. juncea [16], [17].
In B. napus, QTLs for seed oil content have been identified mainly by using DH populations, and 3–27 QTLs with estimated phenotypic variation in the range of 1.2–19.0% were detected in a single B. napus population [4], [18]–[21]. Other types of populations have also been used for B. napus in some cases, for example, intervarietal set of substitution lines [22] and recombinant inbred lines (RILs) [23]. However, the oil content of the parents in these populations showed only small differences, such as 48.0%×47.2% for GS (‘Sollux’בGaoyou’) DH population [4], 41.8%×42.7% for MS (‘Mansholt's Hamburger Raps’בSamourai’) DH population [21], 43.6%×42.0% for TN (‘Tapidor’בNingyou7’) DH population [20], 46.0%×48.0% for DY (‘Darmor-bzh’בYudal’) DH population and 41.2%×46.6% for RNSL (‘Rapid’בNSL96/25’) DH population [19] and 40.4%×37.2% for a RIL (‘GH06’בP174’) population [23]. These experiments were carried out in four, three, four, three, three and seven environments [4], [18]–[21], [23] with 282, 151, 202, 442, 242, 150 and 183 lines, respectively [4], [18]–[21], [23]. More recently, there were QTL analyses for oil content using populations with larger oil content differences in their parents or more environments. Zhao et al. identified nine statistically significant QTLs (SL-QTLs) for seed oil content in 11 trials using the new GS DH map [24], and Sun et al. detected 12 QTLs in three trials using Z5 (‘zy036’ב51070’) DH population with 92 lines whose parents had approximately 10% difference in their seed oil contents [25]. Zhao et al. validated and characterized that the QTL OilA1 could influence oil content in rapeseed within the linkage group A1 using a set of near-isogenic lines (NILs) [26]. Each member of the mapping population might carry different alleles for oil content and represents its own genetic background. However, there were some limiting factors in the populations for detecting QTLs associated with oil content in previous studies: for example, populations were usually not large enough [18], [20], [21], [23], [25], trials were carried out in few environments [4], [19]–[21], [23], [25] or there were only small differences in seed oil content of the two parents [4], [18]–[21], [23], [24].
A high-density genetic linkage map is considered as a key factor for increasing statistical power and precision of detecting QTLs [27]. So far, linkage maps constructed for detecting QTLs associated with oil content in B. napus have been based on a range of marker systems, such as simple sequence repeats (SSRs) [18]–[20], [23]–[25], [28], sequence-related amplified polymorphisms (SRAPs) [18], [23], [24], sequence tagged sites (STSs) [20] and intron fragment length polymorphism (IFLPs) [25]. The number of marker loci in individual genetic linkage maps varied from 125 to 527, covering a total length of 1196.0–2690.0 cM with an average marker interval of 3.5–8.8 cM, respectively [18]–[21], [23]–[25], [28]. Several dense consensus genetic maps have been constructed in B. napus to increase marker density [29], [30]. Lombard and Delourme [31] constructed a consensus map covering a total length of 2429.0 cM by integrating three individual linkage maps. Raman et al. [32] constructed a consensus map, consisting of a 1359 anchored array based on diversity array technology markers, which covered a total of 1987.2 cM. Many QTLs for oil content had been identified in different populations, but the positions of these QTLs differed among various populations and the comparisons of QTLs were tenuous due to the lack of many common markers. Thus it is necessary to construct a consensus map based on individual maps for oil content and to compare the QTLs detected in different populations.
The aims of the present study were as follows: (1) to develop a large segregating DH population with parents showing >10% difference in oil content; (2) to detect QTLs for oil content through a well-constructed linkage map and the phenotypes in eight natural environments; and (3) to construct a consensus linkage map and compare the QTLs for oil content detected with those detected in other studied B. napus populations.
Materials and Methods
Plant Material
The B. napus segregating DH population used in this study was derived from a cross between ‘KenC-8’ and ‘N53-2’. ‘KenC-8’ was the parental of the variety ‘Zayou59’ released in 1996 in China, which was selected from the multi-way hybridized combination, ‘1721-1B’ · ‘start’ב955’ · ‘ChunShan2B’. ‘KenC-8’ is a spring B. napus with seed oil content of approximately 40%. ‘N53-2’ is a DH line developed from the Canadian canola cultivar ‘Midas’ and a Chinese inbred line ‘SE8’. ‘N53-2’ is a winter-type B. napus with seed oil content >50%. A total of 348 DH lines were developed in the year 2007 by microspore culture applied to F1 plants and named the KN DH population.
Field Trials
The field experiments were carried out in eight natural environments at four different locations. The materials were planted in a winter rapeseed area, Dali of Shannxi Province, in northwest China (coded DL) for four years (2008–2009, 2009–2010, 2010–2011 and 2011–2012); in a spring rapeseed area, Sunan of Gansu Province, in northwest China (coded GS) for two years (2010.4–2010.9 and 2011.4–2011.9); and another two semi-winter rapeseed areas, Wuhan (coded WH) and Huanggang (coded HG) of Hubei Province, in central China for one year each (2011–2012 for WH and 2010–2011 for HG). The experiment locations of WH and HG were the experiment bases of Huazhong University of Science and Technology, and DL and GS were the experiment bases of Hybrid Rapeseed Research Center of Shaanxi Province. No specific permissions were required for the field trials. Year–location combinations were treated as microenvironments, and then these microenvironments were divided into three contrasting macroenvironments: spring, semi-winter and winter. The field experiments were in a randomized complete block design with three replications in Dali and with two replications in the other sites. The DH population, together with their parents and F1 hybrids, were sown in double rows for each plot in all locations. There were about 12 plants per row, with a space of 0.4 m between rows and 0.2 m between plants. The field management followed normal agricultural practice. At maturity, five representative plants in the middle of each plot were bulk harvested.
Seed Oil Content Measurement
The seed oil content in all eight trials was measured by nuclear magnetic resonance (NMR) using the method of Burns et al. [22] with modifications.
Molecular Marker Assays
The genotypes of the KN DH population were analyzed using mainly two types of molecular markers: SSRs and SRAPs. In addition, several STSs markers and IFLPs primer pairs were also used to construct the genetic linkage map.
Primer sequence of SSR markers were obtained from various sources: SSR primer pairs prefixed “FITO” were developed by Iniguez-Luy et al. [33]; “CB” and “BRAS” were published by Piquemal et al. [34]; “Na”, “Ol” and “Ra” were developed by Lowe et al. [35]; “CNU” and “niab” were obtained from the Brassica rapa Genome Project (BrGP, http://www.brassica-rapa.org/); “sN”, “sR” and “sS” were provided by Agriculture and Agri-Food Canada (AAFC); “MR” were published by Uzanova and Ecke [36]; “B0”, “H0” and “S0” were developed by Ding et al. [37]; “BnGMS” were developed by Cheng et al. [38]; and “HAU” and “SA” were obtained from private communications. STS markers prefixed “IGF” were published by Qiu et al. [20]; “ANL2” and “CHS” by Li et al. (2006) [39]; ZAAS326 by Zhao et al. [24]; and STS primer pairs prefixed “SF” and “BrSF” as well as IFLP primer pairs GIFLP046 were developed by Sun et al. [25].
The sequence of SRAP markers were from the description of Li and Quiros [40]. There were 32 forward and 21 reverse primers employed (Table S1), resulting in 672 primer combinations. The forward primers were fluorescently labelled with a blue-color dye set (Applied Biosystems), namely, FAM. The SRAP products were separated with Applied Biosystems 3730xl DNA Analyzers with size standards ROX-500 (Applied Biosystems), and GeneMapper Software v3.7 (Applied Biosystems) was used to analyze the results. Each of the polymorphic loci was considered as a dominant marker [18]. The polymorphic primer pairs were named by combining the names of the forward and reverse primers, with a number to indicate the base pair of the polymorphic bands (e.g. e1m5-136).
If SSR, STS or IFLP primer pairs generated more than one polymorphic loci, small letters were used to distinguish the different loci following the marker name. For instance, two genetic loci named BRAS102a on chromosome A2 and BRAS102b on chromosome C2 were both generated from the same primer marker BRAS102.
Linkage Map Construction
The genetic linkage map was constructed by using JoinMap software Version 4.0 [41]. Markers with a mean chi-square value ≥3.0 were excluded in all genetic groups to ensure the markers mapped to linkage groups with a fairly correct order. The threshold for ‘goodness-of-fit’ was set to ≤5.0 with a recombination frequency of <0.4 and LOD scores >1.0. Centimorgan distances were calculated by the Kosambi function for map distance [42]. Finally, all markers in the KN DH map were examined by chi-square test for ‘goodness-of-fit’ to the expected 1∶1 (P<0.01) segregation ratio.
QTL Analysis and Meta-analysis
QTL analysis was performed by using the software Windows QTL Cartographer 2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) [43], [44]. Composite interval mapping (CIM) model was used to estimate putative QTLs with additive effect. A walking speed was set to 2 cM and a window size of 10 cM with five background cofactors. Significance levels for the LOD scores were determined by 1000-permutation test based upon a 5% experiment-wise error rate [45]. Thus, LOD of 2.94–3.10 was used to identify SL-QTLs in each environment, respectively. To avoid missing QTLs with small genetic effects, QTLs that appeared repeatedly in at least two environments at LOD <2.94–3.10 but >2.0 were considered as micro-real QTLs (MR-QTLs) [46]. The overlapping QTLs with LOD >2.0 and <2.94–3.10, as well as all SL-QTLs, were termed identified QTLs [47].
QTLs for oil content, which were detected in multi-environments located in the same region with overlapping confidence interval, may have been one single QTL [48]. Identified QTLs with overlapped confidence intervals were integrated into consensus QTLs using a meta-analysis method with BioMercator2.1 software [48], [49], which has been successfully used in B. napus [47], [50], [51]. The process was according to the description of Shi et al. [47].
The QTLs were named as described by McCouch et al. [52] with minor modifications. A designation begins with QTL abbreviation “qOC” (q, QTL; OC, oil content) suffixed with the linkage group (A1-A10, C1-C9), a hyphen (-), and finally the serial number of QTLs in the linkage group (e.g. qOC-A2-1).
The Consensus Map Construction and QTL Comparison for Oil Content between Different Populations
To determine whether QTLs for oil content detected in the KN DH population were new QTLs, they were compared to QTLs from other populations in previous studies. Comparison of QTLs detected in different populations was carried out using the BioMercator2.1 software [48], [49], including a RIL population [23] and several DH populations: GS/05 [4], GS/12 [24], DY and RNSL [19], Z5 [25] and TN [20]. Although the GS/05 and GS/12 maps were constructed from the same DH population (“Sollux”דGaoyou”), the two maps were considered as different maps because they were constructed by different markers and QTLs were detected in different environments.
A ‘two-round’ strategy of QTL comparison was adopted [47], [50]. In the first round, QTLs identified in different populations were collected, and those QTLs in the same population with overlapped confidence intervals were integrated into consensus QTLs using QTL meta-analysis. The consensus QTLs were named with the population abbreviation followed with ‘qOC’, a hyphen (-) and the linkage group. If more than one consensus QTLs were found in a linkage group, a serial number of the QTL was added (e.g. DY-qOC-A2-2). It should be noted that those consensus QTLs in the KN DH population were named with the population abbreviation ‘KN’ followed with the consensus QTLs names for QTL comparison (e.g. KN-qOC-A1-1). In the second round, the markers in homologous chromosomes were projected from other maps on the reference map to construct a consensus map based on the common markers (sharing the same name) [48], [49]. Then the consensus QTLs detected in different populations were aligned to the consensus map.
Results
Seed Oil Content Analysis
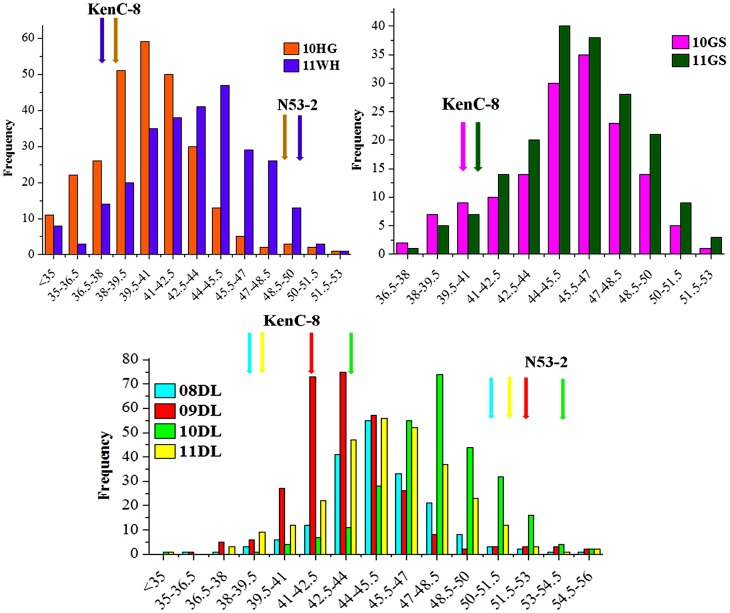
The frequency distributions of oil content in the KN DH population as well as the two parents in the eight microenvironments covering three macroenvironments were summarized in Fig. 1. Because of a strong requirement for vernalization, the high-oil parent ‘N53-2’ and some DH lines did not flower in the spring rapeseed area (10GS and 11GS) and these lines were treated as missing data.
Figure 1. Phenotypic variation in seed oil content in the parents and KN DH population.
(A), (B) and (C) indicated the frequency distribution of seed oil content in semi-winter, spring and winter macroenvironment in different years.
There was a wide range of oil contents in all trials in this study, which showed approximately transgressive and continuous distribution as expected for characters displaying quantitative trait segregation, suggesting polygenic effects of oil content (Fig. 1). The two parents ‘N53-2’ and ‘KenC-8’ showed >10% difference in seed oil contents in six field trials, but not for trials 10GS and 11GS (Table 1). The average oil content of the KN DH population showed significant differences across trials, ranging from 42.0% in trial 10HG to 47.6% in 10DL (Table 1). In general, seed oil content in the winter macroenvironment was higher than in the semi-winter macroenvironment for both parents and DH lines (Table 1 and Fig. 1). In the same macroenvironment, oil content in different trials might also showed large differences; for example, the mean oil content for both parents and DH lines in 10DL was higher than in 09DL. Those findings suggested that the average seed oil content in B. napus was significantly influenced by environment.
Table 1. Means and ranges for seed oil content of KN DH population evaluated in eight microenvironments.
| Trialsa | 08DL | 09DL | 10DL | 10GS | 10HG | 11DL | 11GS | 11WH | |
| KenC-8 | Meanb | 39.4±0.86 | 41.6±0.17 | 43.1±0.81 | 40.5±1.75 | 38.2±1.63 | 39.0±0.23 | 40.9±1.32 | 37.3±1.65 |
| N53-2 | Mean | 50.7±0.61 | 51.7±0.58 | 53.8±0.12 | 48.6±0.75 | 51.4±1.28 | 49.5±1.44 | ||
| DH | Mean | 44.8±0.17 | 44.6±0.13 | 47.6±0.17 | 46.0±0.14 | 42.0±0.17 | 43.6±0.15 | 45.9±0.16 | 43.2±0.22 |
| Max | 51.7 | 52.8 | 54.8 | 51.4 | 51.5 | 53.2 | 52.8 | 51.5 | |
| Min | 38.2 | 37.0 | 34.5 | 39.4 | 33.2 | 36.0 | 38.7 | 34.1 |
Oil content was evaluated in eight microenvironments, where the numbers indicate the year and the letters indicate the location.
Mean value ± SE.
Linkage Map Construction
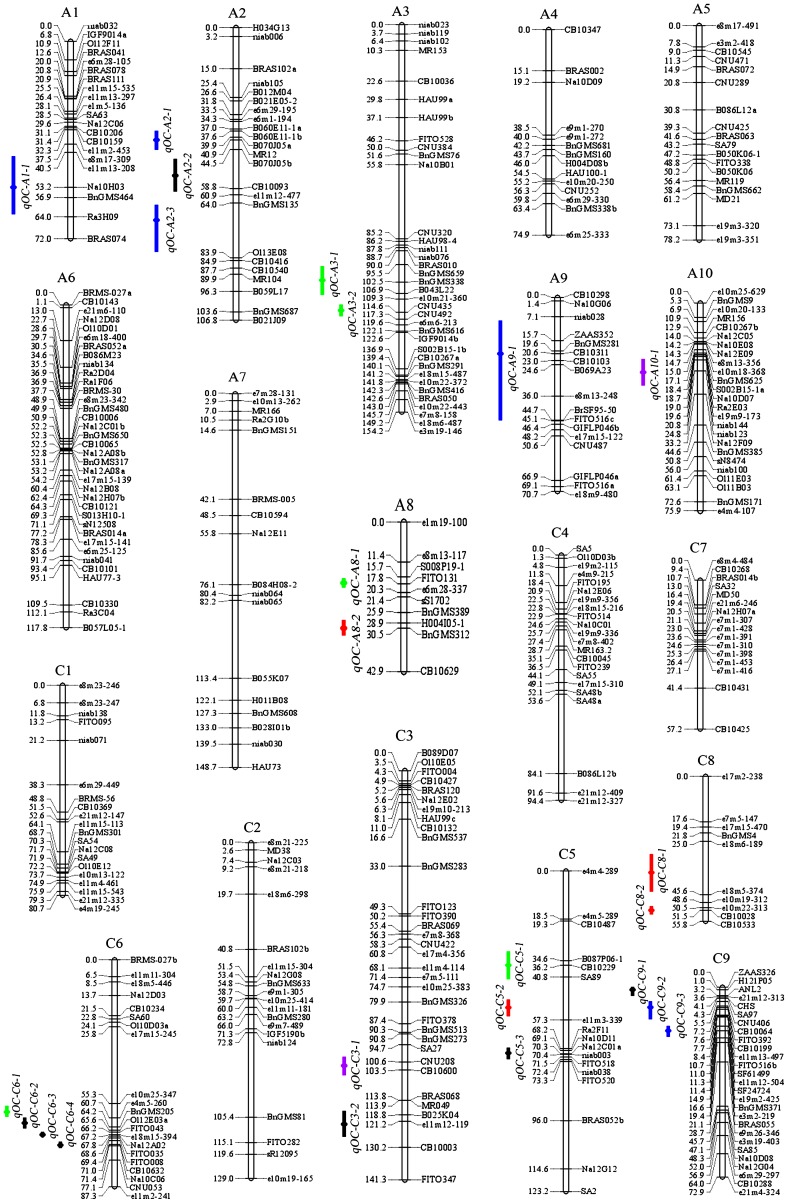
In total, 552 of 1630 molecular markers were identified as polymorphic between the two parents and were subsequently used for genetic linkage map construction using JoinMap software Version 4.0 [41]. Finally, a framework of the genetic linkage map was constructed with 403 of these markers, including 275 SSRs, 117 SRAPs, 10 STSs and 1 IFLP (Fig. 2 and Table 2). As most of the SSR and STS markers were assigned to public linkage maps, they were treated as anchor markers. The 403 markers were assigned to 19 linkage groups, named A1–A10 in the A genome and C1–C9 in the C genome according to new guidelines nomenclature for B. napus linkage groups (http://www.brassica.info/resource/maps/lg-assignments.php). The genetic linkage map had a total length of 1783.9 cM with an average marker interval of 4.4 cM according to the Kosambi function. Each length of the 19 linkage groups ranged from 42.9 (A8) to 154.2 cM (A3), with an average length of 93.9 cM (Fig. 2 and Table 2). The average linkage group lengths of A and C genomes were very similar, with 94.2 and 93.5 cM, respectively. In A genome, there were 217 markers in total and the number in single chromosome ranged from 10 (A8) to 36 (A3 and A6) with an average of 21.7; while 186 markers were located in C genome and the number of markers varied from 10 (C8) to 33 (C3) with an average of 20.7 in single chromosome. This suggested that A and C genomes had similar polymorphism in the KN DH population. Chromosome C9 had the highest marker density (one marker per 2.7 cM), and chromosome A7 had the lowest (one marker per 8.7 cM) (Fig. 2 and Table 2).
Figure 2. Genetic linkage map and the locations of QTLs for seed oil content in KN map.
The 19 linkage groups were represented by vertical bars, designated as A1-A10 in the A genome and C1-C9 in the C genome based on multiple anchor markers located on each chromosome. The loci name were listed on the right of the linkage groups, while position of loci were showed on the left side of linkage groups, which given in Centimorgan (cM). The consensus QTLs associated with oil content in different environments were indicated by bars with various backgrounds on the left of each linkage group. (Blue bar, microenvironment specific QTLs; green bar, one macroenvironment specific QTLs; black bar, QTLs detected in two macroenvironments, red bar, QTLs detected in three macroenvironments; purple bar, MR-QTLs).
Table 2. Distribution of 403 markers on 19 linkage groups in the KN DH map.
| Linkage | No. of markers | Ratio% | Length | Density | No. of markers skewed to | ||||||||
| Group | SRAP | SSR | STS | Total | SRAP | SSR | STS | KenC-8 | N53-2 | Total | Ratio% | ||
| A1 | 7 | 13 | 1 | 21 | 33.3 | 61.9 | 4.8 | 72.0 | 3.4 | 12 | 0 | 12 | 57.1 |
| A2 | 3 | 20 | 0 | 23 | 13.0 | 87.0 | 0 | 106.8 | 4.6 | 0 | 1 | 1 | 4.3 |
| A3 | 8 | 27 | 1 | 36 | 22.2 | 75.0 | 2.8 | 154.2 | 4.3 | 0 | 14 | 14 | 38.9 |
| A4 | 5 | 9 | 0 | 14 | 35.7 | 64.3 | 0 | 74.9 | 5.4 | 0 | 12 | 12 | 85.7 |
| A5 | 4 | 14 | 0 | 18 | 22.2 | 77.8 | 0 | 78.2 | 4.3 | 0 | 2 | 2 | 11.1 |
| A6 | 6 | 30 | 0 | 36 | 16.7 | 83.3 | 0 | 117.8 | 3.3 | 0 | 8 | 8 | 22.2 |
| A7 | 2 | 15 | 0 | 17 | 11.8 | 88.2 | 0 | 148.7 | 8.7 | 2 | 1 | 3 | 17.6 |
| A8 | 3 | 7 | 0 | 10 | 30.0 | 70.0 | 0 | 42.9 | 4.3 | 4 | 0 | 4 | 40.0 |
| A9 | 3 | 11 | 2 | 17 | 17.6 | 64.7 | 11.8 | 70.7 | 4.2 | 3 | 0 | 3 | 17.6 |
| A10 | 6 | 19 | 0 | 25 | 24.0 | 76.0 | 0 | 75.9 | 3.0 | 0 | 24 | 24 | 96.0 |
| C1 | 10 | 10 | 0 | 20 | 50.0 | 50.0 | 0 | 80.7 | 4.0 | 0 | 12 | 12 | 60.0 |
| C2 | 9 | 10 | 1 | 20 | 45.0 | 50.0 | 5 | 129.0 | 6.5 | 0 | 17 | 17 | 85.0 |
| C3 | 7 | 26 | 0 | 33 | 21.2 | 78.8 | 0 | 141.3 | 4.3 | 1 | 11 | 12 | 36.4 |
| C4 | 9 | 13 | 0 | 22 | 40.9 | 59.1 | 0 | 94.4 | 4.3 | 0 | 6 | 6 | 27.3 |
| C5 | 3 | 14 | 0 | 17 | 17.6 | 82.4 | 0 | 123.2 | 7.2 | 1 | 3 | 4 | 23.5 |
| C6 | 7 | 14 | 0 | 21 | 33.3 | 66.7 | 0 | 87.3 | 4.2 | 0 | 14 | 14 | 66.7 |
| C7 | 9 | 7 | 0 | 16 | 56.3 | 43.8 | 0 | 57.2 | 3.6 | 13 | 0 | 13 | 81.3 |
| C8 | 7 | 3 | 0 | 10 | 70.0 | 30.0 | 0 | 55.8 | 5.6 | 8 | 0 | 8 | 80.0 |
| C9 | 9 | 13 | 5 | 27 | 33.3 | 48.1 | 18.5 | 72.9 | 2.7 | 1 | 19 | 20 | 74.1 |
| Total | 117 | 275 | 10 | 403 | 29.0 | 68.2 | 2.5 | 1783.9 | 4.4 | 45 | 144 | 189 | 46.9 |
A large proportion (46.9%, 189/403) of the mapped loci showed segregation distortion in the KN DH map, in which 76.2% deviated toward the high-oil parent ‘N53-2’ and 23.8% toward the low-oil parent ‘KenC-8’. These distorted markers were distributed across all 19 chromosomes and tended to cluster on A3, A10, C2, C9 and C6, especially on A10 with 96.0% of its markers showing distorted segregation (Table 2). The skewed loci on linkage groups A1, C7 and C8 favored the parent ‘KenC-8’ allele, and linkage groups A3, A4, A10, C1, C2, C3, C6 and C9 comprised loci favoring the parent ‘N53-2’ allele (Table 2). In addition, no more than five skewed loci were distributed on chromosomes A2, A5, A7, A8, A9 and C5.
SL-QTL and MR-QTL Detection and Meta-analysis for Oil Content
QTL for oil content were analyzed in each experiment by CIM approach, and identified QTLs were integrated into consensus QTLs using a meta-analysis method. Detailed information of identified QTLs was given in Table S2 and consensus QTLs were summarized in Table 3 and Fig. 2.
Table 3. Consensus QTLs for seed oil content in the KN DH population.
| QTL | Chromosome | Position(cM) | LOD | Additive | R2 (%) | CIa | Environmentb |
| qOC-A1-1 | A1 | 53.2 | 3.99 | −0.80 | 11.74 | 42.0–62.9 | 10GS |
| qOC-A2-1 | A2 | 41.0 | 3.66 | −0.60 | 4.42 | 37.6–44.5 | 10HG |
| qOC-A2-2 | A2 | 53.9 | 3.35–5.15 | −0.92–−0.60 | 4.74–10.26 | 48.1–59.6 | 10HG/11GS |
| qOC-A2-3 | A2 | 70.1 | 4.03 | −0.99 | 11.19 | 64.8–81.6 | 11GS |
| qOC-A3-1 | A3 | 101.8 | 2.98–3.41 | 0.46–0.57 | 3.40–3.74 | 96.2–107.5 | 09DL/11DL |
| qOC-A3-2 | A3 | 113.8 | 2.15–3.28 | 0.40–0.55 | 2.67–5.86 | 111.6–116.1 | 08DL/09DL/11DL |
| qOC-A8-1 | A8 | 17.6 | 2.77–4.28 | 0.53–0.89 | 3.39–5.90 | 16.4–18.9 | 11WH/10HG |
| qOC-A8-2 | A8 | 30.4 | 3.97–7.09 | 0.57–0.82 | 4.81–8.73 | 28.2–32.6 | 10HG/09DL/10DL/11DL/11GS |
| qOC-A9-1 | A9 | 20.6 | 3.22 | 0.58 | 3.55 | 8.6–44.7 | 11DL |
| qOC-A10-1 | A10 | 25.2 | 2.22–2.70 | −0.84–−0.62 | 3.41–5.36 | 20.5–30.0 | 11GS/08DL/11WH |
| qOC-C3-1 | C3 | 102.0 | 2.03–2.55 | 0.55–0.67 | 2.64–5.69 | 98.9–105.2 | 11WH/10GS/10DL |
| qOC-C3-2 | C3 | 122.1 | 2.95–6.09 | 0.53–0.68 | 3.43–7.48 | 117.6–126.5 | 10HG/11DL/09DL/10DL |
| qOC-C5-1 | C5 | 36.3 | 6.06–6.28 | 0.63–0.76 | 6.79–11.36 | 31.1–41.5 | 08DL/09DL |
| qOC-C5-2 | C5 | 52.6 | 2.75–8.23 | 0.63–0.98 | 5.18–17.88 | 49.4–55.8 | 08DL/11WH/09DL/10HG/11GS |
| qOC-C5-3 | C5 | 70.0 | 2.18–6.76 | 0.70–0.90 | 3.71–10.58 | 67.9–72.2 | 11DL/10GS/11DL/11GS |
| qOC-C6-1 | C6 | 58.4 | 4.58–5.72 | 0.76–0.83 | 7.90–8.67 | 56.4–60.4 | 10DL/08DL |
| qOC-C6-2 | C6 | 62.7 | 2.59–4.76 | 0.49–0.83 | 3.51–6.85 | 60.7–64.7 | 10HG/09DL |
| qOC-C6-3 | C6 | 67.2 | 2.99–7.00 | 0.70–0.98 | 4.26–8.57 | 66.4–68.1 | 11DL/08DL/10HG/11WH |
| qOC-C6-4 | C6 | 71.2 | 3.13–8.29 | 0.53–1.01 | 4.16–11.08 | 70.0–72.3 | 09DL/11DL/11WH/10DL |
| qOC-C8-1 | C8 | 37.1 | 2.07–4.21 | 0.68–0.85 | 4.78–6.36 | 30.0–44.3 | 11DL/11GS/11WH |
| qOC-C8-2 | C8 | 51.5 | 2.56–3.53 | 0.41–0.73 | 2.69–8.01 | 49.9–53.1 | 11DL/10GS/09DL/11WH |
| qOC-C9-1 | C9 | 1.0 | 2.54–5.51 | 0.56–0.63 | 4.61–6.19 | 0.0–3.3 | 08DL/09DL/11GS |
| qOC-C9-2 | C9 | 7.7 | 5.90 | 0.65 | 6.60 | 5.5–12.1 | 09DL |
| qOC-C9-3 | C9 | 16.6 | 3.33 | 0.49 | 3.81 | 14.9–19.0 | 09DL |
The 2-LOD confidence interval of QTLs.
The environment in which QTLs were detected. DL: Dali, a winter rapeseed area; GS: Gansu, a spring rapeseed area; WH and HG: Wuhan and Huanggang, both are semi-winter type rapeseed growing area; 08, 09, 10 and 11 denote the year of 2008, 2009, 2010 and 2011, respectively. Environment with bold indicated that the QTLs below the threshold LOD score but with LOD >2.0.
A total of 63 identified QTLs for oil content were detected in eight microenvironments, singly explaining 2.64–17.88% of the estimated phenotypic variation (Table S2). Further analysis revealed that a large proportion of the identified QTLs (42/63) were detected in C genome, with only 21 in A genome. The number of identified QTLs detected in different microenvironments also showed great differences; for example, 11 and 12 QTLs were detected in the 11DL and 09DL trials, respectively, whereas only four were detected in 10GS (Table S2).
The 63 identified QTLs were classified into two types: (i) 57 overlapping QTLs and (ii) six non-overlapping SL-QTLs. The 57 identified QTLs with overlapping confidence intervals were integrated into 18 consensus QTLs by BioMercator2.1 software (Table S2 and Fig. 2). As a result, the average confidence interval of single QTL was reduced from 13.1 to 6.3 cM. Among these consensus QTLs, two were detected in five microenvironments (qOC-A8-2 and qOC-C5-2), five in four microenvironments (qOC-C3-2, qOC-C5-3, qOC-C6-3, qOC-C6-4 and qOC-C8-2) and the remaining 11 QTLs in two to three microenvironments (Table 3 and Table S2). The genes associated with these QTLs controlling seed oil content may be more structurally important and less affected by environment. The consensus QTLs for oil content could be divided into different types based on the macroenvironments in which they were detected; for example, seven were detected in two different macroenvironments, including one expressed in both spring and semi-winter macroenvironments (qOC-A2-2), two were significant in both spring and winter macroenvironments (qOC-C5-3 and qOC-C9-1) and four were detected in both semi-winter and winter macroenvironments (qOC-C3-2, qOC-C6-2, qOC-C6-3 and qOC-C6-4). Furthermore, six were consistently expressed in all three macroenvironments (qOC-A8-2, qOC-A10-1, qOC-C3-1, qOC-C5-2, qOC-C8-1 and qOC-C8-2). Four QTLs were only repeatedly detected in the DL microenvironment in different years (qOC-A3-1, qOC-A3-2, qOC-C5-1 and qOC-C6-1), which were recognized as winter macroenvironment specific QTLs; while one QTL was only repeatedly detected in 11WH/10HG (qOC-A8-1) and considered a semi-winter macroenvironment specific QTL. QTLs detected in multiple macroenvironments are of great importance for breeders to select materials with wide adaptability in different macroenvironments. When doing marker-assisted breeding (MAS) for special geographical region cultivars, macroenvironment specific QTLs should be paid more attention. In addition, the six non-overlapping SL-QTLs were only expressed in a specific microenvironment and were considered as microenvironment specific QTLs (Table 3 and Table S2), these QTLs were also considered to be consensus QTLs, distributed over five linkage groups (A1, A2, A3, A9 and C9). No microenvironment specific QTLs were found in 08DL, 10DL and 11WH. Two microenvironment specific QTLs (qOC-A1-1 and qOC-A2-3) were strongly expressed in 10GS and 11GS, both were belonged to spring macroenvironment, and explained 11.74 and 11.36% of phenotypic variation, respectively. The two QTLs might be worthy of attention when doing MAS for spring geographical region cultivars.
The abovementioned 24 consensus QTLs were located on 11 chromosomes, including A1, A2, A3, A8, A9, A10, C3, C5, C6, C8 and C9, and each explained a mean of 3.55–11.74% of the phenotypic variation, respectively (Table S2). Two consensus QTLs (qOC-A10-1 and qOC-C3-1), both with LOD value below the significance levels but repeatedly detected in three microenvironments, were regarded as MR-QTLs. The low-oil content parent ‘KenC-8’ contributed favorable alleles at five loci, which had negative additive effects and were all located on A genome, including three microenvironment-specific QTLs (qOC-A1-1, qOC-A2-1 and qOC-A2-3), one MR-QTL (qOC-A10-1) and the QTL qOC-A2-2. The remaining 19 QTLs all had positive additive effects, indicating that they came from the high-oil parent ‘N53-2’.
Consensus Map Construction and QTL Comparison for Oil Content between Different Populations
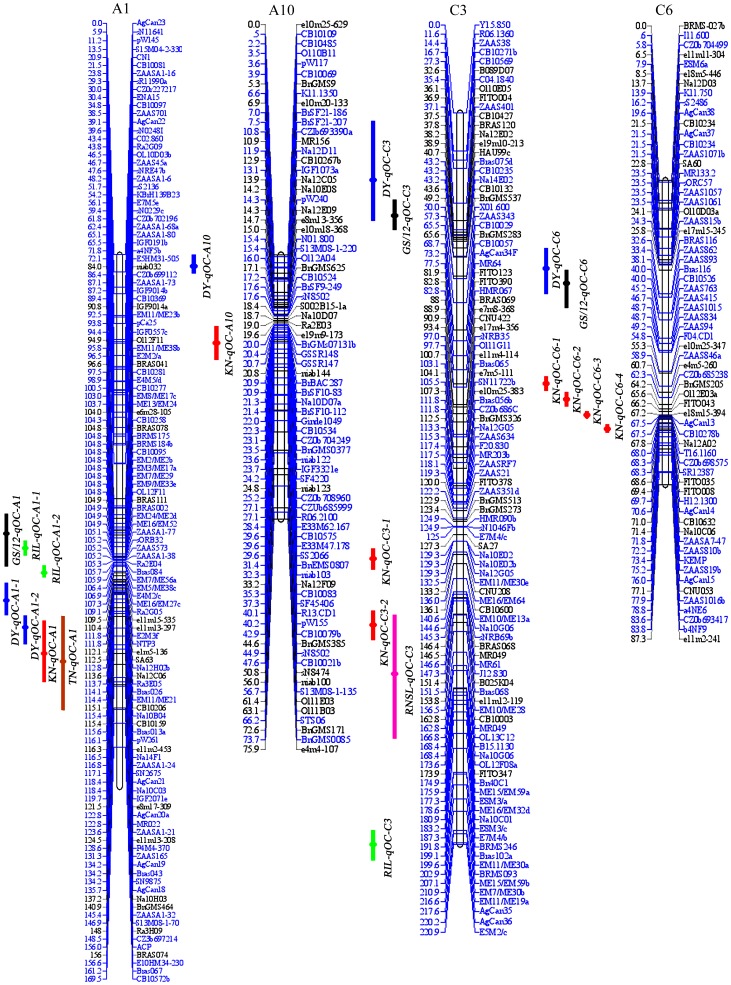
According to the reported QTLs for oil content in B. napus, seven populations (eight maps) were selected for comparison, including KN, DY, RNSL, TN, Z5, GS/05 and GS/12 DH population maps and a RIL population map. To exactly compare the results from different populations, QTLs detected in a single population clustered in the same region were firstly integrated to consensus QTLs by BioMercator 2.1 software. Detailed information of those consensus QTLs in different populations was summarized in Table S3. The KN map was treated as the reference map in most linkage groups, and the markers and QTLs were projected from other maps onto the KN map except for A8 and C8. On linkage groups A8 and C8, the markers and QTLs were projected from other maps onto the TN map, because few common markers were available between KN and other populations. The comparison of markers and QTLs on linkage groups A4, A9, C4 and C7 were tenuous due to the lack of common markers between different populations, and no markers and QTLs were projected from other maps onto the KN map for these linkage groups. Individual component maps were aligned to the reference linkage map to construct a consensus map. Finally, a total of 1335 markers were integrated in the consensus map. The consensus map consisted of 19 linkage groups, covering 2395.2 cM of the total genome with an average marker interval of 1.8 cM (Fig. 3 and Table S4). The length ranged from 57.2 (C7) to 220.9 cM (C3) for each individual chromosome, with an average length of 126.1 cM. Chromosome A1 had the highest marker density (one marker per 1.0 cM) and chromosome A8 had the lowest marker density (one marker per 5.9 cM) (Fig. 3 and Table S4).
Figure 3. The consensus map and QTLs for oil content detected in different populations.
Markers with blue color indicated these makers were projected from other maps on the KN map, via application of the homothetic projection based on common markers by BioMercator 2.1 software. QTLs for oil content detected in different populations were discriminated with different color bars (blue bar, QTLs detected in DY population; Orange bars, TN population; black bars, GS/12 population; green bars, RIL population; pink bars, RNSL population; red bars, KN population).
A total of 99 consensus QTLs were detected in the abovementioned seven populations (eight maps) (Table S3). The number of consensus QTLs in different populations ranged from seven (TN and GS/05 populations) to 24 (KN population). Of 16 consensus QTLs, 15 were projected from the DY map onto the consensus map; however, only two of seven consensus QTLs was projected from the GS/05 map onto the consensus map. Among the 24 consensus QTLs detected in the KN DH population, 20 consensus QTLs were aligned to the consensus map. Except for the 16 consensus QTLs distributing on chromosomes A4, A9, C4 and C7 in all eight maps, 69 of the remaining 83 consensus QTLs were successfully projected from individual maps onto the consensus map (Table 4).
Table 4. Comparison of consensus QTLs for seed oil content detected in different populations in B. napus.
| QTLa | Chrb | SG/12 | SG/05 | DY | KN | Z5 | TN | RNSL | RIL |
| HO×HOc | HO×HO | HO×MO | HO×LO | HO×LO | MO×MO | MO×LO | LO×LO | ||
| qA1-1 | A1 | GS/12-qOC-A1-1 | RIL-qOC-A1-1 | ||||||
| qA1-2 | A1 | RIL-qOC-A1-2 | |||||||
| qA1-3 | A1 | DY-qOC-A1-1 | |||||||
| qA1-4 | A1 | DY-qOC-A1-2 | KN-qOC-A1-1 | TN-qOC-A1 | |||||
| qA2-1 | A2 | DY-qOC-A2-1 | |||||||
| qA2-2 | A2 | KN-qOC-A2-1 | |||||||
| qA2-3 | A2 | KN-qOC-A2-2 | |||||||
| qA2-4 | A2 | DY-qOC-A2-2 | KN-qOC-A2-3 | ||||||
| qA2-5 | A2 | DY-qOC-A2-3 | |||||||
| qA3-1 | A3 | RNSL-qOC-A3 | RIL-qOC-A3 | ||||||
| qA3-2 | A3 | DY-qOC-A3 | KN-qOC-A3-1 | Z5-qOC-A3-1 | |||||
| Z5-qOC-A3-2 | |||||||||
| qA3-3 | A3 | KN-qOC-A3-2 | |||||||
| qA5-1 | A5 | GS/12-qOC-A5-1 | |||||||
| qA5-2 | A5 | GS/12-qOC-A5-2 | DY-qOC-A5 | ||||||
| qA6-1 | A6 | DY-qOC-A6-1 | |||||||
| qA6-2 | A6 | Z5-qOC-A6-1 | RNSL-qOC-A6 | ||||||
| Z5-qOC-A6-2 | |||||||||
| qA6-3 | A6 | DY-qOC-A6-2 | |||||||
| qA7-1 | A7 | GS/05-qOC-A7 | |||||||
| qA7-2 | A7 | GS/12-qOC-A7 | RNSL-qOC-A7 | RIL-qOC-A7 | |||||
| qA8-1 | A8 | TN-qOC-A8 | RNSL-qOC-A8 | ||||||
| qA10-1 | A10 | DY-qOC-A10 | |||||||
| qA10-2 | A10 | KN-qOC-A10-1 | |||||||
| qC1-1 | C1 | RNSL-qOC-C1 | |||||||
| qC1-2 | C1 | GS/05-qOC-C1 | |||||||
| qC2-1 | C2 | DY-qOC-C2-1 | |||||||
| qC2-2 | C2 | GS/12-qOC-C2-1 | DY-qOC-C2-2 | Z5-qOC-C2 | TN-qOC-C2 | ||||
| qC2-3 | C2 | GS/12-qOC-C2-3 | |||||||
| qC3-1 | C3 | GS/12-qOC-C3 | DY-qOC-C3 | ||||||
| qC3-2 | C3 | KN-qOC-C3-1 | |||||||
| qC3-3 | C3 | KN-qOC-C3-2 | RNSL-qOC-C3 | ||||||
| qC3-4 | C3 | RIL-qOC-C3 | |||||||
| qC5-1 | C5 | KN-qOC-C5-1 | |||||||
| qC5-2 | C5 | KN-qOC-C5-2 | |||||||
| qC5-3 | C5 | KN-qOC-C5-3 | |||||||
| qC5-4 | C5 | DY-qOC-C5 | |||||||
| qC6-1 | C6 | GS/12-qOC-C6 | DY-qOC-C6 | ||||||
| qC6-2 | C6 | KN-qOC-C6-1 | |||||||
| qC6-3 | C6 | KN-qOC-C6-2 | |||||||
| qC6-4 | C6 | KN-qOC-C6-3 | |||||||
| qC6-5 | C6 | KN-qOC-C6-4 | |||||||
| qC8-1 | C8 | GS/12-qOC-C8-1 | |||||||
| qC8-2 | C8 | RNSL-qOC-C8 | |||||||
| qC8-3 | C8 | GS/12-qOC-C8-2 | |||||||
| qC8-4 | C8 | GS/12-qOC-C8-3 | |||||||
| qC9-1 | C9 | KN-qOC-C9-1 | |||||||
| qC9-2 | C9 | KN-qOC-C9-2 | Z5-qOC-C9-1 | ||||||
| qC9-3 | C9 | KN-qOC-C9-3 | Z5-qOC-C9-2 | ||||||
Consensus QTLs detected in different populations clustered in the same regions on the consensus map were considered to be one QTL, named a designation begins with abbreviation “q” suffixed with the linkage group (A1-A10, C1-C9), a hyphen (-), and finally the serial number of the QTL in the linkage group. As a result, 69 consensus QTLs aligned to the consensus map were integrated into 47 new QTLs.
Chromosome.
The types of population based on their parents oil content. LO: low oil content cultivar; MO middle oil content cultivar; HO: high oil content cultivar. QTLs with bold indicated that these consensus QTLs were detected in the KN DH population equivalent to the QTLs showed in Table 3.
A total of eight consensus QTLs were detected in TN, DY, RNSL, RIL, GS/05 and GS/12 populations on chromosome A1, and seven of those QTLs were aligned to the consensus map, except for one QTL in GS/05 population because of a lack of common markers (Fig. 3 and Table 4). One QTL on chromosome A1 (KN-qOC-A1-1) of the KN population was co-localized with QTLs TN-qOC-A1 and DY-qOC-A1-2, but differed from the other four QTLs in DY, RIL and GS/12 populations. QTLs on chromosome A2 were only detected in KN, DY and Z5 populations, and one QTL KN-qOC-A2-3 was co-localized with QTL DY- qOC-A2-2; the other two QTLs (KN-qOC-A2-1 and KN-qOC-A2-2) were new QTLs, while the two QTLs in Z5 population were not aligned to the consensus map (Table S4). Totally, seven QTLs on chromosome A3 were successfully projected from DY, RNSL, RIL and Z5 populations onto the consensus map. One QTL on A3 (KN-qOC-A3-1) in KN population overlapped with QTLs Z5-qOC-A3-1, Z5-qOC-A3-2 and DY-qOC-A3, but differed from QTLs RIL-qOC-A3-1 and RNSL-qOC-A3-1; and the other QTL KN-qOC-A3-2 was a new QTL. For the QTLs on chromosome A8, RNSL-qOC-A8 was considered the same as QTL TN-qOC-A8 with overlapping confidence interval. Two QTLs (KN-qOC-A8-1 and KN-qOC-A8-2) were detected on chromosome A8 in the KN DH population, and KN-qOC-A8-1 was potentially co-localized with QTLs TN-qOC-A8 and RNSL-qOC-A8 based on the only common marker (sS1702). KN-qOC-A8-2 was potentially a new QTL. QTLs on chromosome A10 were only detected in KN and DY populations, and the MR-QTL KN-qOC-A10-1 in KN population was considered a new QTL when compared with the QTL in DY population (DY-qOC-A10).
An interesting finding was the large number of new consensus QTLs (nine QTLs) for oil content in the C genome (Table 4), which has been rarely reported in previous studies. One QTL on chromosome C3 (KN-qOC-C3-1) in KN population was a new QTL compared with QTLs in DY, RNSL, RIL and GS/12 populations, and another QTL KN-qOC-C3-2 was co-localized with QTL RNSL-qOC-C3 (Fig. 3). Three new QTLs (KN-qOC-C5-1, KN-qOC-C5-2 and KN-qOC-C5-3) were identified on chromosome C5 in KN population compared with QTLs in DY population. There were four QTLs (KN-qOC-C6-1, KN-qOC-C6-2, KN-qOC-C6-3 and KN-qOC-C6-4) detected in KN population on chromosome C6, which differed from QTLs in DY (DY-qOC-C6) and GS/12 (GS/12- qOC-C6) populations (Fig. 3), and no QTL were found in other populations, suggesting that these were all new QTLs. QTLs on chromosome C8 were consistently detected in KN, GS/05, RNSL, Z5 and GS/12 populations, but the QTLs in KN (two QTLs), GS/05 (one QTL) and Z5 (one QTL) populations were not compared because no common markers were available. QTLs on chromosome C9 were seldom detected in previous studies except for two QTLs in Z5 population. KN-qOC-C9-1 on chromosome C9 in KN population was a new QTL; and the other two QTLs, KN-qOC-C9-2 and KN-qOC-C9-3, were co-localized with QTLs Z5-qOC-C9-1 and Z5-qOC-C9-2 in Z5 population, respectively (Table 4 and Table S4). Compared with the abovementioned previous studies, 14 of the 24 consensus QTLs in KN population were considered as potential new QTLs in the present study. So far, QTLs for oil content have been observed in all 19 linkage groups, but the QTLs were not well distributed between linkage groups, and the different types of populations showed different capacities for detecting QTLs. Some QTLs were repeatedly detected in different populations and formed QTL clusters in some chromosome regions, while some QTLs were only detected in specific populations. Those QTLs repeatedly detected in different populations offer the possibility of fine mapping and map-based cloning of genes contributing to seed oil content in B. napus. This method provided a new clue for comparing QTLs detected in different populations.
Discussion
Seed oil content is an important trait for B. napus, controlled by complex genetic architecture and influenced by environment. High-oil cultivars may contain excellent alleles for high oil content and using high-oil rapeseed cultivars to construct populations may give higher resolution for QTL screening [25]. Many DH populations have been constructed for QTL analysis of seed oil content in B. napus [4], [19]–[25], with the differences in seed oil content between parents in the range of 0.8–5.9% and population sizes of 150–442 lines; except for the Z5 population with approximately 10% difference in seed oil content between parent but with only 92 DH lines. A DH population (KN DH) was constructed in the present study, in which the parents showed significant differences in oil content, with 11.3, 10.1, 10.7, 10.4, 12.4 and 12.2% differences in 08DL, 09DL, 10DL, 10HG, 11DL and 11WH trials, respectively. In addition to an excellent performance in oil content, the KN DH population consisted of 348 lines and to our knowledge is the largest B. napus DH population with >10% difference in oil content of parents.
The New Constructed Larger DH Population and Linkage Map were Suitable for Detecting QTLs Associated with Oil Content
A DH genetic linkage map covering a total length of 1783.9 cM with an average marker interval of 4.4 cM was constructed using the KN DH population. The marker order in the KN linkage map was in good agreement with an international TN DH reference map for B. napus (http://brassica.nbi.ac.uk/) that has been widely used for mapping different agronomic traits [20], [46], [47], [50], [51]. A high percentage of markers (46.9%) in the KN DH map showed segregation distortion, as also observed in other B. napus DH populations, such as Z5 [25], MS [21], RNSL [19], DY [19] and GS/12 [24] populations with 35.9, 11.2, 20, 35.7 and 48% of markers with segregation distortion, respectively. Segregation distortion has been identified as a strong evolutionary force [53]. The unequal segregation of alleles results from a variety of different mechanisms, such as segregation distortion regions on chromosomes [54], [55], genetic hitchhiking effect [56], [57], strong zygotic selection, certation and gamete selection [58]. Interestingly, the skewed loci on chromosome A1 favored the low-oil parent ‘KenC-8’ alleles and the low-oil QTL (qOC-A1-1) with negative additive effect was also located on the region of skewed loci clustering. The skewed loci on chromosomes A10, C6 and C9 favored the high-oil parent ‘N53-2’ alleles and QTLs on these chromosomes were all high-oil QTLs with positive additive effect distributed over the region of skewed loci clusters. Previously, Zhao et al. [4] identified 35.2% of the mapped makers with significant deviations and two QTLs for oil content located on the region of skewed loci clusters in GS/05 DH population. Zhao et al. [24] identified 48.0% of mapped makers with distorted segregation in the GS/12 DH population and three QTLs situated in the genomic regions with distorted segregation of the marker loci. These findings suggested that segregation distortion was partially associated with QTLs for oil content in DH populations constructed from high-oil B. napus cultivars; however, the relationships with other types of populations require further research.
A large population, a high-density genetic map and replicated experiments in multiple environments are considered as three key factors for increasing statistical power and precision in detecting QTLs [27]. Consensus QTLs for oil content detected in previous studies were 7, 16, 11, 7, 10, 12 and 12 in TN, DY, RNSL, GS/05, RIL, Z5, GS/12 populations, respectively, and the DY population showed the most consensus QTLs (16 QTLs) with the largest population size of 442 lines (Table S3). Comparison of QTLs between the Z5 and KN populations supported the view that a larger mapping population enabled higher capacity to identify QTLs and correctly estimate the magnitude of their effects. Both of the populations showed approximately 10% difference in their parents’ seed oil contents, but the KN (348 lines) was about four times larger than the Z5 population (92 lines), as a result, the number of consensus QTLs detected in the KN population (24 QTLs) was twice that in the Z5 population (12 QTLs). Sun et al. [25] reported that the proportion of phenotypic variation explained by an individual QTL in Z5 population was higher than in most other reported populations, which were in the range of 9.15–24.56%. As the Z5 population was smaller than most other populations, the phenotypic variation explained by an individual QTL might be overestimated, whereas that in KN population was in the range of 2.64–17.88%. This phenomenon was also found in other studies, for example, Bradshaw et al. [59] reported that double the number of QTLs for 12 floral traits was detected when the F2 sample size increased from 93 to 465 plants, and the lowest phenotypic variation explained by an individual QTL decreased from 18.7 to 3.3% in monkey flower (Mimulus). In a simulation study, Li et al. [60] pointed out that as the population size increased from 100 to 500, the estimated QTL position and the effect asymptotically approached their true values. These results indicated the importance of population size in QTL mapping, and the use of large populations could distinguish a higher level of allelic variation and improve mapping efficiency.
Mapping populations constructed from parents with great differences in seed oil content could enable the identification of QTL associated with large phenotypic differences in seed oil content in B. napus [25]. The parents in most studied populations showed only small differences (usually <5%) in seed oil content except for Z5 [19]–[24]. The QTLs obtained in GS/05 and GS/12 (282 lines) population were compared with QTLs detected in KN population, as the GS population had the most similar population size to the KN population but the two populations were significantly different in their parents’ seed oil contents. The results revealed that the consensus QTLs detected in KN population (24 QTLs) were three and two times that detected in the GS/05 (seven QTLs) and GS/12 (12 QTLs) populations, respectively (Table S3). In addition, although the DY (442 lines) was larger than the KN population (348 lines), there were less QTLs detected in DY population (16 QTLs) associated with seed oil content because two parents used in DY population only showed approximately 2% difference in oil content. The results indicated that populations with significant differences in their parents’ seed oil contents might have more different alleles resulting in increasing oil content and could give a higher power of detection for QTLs. Exploring the genetic diversity in varieties’ genetic backgrounds such as in the KN population would be helpful to identify more effective alleles for increasing oil content.
QTLs Comparison and New QTLs Detection for Oil Content in the KN Population
Generally speaking, consensus maps can greatly increase map resolution, and might be a powerful tool to survey the genetic diversity of loci/alleles underlying complex traits, develop molecular breeding and map-based cloning of genes. So far, consensus maps have been constructed for many plants, such as barley [61]–[64], bread wheat [65], sorghum [66], sunflower [67], cowpea [68] and rye [69]. However, few consensus maps have been constructed in B. napus. Lombard and Delourme and Raman et al. [31], [32] constructed consensus maps consisting of 540 and 1359 marker loci from the integration of three and six DH mapping populations, respectively, and covered a total of 2429.0 and 1987.2 cM. In order to compare the difference in QTLs for oil content detected in the KN with other populations, a consensus genetic map was constructed based on the common marker loci from different populations. Lombard and Delourme [31] predicted that the length of the B. napus genome was between 2127 and 2480 cM. By combining the eight individual maps into a consensus map, the length of the KN map increased from 1783.9 to 2395.2 cM with the average marker interval reduced from 4.4 to 1.8 cM, which indicated that this consensus map might have a near-complete coverage of the B. napus genome. This newly constructed consensus map comprised different genetic backgrounds for the eight individual maps derived from the 14 parents involved, and could be of nearly universal use as a reference map for QTL mapping in B. napus. In addition, most of the linkage gaps of >15 cM in the KN map were filled by the addition of 932 molecular markers to the reference map, with the exception of gaps on chromosomes A4, A9, C4, C7 and C8 (Table S4). Compared with consensus genetic maps reported for B. napus, the present consensus map has more markers or greater total length and more complicated genetic backgrounds. This high-density consensus map should facilitate the selection of polymorphic markers in important chromosomal intervals and provides a framework for comparing QTLs associated with oil content detected in different populations.
According to oil content of the parents used to construct the B. napus population for QTL analysis associated with oil content, the parents could be factitiously divided into three types: low-oil (LO, oil content <42.0%), middle-oil (MO, oil content 42.0–47.0%) and high-oil (HO, oil content >47.0%) parents. Thus the segregating populations in the present study were divided into six types based on parents’ oil contents: HO×HO (SG/05, SG/12), HO×MO (DY), HO×LO (Z5), MO×MO (TN), MO×LO (RNSL) and LO×LO (RIL) populations. Projecting the QTLs from different population maps on the consensus map showed that 14 of 24 QTLs identified in the KN DH population were new QTLs, including five new QTLs in A genome and nine in C genome. All of the six types of populations had QTLs on chromosomes A1 and C3, two QTLs detected in the KN population on C3 were new QTLs, and one QTL on A1 was co-localized with QTLs in TN and DY populations (Table 4). QTLs on chromosomes A3 and A8 were detected in four of the population types (HO×MO, HO×LO, MO×LO and LO×LO), and one of two QTLs on A3 (KN-qOC-A3-2) detected in the KN population was a new QTL and one of two QTLs on A8 (KN-qOC-A8-2) was a potential new QTL.
Zhao et al. [24] showed that QTLs for oil content on A1 and C3 could be detected in all six maps used in their comparison. Delourme et al. [19] reported that QTLs for oil content on A1 and A3 were detected in five and six populations, respectively. The results revealed that QTLs for oil content on these chromosomes appeared to be more consistent and could be detected in various gene pools. The reason might be in part that those QTLs affecting oil content were more structurally important and less affected by environment. However, QTLs on some chromosomes might only be specific to one or more particular genetic backgrounds. QTLs on chromosome C6 were detected in HO×HO (GS/12), HO×MO (DY) and HO×LO (KN); and QTLs on chromosomes A2, A10 and C5 were only detected in the HO×MO (DY) and HO×LO (KN and/or Z5) populations, which had one parent that was a special high-oil cultivar (Table 4), indicating that high-oil rapeseed cultivars may contain excellent alleles for oil content on those chromosomes. Our QTLs on A2, A10, C5 and C6 were all new QTLs (10 QTLs) except for one QTL on chromosome A2 (KN-qOC-A2-3) that co-localized with QTL in DY population (DY-qOC-A2-2). Sun et al. [25] reported that they had first mapped two QTLs on chromosome C9 in Z5 population. One of our QTLs (KN-qOC-C9-1) on chromosome C9 was a new QTL and the other two QTLs were co-localized with QTLs detected in Z5 population. Since the QTLs on chromosome C9 were only detected in KN and Z5 populations, and both were HO×LO population and had a high-oil parent with seed oil content of approximately 50%, it appeared that the alleles increasing seed oil content on chromosome C9 were only expressed in cultivars with particularly high oil contents. These findings revealed a complex genetic determinism underlying seed oil content among various B. napus cultivars. Many other quantitative traits have also showed that some QTLs could be detected in various gene pools, while others might only be specific to particular genetic backgrounds. For example, QTLs for erucic acid on A8 were detected in most populations, but QTLs on A6, C2 and C8 chromosomes were only detected in special individual populations [20], [21], [70]. QTLs for seed yield on A3, A10 and C4 were detected in different populations [18], [71], [72], and QTLs consistently associated with flowering time across populations were repeatedly identified on A2, A3 and C7 [46], [71], [73]. As every segregating population has its own potentiality to reveal genetic limiting factors, it will be possible to combine the alleles increasing seed oil content from different genetic background in MAS.
Supporting Information
The sequence of thirty-two forward and twenty-one reverse primers of SRAP markers.
(XLS)
Identified QTLs and consensus QTLs detected in eight experiments for seed oil content in the KN population.
(XLS)
Detailed information of QTLs and the consensus QTLs identified in eight populations.
(XLS)
The consensus map and the location of consensus QTLs detected in different populations.
(DOCX)
Funding Statement
The work was supported by National Natural Science Foundation of China (31071447), the Key Natural Science Foundation of Shannxi Province (2012JZ3001), the High Technology Program of China (2011AA10A104), the Natural Science Funds for Distinguished Young Scholars of Hubei Province of China (2010CDA097) and New Century Talents Support Program of the Ministry of Education of China (NCET110172). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.The World Bank, authors (2008) Global Economic Prospects 2008: Technology diffusion in the developing world. Washington, DC: World Bank Publications. 40–41.
- 2. Wang H (2004) Strategy for rapeseed genetic improvement in China in the coming fifteen years. Chin J Oil Crop Sci 26: 98–101. [Google Scholar]
- 3. Si P, Mailer RJ, Galwey N, Turner DW (2003) Influence of genotype and environment on oil and protein concentrations of canola (Brassica napus L.) grown across southern Australia. Aust J Agr Res 54: 397–407. [Google Scholar]
- 4. Zhao JY, Becker HC, Zhang DQ, Zhang YF, Ecke W (2005) Oil content in a European×Chinese rapeseed population: QTL with additive and epistatic effects and their genotype-environment interactions. Crop Science 45: 51–59. [Google Scholar]
- 5. Mauricio R (2001) Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nature Reviews Genetics 2: 370–381. [DOI] [PubMed] [Google Scholar]
- 6. Yang X, Guo Y, Yan J, Zhang J, Song T, et al. (2010) Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet 120: 665–678. [DOI] [PubMed] [Google Scholar]
- 7. Mangolin CA, de Souza Jr CL, Garcia A, Garcia AF, Sibov ST, et al. (2004) Mapping QTLs for kernel oil content in a tropical maize population. Euphytica 137: 251–259. [Google Scholar]
- 8. Zhang J, Lu XQ, Song XF, Yan JB, Song TM, et al. (2008) Mapping quantitative trait loci for oil, starch, and protein concentrations in grain with high-oil maize by SSR markers. Euphytica 162: 335–344. [Google Scholar]
- 9. Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, et al. (1999) Genetics of soybean agronomic traits: I. Comparison of three related recombinant inbred populations. Crop Science 39: 1642–1651. [Google Scholar]
- 10. Kabelka EA, Diers BW, Fehr WR, LeRoy AR, Baianu IC, et al. (2004) Putative alleles for increased yield from soybean plant introductions. Crop science 44: 784–791. [Google Scholar]
- 11. Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, et al. (2004) QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet 108: 1131–1139. [DOI] [PubMed] [Google Scholar]
- 12. Mestries E, Gentzbittel L, de Labrouhe DT, Nicolas P, Vear F (1998) Analyses of quantitative trait loci associated with resistance to shape Sclerotinia sclerotiorum in sunflowers (Helianthus annuus L.) using molecular markers. Mol Breeding 4: 215–226. [Google Scholar]
- 13. Mokrani L, Gentzbittel L, Azanza F, Fitamant L, Al-Chaarani G, et al. (2002) Mapping and analysis of quantitative trait loci for grain oil content and agronomic traits using AFLP and SSR in sunflower (Helianthus annuus L.). Theor Appl Genet 106: 149–156. [DOI] [PubMed] [Google Scholar]
- 14. Gomez Selvaraj M, Narayana M, Schubert AM, Ayers JL, Baring MR, et al. (2009) Identification of QTLs for pod and kernel traits in cultivated peanut by bulked segregant analysis. Electron J Biotechn 12: 3–4. [Google Scholar]
- 15. Sarvamangala C, Gowda M, Varshney RK (2011) Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.). Field Crop Res 122: 49–59. [Google Scholar]
- 16. Mahmood T, Rahman MH, Stringam GR, Yeh F, Good AG (2006) Identification of quantitative trait loci (QTL) for oil and protein contents and their relationships with other seed quality traits in Brassica juncea . Theor Appl Genet 113: 1211–1220. [DOI] [PubMed] [Google Scholar]
- 17. Cheung WY, Landry BS, Raney P, Rakow G (1997) Molecular mapping of seed quality traits in Brassica juncea L. Czern. and Coss. In International Symposium Brassica 97, Xth Crucifer Genetics Workshop 459: 139–148. [Google Scholar]
- 18. Chen G, Geng JF, Rahman M, Liu XP, Tu JX, et al. (2010) Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 175: 161–174. [Google Scholar]
- 19. Delourme R, Falentin C, Huteau V, Clouet V, Horvais R, et al. (2006) Genetic control of oil content in oilseed rape (Brassica napus L.). Theor Appl Genet 113: 1331–1345. [DOI] [PubMed] [Google Scholar]
- 20. Qiu D, Morgan C, Shi J, Long Y, Liu J, et al. (2006) A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor Appl Genet 114: 67–80. [DOI] [PubMed] [Google Scholar]
- 21. Ecke W, Uzunova M, Leder KW (1995) Mapping the genome of rapeseed (Brassica napus L.). II. Localization of genes controlling erucic acid synthesis and seed oil content. Theor Appl Genet 91: 972–977. [DOI] [PubMed] [Google Scholar]
- 22. Burns MJ, Barnes SR, Bowman JG, Clarke M, Werner CP, et al. (2003) QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 90: 39–48. [DOI] [PubMed] [Google Scholar]
- 23. Yan XY, Li JN, Fu FY, Jin MY, Chen L, et al. (2009) Co-location of seed oil content, seed hull content and seed coat color QTL in three different environments in Brassica napus L. Euphytica. 170: 355–364. [Google Scholar]
- 24. Zhao JY, Huang JX, Chen F, Xu F, Ni XY, et al. (2012) Molecular mapping of Arabidopsis thaliana lipid-related orthologous genes in Brassica napus . Theor Appl Genet 124: 407–421. [DOI] [PubMed] [Google Scholar]
- 25.Sun MY, Hua W, Liu J, Huang SM, Wang XF, et al. (2012) Design of new genome- and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. PLoS one 7(10), e47037. [DOI] [PMC free article] [PubMed]
- 26. Chen Y, Qi L, Zhang X, Huang J, Wang J, et al. (2013) Characterization of the quantitative trait locus OilA1 for oil content in Brassica napus . Theor Appl Genet 126: 2499–2509. [DOI] [PubMed] [Google Scholar]
- 27. Jiang C, Zeng Z (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao JY, Becker HC, Zhang DQ, Zhang YF, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113: 33–38. [DOI] [PubMed] [Google Scholar]
- 29. Delourme R, Falentin C, Fomeju BF, Boillot M, Lassalle G, et al. (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics. 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J, Lydiate DJ, Parkin IA, Falentin C, Delourme R, et al. (2011) Integration of linkage maps for the amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa . BMC Genomics 12: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lombard V, Delourme R (2001) A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet 103: 491–507. [Google Scholar]
- 32. Raman H, Raman R, Kilian A, Detering F, Long Y, et al. (2013) A consensus map of rapeseed (Brassica napus L.) based on diversity array technology markers: applications in genetic dissection of qualitative and quantitative traits. BMC Genomics 14: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iniguez-Luy FL, Lukens L, Farnham MW, Amasino RM, Osborn TC (2009) Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea . Theor Appl Genet 120: 31–43. [DOI] [PubMed] [Google Scholar]
- 34. Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, et al. (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111: 1514–1523. [DOI] [PubMed] [Google Scholar]
- 35. Lowe AJ, Moule C, Trick M, Edwards KJ (2004) Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor Appl Genet 108: 1103–1112. [DOI] [PubMed] [Google Scholar]
- 36. Uzunova MI, Ecke W (1999) Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.). Plant Breeding 118: 323–326. [Google Scholar]
- 37. Ding G, Liao Y, Yang M, Zhao Z, Shi L, et al. (2011) Development of gene-based markers from functional Arabidopsis thaliana genes involved in phosphorus homeostasis and mapping in Brassica napus . Euphytica 181: 305–322. [Google Scholar]
- 38. Cheng XM, Xu JS, Xia S, Gu JX, Yang Y, et al. (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus . Theor Appl Genet 118: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Ma C, Fu T, Yang G, Tu J, et al. (2006) Construction of a molecular functional map of rapeseed (Brassica napus L.) using differentially expressed genes between hybrid and its parents. Euphytica 152: 25–39. [Google Scholar]
- 40. Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica . Theor Appl Genet 103: 455–461. [Google Scholar]
- 41.Van Ooijen JW (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands.
- 42. Kosambi DD (1943) The estimation of map distances from recombination values. Annals Eugenics 12: 172–175. [Google Scholar]
- 43.Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics. North Carolina State University, Raleigh, NC.
- 44. Zen ZB (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Long Y, Shi J, Qiu D, Li R, Zhang C, et al. (2007) Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis . Genetics 177: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi JQ, Li RY, Qiu D, Jiang CC, Long Y, et al. (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus . Genetics 182: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, et al. (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20: 2324–2326. [DOI] [PubMed] [Google Scholar]
- 49. Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155: 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Feng J, Long Y, Shi L, Shi JQ, Barker G, et al. (2012) Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus . New Phytologist 193: 96–108. [DOI] [PubMed] [Google Scholar]
- 51. Zhao ZK, Wu LK, Nian FZ, Ding GD, Shi TX, et al. (2012) Dissecting quantitative trait loci for boron efficiency across multiple environments in Brassica napus . PLoS one 7(9): e45215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, et al. (1997) Report on QTL nomenclature. Rice Genet Newsl 14: 11–13. [Google Scholar]
- 53.Charlesworth B (1988) Driving genes and chromosomes. Nature doi:10.1038/332394a0. [DOI] [PubMed]
- 54. Matsushita S, Iseki T, Fukuta Y, Araki E, Kobayashi S, et al. (2003) Characterization of segregation distortion on chromosome 3 induced in wide hybridization between indica and japonica type rice varieties. Euphytica 134: 27–32. [Google Scholar]
- 55. Sibov ST, De Souza CL, Garcia A, Garcia AF, Silva AR, et al. (2003) Molecular mapping in tropical maize (Zea mays L.) using microsatellite markers. 1. Map construction and localization of loci showing distorted segregation. Hereditas 139: 96–106. [DOI] [PubMed] [Google Scholar]
- 56. Harr B, Kauer M, Schl O Tterer C (2002) Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster . Proc Natl Acad Sci USA 99: 12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith JM, Haigh J (1974) Others (1974) The hitch-hiking effect of a favourable gene. Genet Res 23: 23–35. [PubMed] [Google Scholar]
- 58. Kreike CM, Stiekema WJ (1997) Reduced recombination and distorted segregation in a Solanum tuberosum (2×)×S. spegazzinii (2×) hybrid. Genome 40: 180–187. [DOI] [PubMed] [Google Scholar]
- 59. Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW (1998) Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H, Hearne S, Banziger M, Li Z, Wang J (2010) Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 105: 257–267. [DOI] [PubMed] [Google Scholar]
- 61. Langridge P, Karakousis A, Collins N, Kretschmer J, Manning S (1995) A consensus linkage map of barley. Mol breeding 1: 389–395. [Google Scholar]
- 62. Karakousis A, Gustafson JP, Chalmers KJ, Barr AR, Langridge P (2003) A consensus map of barley integrating SSR, RFLP, and AFLP markers. Crop Pasture Sci 54: 1173–1185. [Google Scholar]
- 63. Varshney RK, Marcel TC, Ramsay L, Russell J, Roder MS, et al. (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114: 1091–1103. [DOI] [PubMed] [Google Scholar]
- 64. Wenzl P, Li H, Carling J, Zhou M, Raman H, et al. (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics 7: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- 66. Mace ES, Rami JF, Bouchet S, Klein PE, Klein RR, et al. (2009) A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers. BMC Plant Biol 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gentzbittel L, Vear F, Zhang Y, Berville A, Nicolas P (1995) Development of a consensus linkage RFLP map of cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 90: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 68. Muchero W, Diop NN, Bhat PR, Fenton RD, Wanamaker S, et al. (2009) A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc Natl Acad Sci USA 106: 18159–18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gustafson JP, Ma XF, Korzun V, Snape JW (2009) A consensus map of rye integrating mapping data from five mapping populations. Theor Appl Genet 118: 793–800. [DOI] [PubMed] [Google Scholar]
- 70. Zhao JY, Dimov Z, Becker HC, Ecke WG, Mollers C (2008) Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol Breeding 21: 115–125. [Google Scholar]
- 71. Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet 113: 597–609. [DOI] [PubMed] [Google Scholar]
- 72. Quijada PA, Maureira IJ, Osborn TC (2004) Confirmation of QTL controlling seed yield in spring canola (Brassica napus L.) hybrids. Mol Breeding 13: 193–200. [Google Scholar]
- 73. Mei DS, Wang HZ, Hu Q, Li YD, Xu YS, et al. (2009) QTL analysis on plant height and flowering time in Brassica napus . Plant Breeding 128: 458–465. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sequence of thirty-two forward and twenty-one reverse primers of SRAP markers.
(XLS)
Identified QTLs and consensus QTLs detected in eight experiments for seed oil content in the KN population.
(XLS)
Detailed information of QTLs and the consensus QTLs identified in eight populations.
(XLS)
The consensus map and the location of consensus QTLs detected in different populations.
(DOCX)