Abstract
The “click chemistry” approach utilizing 5-ethynyl-2′-deoxyuridine (EdU) as a DNA precursor was recently introduced to assess DNA replication and adapted to flow- and imaging-cytometry. In the present study, we observed that EdU, once incorporated into DNA, induces DNA damage signaling (DDS) such as phosphorylation of ATM on Ser1981, of histone H2AX on Ser139, of p53 on Ser15, and of Chk2 on Thr68. It also perturbs progression of cells through the cell cycle and subsequently induces apoptosis. These effects were observed in non-small cell lung adenocarcinoma A549 as well as in B-cell human lymphoblastoid TK6 and WTK1 cells, differing in the status of p53 (wt versus mutated). After 1 h EdU pulse-labeling, the most affected was cells progression through the S phase subsequent to that at which they had incorporated EdU. This indicates that DNA replication using the template containing incorporated EdU is protracted and triggers DDS. Furthermore, progression of cells having DNA pulse-labeled with EdU led to accumulation of cells in G2, likely by activating G2 checkpoint. Consistent with the latter was activation of p53 and Chk2. Although a correlation was observed in A549 cells between the degree of EdU incorporation and the extent of γH2AX induction, such correlation was weak in TK6 and WTK1 cells. The degree of perturbation of the cell cycle kinetics by the incorporated EdU was different in the wt p53 TK6 cells as compared to their sister WTK1 cell line having mutated p53. The data are thus consistent with the role of p53 in modulating activation of cell cycle checkpoints in response to impaired DNA replication. The confocal microscopy analysis of the 3D images of cells exposed to EdU for 1 h pulse and then grown for 24 or 48 h revealed an increased number of colocalized γH2AX and p53BP1 foci considered to be markers of DNA double-strand breaks and enlarged nuclei.
Key terms: click chemistry, DNA strand breaks, p53 activation, Chk2 activation, ATM activation, γH2AX foci, p53BP1 foci, caspase-3 activation, laser scanning cytometry, flow cytometry, confocal microscopy, SBIP methodology
Biomarkers of cellular function applied as supravital probes should have minimal effect on the interrogated cell. This is of particular importance in studies involving cell sorting for evaluation of their functional properties, cloning, or propagation as it is in the case of analysis of stem cells. Several biomarkers used as supravital probes however interact with different cell constituents affecting the studied cells and perturbing their growth. Among such biomarkers are probes of DNA replication. Radioactive precursors of DNA such as tritium [3H]-thymidine or [14C]-thymidine, used in the early studies utilizing autoradiography, have been shown to induce DNA radiation damage, including formation of DNA double-strand breaks (DSBs), and to perturb the cell cycle progression (1–4). The most widely used DNA precursor 5-bromo-2′-deoxyuridine (BrdU) introduced with the down of flow cytometry was also shown to induce cytostatic and cytotoxic effects (5–9). The BrdU-labeled DNA in cells exposed to visible or UV light undergoes photolysis (10) resulting in the formation of DNA double-strand breaks (DSBs) and DNA damage signaling (DDS) (11). Likewise, another halogenated DNA precursor 5-iodo-2′-deoxyuridine is cytotoxic and once incorporated into DNA increases cell sensitivity to ionizing radiation and also to light (11,12).
The “click chemistry” approach utilizing 5-ethynyl-2′-deoxyuridine (EdU) as a DNA precursor was recently introduced to assess DNA replication (13) and adapted to flow and imaging cytometry (14–20). Compared with BrdU, the method offers several advantages such as simplicity and rapidity as well as no need for partial DNA denaturation as the latter is incompatible with immunocytochemical detection of other cell constituents. Given its advantages, it is expected that EdU will be widely used as DNA precursor in many areas of cell biology and medicine. While EdU is an excellent marker of DNA replication, the evidence is forthcoming that its applicability for long-term cell labeling, at least in some cell types, may be problematic because of the effect on cell cycle progression and cytotoxicity (20–22). The findings by Diermeier-Daucher et al. (20) revealed impairment of the cell cycle progression of cells that incorporated EdU that varied significantly depending on the cell type. Ross et al. (21) observed perturbation of the cell cycle progression and cytotoxic effects following incorporation of EdU. Interestingly, in the in vivo studies on mice, administration of EdU was shown to reduce growth of the subcutaneous grafts of human glioblastoma and increased animal survival, without apparent significant toxicity. In light of the evidence that EdU crosses the blood–brain barrier, these findings prompted the authors to propose investigation of EdU as potential therapy for malignant brain tumors (21). Most recently, when this article was in preparation, Kohlmeier et al. (22) reported that depending on the cell type EdU can grossly perturb the cell cycle progression and induce cell death. The most sensitive were mouse embryonic stem cells which become arrested in G2/M phase and underwent apoptosis (22). These authors also observed that incorporation of EdU triggers DDS, manifested as histone H2AX Ser139 phosphorylation (induction of γH2AX) and formation of 53BP1 foci (22).
We previously reported that several“supravital” DNA probes, commonly used to label DNA in live cells, induce DDS detected by activation of Ataxia Telangiectasia mutated (ATM), Chk2 and p53 as well as induction of γH2AX (23). These phosphorylation events detected by cytometry using phosphospecific Abs are considered to be indicative of damage to DNA (24,25). Phosphorylation of H2AX, especially manifested by formation of characteristic γH2AX foci, combined with activation of ATM are likely the reporters of DSBs (26,27). In the present study, we explored whether incorporation of EdU into DNA can also induce DDS that can be detected by cytometry. Specifically, using the multiparameter flow and laser scanning-cytometry combined with confocal microscopy, we attempted to observe a possible correlation between the incorporated EdU and cell cycle phase(s) at which the EdU-induced cell cycle progression was impaired.
We have tested the EdU effects on the non-small cell pulmonary adenocarcinoma A549 cells that express wt p53, used previously by us to assess induction of DDS by oxidative stress or by DNA topoisomerase inhibitors in relation to DNA replication (28,29). In addition, the effect of EdU incorporation was also tested on human B-cell lymphoblastoid leukemic cells using two sister cell lines TK6 and WTK1 derived from the same WIL2 cell line, the TK6 having wt p53 while WTK1 expressing spontaneously mutated p53 (30,31).
Materials And Methods
Cells, Cell Treatment
A549 cells, obtained from American Type Culture Collection (ATCC; Manassas, VA), were grown in Ham’s F-12K Nutrient Mixture (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (GIBCO/BRL; Grand Island, NY) in 25-mL FALCON flasks (Becton Dickinson, Franklin Lakes, NJ) at 37.5°C in an atmosphere of 95% air and 5% CO2. The cells were maintained in exponential and asynchronous phase of growth by repeated trypsinization and reseeding prior to reaching subconfluency. The cells were then trypsinized and seeded at low density (about 5 × 104 cells per chamber for chase up to 5 h, about 2.5 × 104 cells per chamber for chase up to 23 h, and about 1 × 104 cells per chamber for chase up to 47 h) in two-chambered Falcon CultureSlides (Beckton Dickinson). Twenty-four hours after seeding, the cultures were treated with 20 μM EdU (Invitrogen/Molecular Probes, Eugene, OR) for different time intervals, as described in the figure legends. In the pulse-chase experiments, the cells exposed to EdU were rinsed twice with medium, then suspended in the medium and cultured in the absence of EdU for time intervals as indicated in the figure legends. Subsequently, the cells were fixed by transferring slides into Coplin jars containing 1% methanol- free formaldehyde (Polysciences, Warrington, PA) in PBS for 15 min on ice, then rinsed with PBS and stored in 1% BSA in PBS, at 4°C for up to 24 h.
Fluorochrome Cell Labeling
The fixed cells were then washed twice in PBS and treated on slides with 0.1% Triton X-100 (Sigma) in PBS for 15 min, and then in a 1% (w/v) solution of bovine serum albumin (BSA; Sigma) in PBS for 30 min to suppress nonspecific antibody (Ab) binding. The cells were then incubated in 100 μL volume of 1% BSA containing 1:300 dilution of phosphospecific (Ser–139) histone H2AX (γ-H2AX) mouse monoclonal antibody (mAb) (BioLegend, San Diego, CA), or a 1:200 dilution of phosphospecific ATM (Ser–1981) mAb (Millipore, Billerica, MA), or 1:100 dilution of phosphospecific (Ser-15) p53 rabbit polyclonal antibody (Cell Signaling, Danvers, MA), or 1:100 dilution of phosphospecific (Thr-68) Chk2 rabbit polyclonal antibody (Cell Signaling). After overnight incubation at 4°C, the slides were washed twice with PBS and then incubated with the fluorochrome-tagged secondary Abs: 100 μL of 1:100 dilution of AlexaFluor647 (AF647) goat anti-mouse (γH2AX and ATM) or 1:100 dilution of AF633 goat anti-rabbit (p53 and Chk2) (all from Invitrogen/Molecular Probes, Eugene, OR), respectively, for 45 min at room temperature in the dark. The Click-iT™ EdU AF488 imaging kit (Invitrogen/Molecular Probes) was used to detect EdU incorporation. The cells were then counterstained with 2.8 μg/mL 4,6-diamidino-2-phenylindole (DAPI; Sigma) in PBS for 15 min to be analyzed by laser scanning cytometry (LSC). TK6 and WKT1 cells, kindly provided by Dr. Liber (30), were maintained in RPMI 1640 medium (GIBCO/Life Technologies, Grand Island, NY) supplemented with l-glutamine (2 mM) and fetal bovine serum (10%), as described before (32). The cells, in exponential phase of growth, were treated with 20 μM EdU for 1 h, then rinsed twice with medium, re-suspended in medium and cultured for additional 23 h. Their subsequent treatment (fixation, labeling with Abs, staining of EdU and with DAPI) was similar as in the case of A549 cells except it was performed in suspension. In the case of detection of apoptosis, the TK6 and WTK1 cells were labeled with the primary activated (cleaved) caspase-3 rabbit Abs (Promega, WI) at 1:100 titer, followed by AF633-tagged goat anti-rabbit (1:100) Ab (Invitrogen/Molecular Probes).
Analysis of Cellular Fluorescence
A549 cells
Cellular immunofluorescence representing the binding of the respective phosphospecific Abs, EdU incorporation, as well as the blue emission of DAPI-stained DNA was measured by LSC (33) (iCys; CompuCyte, Westwood, MA) utilizing standard filter settings; fluorescence was excited with helium neon (633 nm, for detection of phosphospecific Abs), 488-nm argon (for EdU), and violet (405 nm, for DAPI) lasers. Intensities of maximal pixel and integrated fluorescence were measured and recorded for each cell. At least 3,000 cells were measured per sample. Gating analysis was carried out as described in the figure legends.
TK6 and WTK1 cells
Intensity of cellular fluorescence was measured using a MoFlo XDP (Beckman-Coulter, Brea, CA) high-speed flow cytometer/sorter. DAPI fluorescence was excited with the UV laser (355 nm), AF488 with the argon ion (488 nm) laser, and AF647 with the red 633 nm laser. Ten thousand cells were measured per each sample. All experiments were repeated at least three times, representative data are presented. Other details are presented in the figure legends.
Confocal Microscopy
A549 cells were grown under standard conditions, on round coverslips (Menzel Gläser, Braunschweig, Germany) placed in Petri dishes. EdU was added to culture medium to a final concentration of 10 μM, the medium was replaced after 1 h and cells were incubated under standard conditions for 6, 24, or 48 h after. After fixation, in the first step, EdU was fluorescently labeled with Atto 488 (Atto-Tec, Germany) (green) according to manufacturer’s instructions. Subsequently, γH2AX and 53BP1 were labeled by immunofluorescence. Prior to use, antibodies were centrifuged at 14,000 rpm for 5 min to remove protein aggregates. Primary mouse anti-γH2AX (Millipore 05–636, dilution 1:350), rabbit polyclonal anti-53BP1 (Abcam, ab 21083, dilution 1:250), secondary anti-mouse antibody conjugated with AF568 (Invitrogen alexa 633 anti-mouse (A11004) - red), and anti-rabbit conjugated with AF633 (Invitrogen alexa 633 anti-rabbit (A21071) for convenience represented in images by blue color) were used (Fig. 5). Coverslips with the stained cell cultures were mounted in custom-made sample holders and 3D image stacks were collected using Leica SP5 confocal imaging system. The following instrumental parameters were used: excitation wavelength 488, 561, and 633nm, for Atto 488, AF568, and AF633, respectively. EdU and 53BP1 signals were recorded simultaneously with no detectable channel cross-talk. Subsequently, the γH2AX signal was collected. The images were deconvolved using Huygens software (SVI, The Netherlands). Maximum value projections representing the central slices of the thickness of approximately 2 μm are shown in the figure.
Figure 5.
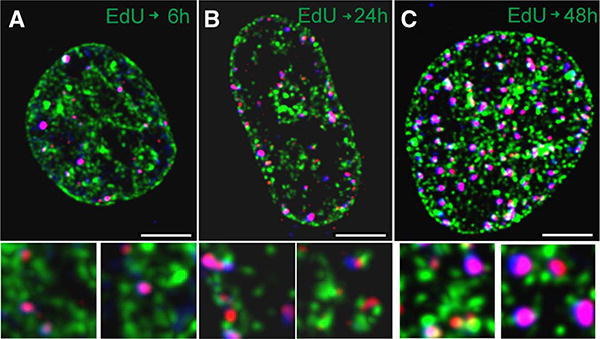
Confocal images (central slices) of nuclei of cells exposed to 10 μM EdU for 1 h and then grown in absence of EdU for 6 h (A), 24 h (B), or 48 h (C). The images show the incorporated EdU (green), γH2AX (red), and 53BP1 (blue). Thus, the foci containing both γH2AX (red) and 53BP1 (AF633, for convenience represented by in blue) appear purple. The size marker is 5 μm. The bottom panels (field of view 3 3 3 μm each) show the γH2AX and/or p53BP1 foci that colocalize and are in close proximity to sites of EdU incorporation, selected from the respective cell images (enlarged).
Results
Figure 1 illustrates the effect of continuous exposure of A549 cells to 20 μM EdU for up to 6 h on the level of phosphorylation of histone H2AX on Ser139 and ATM on Ser1981. It should be noted that the constitutive DDS seen as the background level of γH2AX expression and activation of ATM in the untreated normal and tumor cells (Ctrl) is primarily reporting oxidative DNA damage induced by endogenous oxidants, by-products of aerobic respiration (34–36). The data show the increase in expression of γH2AX and ATM-S1981P occurring predominantly in the S-phase cells, progressive with the time of exposure to the precursor. Compared with the untreated cells, the level of expression of γH2AX and ATM-S1981P in the cells treated with EdU for 6 h was increased by 88% and 116%, respectively. Interestingly, the increase in expression of γH2AX and ATM-S1981P appears to be more pronounced in the late compared with the early S phase cells. Treatment of A549 cells with BrdU for up to 8 h under identical conditions as with EdU shows no effect in terms of induction of ATM activation, H2AX phosphorylation, or cell cycle distribution as measured by DNA content frequency histograms (Supporting Information Fig. 1).
Figure 1.
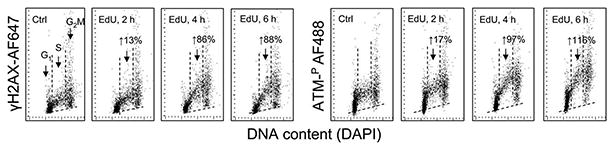
Effect of continuous exposure of A549 cells to 20 μM EdU on the level of expression of γH2AX and ATM-Ser1981P. Exponentially growing cells were untreated (Ctrl) or treated with EdU for 2, 4, or 6 h. The presence of γH2AX and ATM-Ser1981P was detected immunocytochemically with phosphospecific Abs and intensity of cell fluorescence measured by laser scanning cytometry (LSC). The bivariate distributions illustrate expression of these phosphoproteins in relation to cellular DNA content (cell cycle phase). The skewed dashed lines show the upper threshold of nonspecific fluorescence of the cells treated with the secondary Ab only, respectively. The figures above the respective arrows indicate the percent increase in the mean values of S-phase cells of γH2AX and ATM-Ser1981P fluorescence in the EdU-treated cells with respect to the untreated (Ctrl) cells.
The effects of 1 h pulse labeling of A549 cells with EdU on the expression of γH2AX after 5 h, 23 h, and 47 h following the pulse are shown in Figure 2. The “paint-a-gate” analysis of these data demonstrates a relationship between the incorporation of EdU and induction of γH2AX. The cells that incorporated EdU during the pulse were gated and electronically colored red (B– D). Two subpopulations of EdU-positive cells can be distinguished 5 h after pulse-labeling (B). Since duration of G2 plus M is about 3 h the “a” subpopulation with the DNA content that of G1 and intensity of EdU labeling approximately half of that of the subpopulation “b” likely represents the cells that incorporated EdU during the pulse and then divided and reentered G1, “diluting” their EdU content by half. The subpopulation “b” of the EdU labeled cells in all probability represents cells that are still progressing through S, G2, and perhaps M. As is evident from the DNA frequency histogram, the cell progression through G2/M is distinctly slowed down as reflected by the increased proportion of the EdU-labeled cells within the G2/M peak (panel B, inset). Clearly, 5 h after the pulse, the incorporation of EdU inhibits the movement of cells through G2/M. However, the expression of γH2AX by the EdU-labeled cells is not increased at that time (F). This is also reflected by the very weak correlation (r = 0.11) between the extent of EdU incorporation and expression of γH2AX (panel I).
Figure 2.
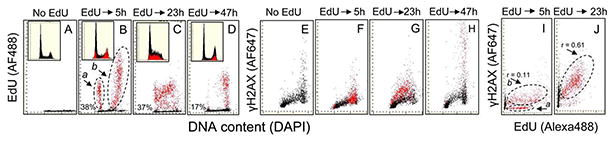
Induction of γH2AX in A549 cells 5 h, 23 h, and 47 h after 1 h pulse-labeling with EdU. Exponentially growing cells were treated for 1 h with 20 μM EdU, rinsed and then incubated in the EdU-free medium. (A–D) Incorporation of EdU, (E–H) expression of γH2AX, both in relation to DNA content (cell cycle phase), and (I and J) a correlation between incorporation of EdU and induction of γH2AX. Insets show the DNA content frequency histograms from the respective cultures. For the “paint-a-gate” analysis, the cells that incorporated EdU are colored red. See the text for further explanation. [Color figure can be viewed in the online issue which is available at at wileyonlinelibrary.com]
Twenty-three hours after the pulse, most of the EdU labeled cells appear to be already in the S phase of the cycle reentering S after the division; some cells are still in G1. The DNA content histogram showing the increased frequency of the EdU-labeled cells in S-phase reveals that progression of these cells through that phase is slowed down (C). However, unlike 5 h after the pulse, the EdU-labeled cells show distinctly elevated expression of γH2AX compared to the EdU-negative cells 23 h after the pulse (G). Also, a correlation between the extent of EdU incorporation and expression of γH2AX is stronger (r = 0.61; J). Forty-seven hours after the pulse, nearly all EdU-labeled cells are in G2/M (D). They also show markedly elevated expression of γH2AX (H).
Of note, the percentage of EdU-labeled cells 5 h, 23 h, and 47 h after the pulse, detected as 38%, 37%, and 17%, respectively, showing a decline, is consistent with the observed slowdown in progression of these cells through the cycle. Specifically, the EdU-unlabeled cells progress the cycle faster and divide more frequently than the labeled cells thereby resulting in the decrease in proportion of the latter with the time of culturing. This is particularly evident between 23 h and 47 h after the pulse when the EdU-labeled cells appear to be traversing S phase at a slow pace (Fig. 2C) and their proportion falls from 37% to 17% of the total cell population (Fig. 2D). However, in addition to the perturbed cell cycle progression, some EdU-labeled A549 cells die, detach from the slide, and thereby cannot be detected. Apoptotic cells are known to detach from slides and flow in the culture medium (37,38) and such cells were observed 47 h after pulse of EdU. Furthermore, the appearance of apoptotic (having activated caspase-3 and degraded DNA) cells was detected after pulse-labeling with EdU in cells growing in suspension, with more TK6 (11%, 27%) than WTK1 (6%, 20%) cells showing the signs of demise, after 23 h and 47 h, respectively (see further in the article). The cytotoxic effects of EdU upon pulse-incorporation and the long-term culturing of A549 and other cells are consistent with the observation of Kohlmeier et al. (22). Interestingly, there is no evidence of the presence of the EdU-labeled cells in S phase after 47 h as nearly all such cells are in G2M (Figs. 2 and 3).
Figure 3.
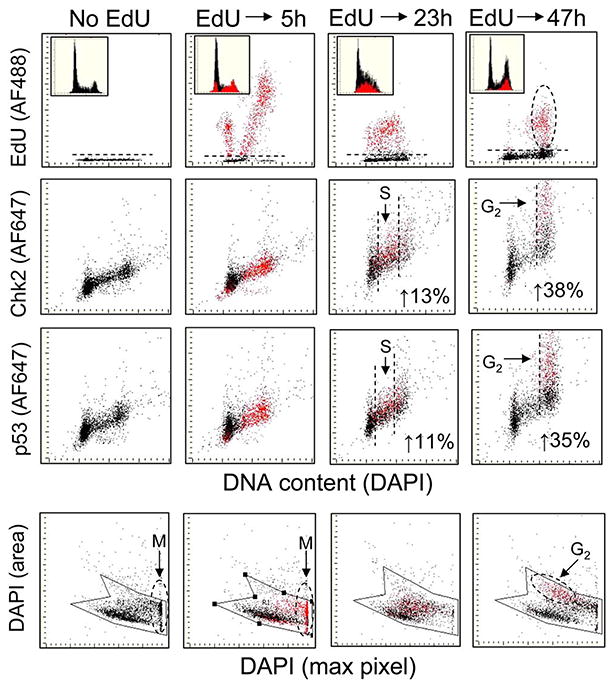
The effect of EdU pulse labeling of A549 cells on phosphorylation of Chk2-Thr68 and p53-Ser15. Exponentially growing cells were treated for 1 h with 20 μM EdU, rinsed and then cultured in absence of EdU for 5 h, 23 h, or 47 h. As in Figure 2, the cells that incorporated EdU were colored red for the “paint-a-gate” analysis. The top three rows of panels show effects of the incorporated EdU on cell cycle progression and a correlation between the incorporated EdU versus induction of Chk2–Thr68 and p53–Ser15 phosphorylation. The bottom row of panels reveals the effect of incorporated EdU on morphometric features of cell nuclei assessed by LSC that allow one to distinguish between G2 and mitotic (M) cells (35,36). The figures in the individual panels indicate the level of fluorescence intensity of the Chk2– Thr68P or p53–Ser15P expressed as a percent increase of the EdU-labeled cells above that of the EdU-negative cells for the selected populations of the S-phase cells in cultures incubated for 23 h and G2M cells in cultures incubated for 47 h after the pulse labeling. See the text for further explanation. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Because the data shown in Figure 2 indicated that the cells having DNA with incorporated EdU were being arrested, at least transiently, in G2M (panels B, D, insets) we assessed possible involvement of phosphorylation of Thr68 on Chk2 and Ser15 on p53, the potential mediators of G2 arrest in response to DNA damage (39,40). The data shown in Figure 3 reveal that that indeed Chk2 and p53 become activated in the cells with the EdU-tagged DNA, particularly 47 h after the pulse of EdU. The top row of panels in this figure illustrating incorporation of EdU versus DNA content reveal similar effect of EdU on progression of cells through the cell cycle, as in the previous experiment (Fig. 2). Of notable feature is the apparent accumulation of cells in S and G2M 23 h and 47 h after the pulse, respectively. The paint-a-gate analysis to assess a correlation between EdU incorporation and activation of Chk2 or p53 made it possible to select the EdU-positive and EdU-negative cells and obtain the mean values of Chk2–Thr68P and p53–Ser15P for each of the population. There is no evidence of an effect of the incorporated EdU on Chk2 or p53 phosphorylation during the initial 5 h of cells growth following pulse of the precursor. However, a minor increase in the level of phosphorylation of Chk2 and p35 in the S phase cells is apparent after 23 h, as the red-colored S-phase cells appear slightly above the black and their mean intensity of Chk2-Thr69P and p53-Ser15P fluorescence is 13% and 11% higher than that of the EdU-negative cells, respectively. A more distinct increase in intensity of Chk2 and p53 phosphospecific Ab fluorescence is evident in the G2M phase cells. Although there is an overlap between the EdU-labeled and unlabeled G2/M cells, the cell population with the increased Chk2–Thr68P or p53–Ser15P (above the control, “No EdU”) clearly shows a predominance of the EdU-labeled cells. The mean values of the EdU-labeled G2M cells are 38% and 35% higher than that of the unlabeled cells for Chk2–Thr68P and p53–Ser15P, respectively.
Analysis of bivariate distributions of fluorescence intensity of maximal pixel of DAPI (reporting degree of compactness of nuclear chromatin) versus area of DAPI fluorescence (reporting projected area of chromatin) provided by LSC allows one to distinguish mitotic (M) from G2 cells (41,42). Mitotic cells (M) are characterized by high intensity of maximal pixel and minimal area of the DNA-associated fluorescence (DAPI) as shown in the bottom row of panels Figure 3. The data in this figure reveal that 5 h after the EdU pulse the EdU-labeled cells progress through M. However, 47 h after the pulse, the EdU-labeled cells show low intensity of DAPI maximal pixel and larger DAPI area, typical of G2 cells (33,41,42). Collectively, the results indicate that 5 h after the pulse the EdU-labeled cells do progress rather unperturbed through M. Twenty-three hours after the pulse, the EdU-labeled cells following division are in the subsequent cycle and while progressing through S activate, at a modest degree, Chk2 and p53. However, 47 h after the pulse many of them are arrested in G2. Their arrest in G2 is consistent with activation of Chk2 and p53, likely mediated by these phosphoproteins (39).
We have also tested the effect of incorporation of EdU on human B-cell lymphoblastoid cell lines TK6, a wt p53 line and WTK1, a homozygous mutant of p53 (30,31). These cell lines are characterized by different levels of constitutive H2AX phosphorylation, with the p53 mutant (WTK1) having distinctly lower expression of γH2AX than the wt TK6 cells (31). It should be noted that duration of the cell cycle of both TK6 and WTK1 cells is nearly twice shorter than that of A549 cells (31) and thus, in relation to the cell cycle length the 23 h postpulse time span would be rather comparable to 47 h than to 23 h of the A549 cells. Figure 4 illustrates perturbation of progression through the cell cycle of these cells as well as the induction of γH2AX in response to incorporation of EdU 23 h after the pulse. The data show that incorporation of EdU has much different effect on both, the cell cycle and γH2AX expression in TK6 compared to WTK1 cells. As is evident in the panels’ insets showing DNA content histograms of the TK6 cells, the EdU-labeled cells are arrested in G2M and S. However, the presence of EdU-labeled cells in G1 indicates that following the pulse they were able to divide and reenter G1. The EdU-labeled cells in S and G2M most likely are the cells that were arrested or slowed down in progression through these phases. It is unclear from these data whether they were arrested or slowed down in the same cell cycle in which were labeled or after a division, in the subsequent cycle.
Figure 4.
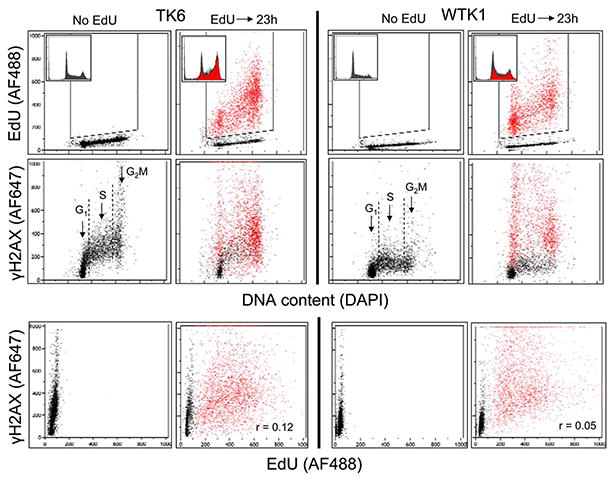
Effect of incorporation of EdU on cell cycle progression and expression of γH2AX in cells of the TK6 and WTK1 lines. Exponentially growing human lymphoblastoid TK6 and WTK1 cells were untreated (Ctrl) or exposed for 60 min to 20 μM EdU followed by 23 h incubation in EdU-free medium (EdU). By the “paint-a-gate” analysis the EdU incorporating cells were colored red. While the top two panels present the EdU incorporation and γH2AX expression vis-a-vis the cell cycle phase (DNA content), the bottom panels show a correlation between EdU incorporation and γH2AX expression in individual cells. The DNA content frequency histograms from the respective cultures are included as insets in the top panels. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The data showing response of WTK1 cells to EdU pulse indicate that 23 h after the pulse larger proportion of EdU labeled cells was able to divide and reenter G1, compared to TK6 cells. However, in analogy to TK6, the increased frequency of WTK1 cells in S- and G2M- after 23 h provides evidence that progression of the EdU-labeled cells was also slowed down in these phases of the cell cycle, compared to control.
The induction of γH2AX by the incorporated EdU shows also different pattern in TK6 versus WTK1 cell lines. First, consistent with our prior findings (31), the level of constitutive expression of this phosphoprotein is distinctly lower in the untreated WTK1 than in TK6 cells (No EdU). After pulse of EdU, the increased expression of γH2AX in the TK6 is seen essentially only in the EdU-labeled G1 cells, whereas the S and G2M cells have approximately similar level of γH2AX in both the EdU-treated and Ctrl cultures. This is clearly not the case in the WTK1 line, where the EdU-labeled cells show dramatic increase in expression of γH2AX in all phases of the cell cycle compared to either the EdU untreated or treated but EdU-negative cells. However, there is relatively weak correlation between the extent of EdU incorporation and the degree of induction of γH2AX in individual cells as shown by the low correlation coefficients r = 0.12 and r = 0.05 for TK6 and WTK1 cells, respectively (Fig. 4, bottom panels).
The LSC data were further confirmed by confocal imaging of A549 cells in cultures exposed to 10 μM EdU for 1 h and fixed 6 h, 24 h, or 48 h later (Fig. 5). Cells that experienced EdU for 1 h followed by 6 h incubation show characteristic γH2AX foci, often located in, or close to, the regions labeled with EdU (Fig. 5A). Many of these γH2AX foci contain 53BP1. Most likely, thus, they represent DSBs (22). In cultures exposed to EdU for 1 h and then grown in the absence of EdU for 24 h (Fig. 5B), many cells have significantly enlarged nuclei, with a large number of γH2AX foci, again some of them also labeled with anti-53BP1 antibody. A 48 h growth following 1 h exposure to EdU results in death of many cells which detach from the coverslips. The cell’s image in Figure 5C represents surviving cells that are still attached to substratum. These cells exhibit enlarged nuclei with a numerous γH2AX foci, containing also 53BP1.
Discussion
As mentioned, DNA precursors harboring either radioactive or halogenate tracers such as BrdU, IdU, or CldU have side effects. Particularly under conditions of long-term labeling, the tracers may disturb cell cycle progression or even be cytotoxic. This may lead to significant bias when such probes are used in studies to assess cell sensitivity to other agents. EdU is not an exception and after incorporation into DNA it perturbs the cell cycle progression to even a greater degree than for example BrdU. Consistent with the previous reports (19–22), we presently observe that responses to EdU, in terms of the cell cycle perturbation, vary depending on the cell type. In addition, our data provide evidence that parallel to the cell cycle effect, EdU once incorporated into DNA, induces DNA damage signaling that manifests as induction of γH2AX and activation of ATM. These data are in support of the findings by Kohlmeier et al. (22) that EdU triggers DDS and depending on the cell type can grossly perturb the cell cycle progression (22).
Our data show that in the case of human non-small cell pulmonary carcinoma A549 cells, the activation of ATM and phosphorylation of H2AX can be seen already after 2 h of constant exposure to the precursor (Fig. 1). After 4 h and 6 h, the cells that are more advanced through S phase (late S-phase) show more pronounced effect compared with the early S-phase cells (Fig. 1). Thus, in the continuous presence of this precursor, the DNA damage signaling increases with the time of DNA replication, likely with the extent of EdU incorporated into DNA. In contrast to EdU, the incorporation of BrdU under the same conditions as EdU induces neither a detectable increase in ATM activation nor H2AX phosphoryl-ation (Supporting Information Fig. 1).
Perturbation of the cell cycle progression in A549 cells that incorporated EdU and its correlation with DDS is much more apparent in the pulse-chase experiments (Fig. 2). The data shown in this figure reveal that the progression through S phase initially, during the cycle at which the EdU pulse was applied (5 h), appears to be unperturbed and no significant γH2AX is induced (Fig. 2F). However, the G2 checkpoint becomes activated, as evidenced by accumulation of cells in G2M (B). These cells (similar as the ones seen in G2M after 47 h; D) were arrested in G2 and not in mitosis. This was ascertained both by their imaging on LSC and their testing for his-tone H3-Ser10 phosphorylation, the marker of mitotic cells (42), of which they were both negative (not shown). Thus, the cells having the EdU-labeled DNA while progressing through G2 appear to trigger activation of Chk2 which leads to their slowed progression though this phase.
The data in Figure 2 also show accumulation of the EdU-labeled cells in the S phase in the cell cycle subsequent to the pulse (C, inset) which indicates on their protracted progression through that phase. The rate of DNA replication thus is distinctly slower when the template for the replication contains EdU incorporated in the places of dT. Such defective DNA replication triggers DDS as shown by the induction of γH2AX in the EdU-positive S-phase cells (G), and by relatively strong correlation between the level of EdU incorporation and expression of γH2AX (J).
In addition to analysis of the effect of EdU on induction of γH2AX, we have also tested its effect on phosphorylation of Chk2 and p53, the events that may mediate the observed accumulation of cells in G2M; the data are shown in Figure 3. Phosphorylation of Chk2 and p53 triggers variety of responses resulting in inhibition of cell cycle progression particularly mediated by activation of Cdc25 phosphatases that lead to arrest in G2(43,44), which is consistent with our findings.
Incorporation of EdU perturbs the cell cycle progression and induces γH2AX also in human B-cell lymphoblastoid leukemic cells, and these effects are different in the cells that have wt p53 (TK6) as compared with the mutated p53 (WKT1) cells. Since these cell lines are derived from the same WIL2 line (30) and the presumed sole difference is mutation of p53, most likely their different response to EdU is due to the status of p53 tumor suppressor gene. Specifically, the data indicate that during the same cycle at which the cells were exposed to the EdU, the incorporated precursor more strongly retarded progression through S and G2M of the wt p53 TK6 (Fig. 3B) than of the mutated p53 WKT1 cells (Fig. 3D). In both cell lines, however, the induction of γH2AX was much more pronounced during the cycle subsequent to that at which the pulse-incorporation took place.
The presence of γH2AX foci in close proximity or colo-calized with p53BP1 (Fig. 5) reveals formation of DSBs (46,47). High frequency of the co-localized γH2AX and BP1 foci was particularly evident 48 h after pulse-labeling with EdU. This may indicate beginning of DNA fragmentation in cells undergoing apoptosis in which activation of caspase-3 seen in TK6 and WTK1 cells after 48 h (Figure 6) leads to proteolysis of the inhibitor of caspase-activated DNase (CAD), activation the latter and thus generation of DSBs (48,49). However, the presence of colocalized γH2AX and BP1 is also seen 6 h and 24 h after cells pulse-exposure to EdU which suggests formation of DSBs unrelated to apoptosis but in direct response to the incorporated EdU.
Figure 6.
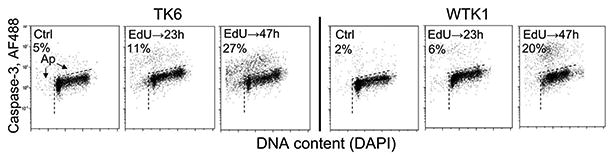
Extent of apoptosis in cultures of TK6 and WTK1 cells 23 h and 47 h following EdU pulse labeling. Exponentially grown TK6 and WTK1 cells were exposed for 1 h to 20 μM EdU, rinsed and subsequently incubated for 23 h or 47 h. Apoptotic cells were discerned as having activated (cleaved) caspase-3 (above the dashed broken line) and/or decreased (“sub-G1”) DNA content (left to the dashed vertical line) (37,38,45). Their percent is shown in the respective panels.
The question may be asked what means have to be taken to escape possible deleterious effects of EdU incorporation manifesting as perturbed progression through the cell cycle and/or DNA damage response. Since most of these effects occur at the time of DNA replication subsequent to the S phase at which EdU has been used as the precursor, efforts have to be made to limit length of time of the experiment that it would not extent to the next S phase following the initial EdU incorporation. Alternatively, for long-term experiments BrdU, known to have less pronounced cytostatic/cytotoxic effects, has to be used as DNA precursor. Its detection, with no need for partial DNA denaturation, can be achieved using the Strand Break Induction by Photolysis (SBIP) methodology (10,50,51).
In conclusion, our findings indicate that EdU has no detectable impact on DNA replication during the initial 6 h of incubation, when the template is innate (dT). Yet, even at that time DDS is triggered above the level of that seen in EdU-untreated cells. However, in long-term experiments following 1 h pulse-labeling with EdU, when DNA template has already dE replacing dT, the replication rate is distinctly slowed down and DDS is triggered at a greater intensity. DDS is still more elevated in the cells that already have traversed the S phase replicating DNA using the EdU-labeled (dE) template. Their subsequent arrest in G2 is likely a consequence of p53 and Chk2 activation. p53 activation may also contribute to induction of apoptosis seen 24 h and 48 h after the pulse. DSBs appear to be present already 6 h after pulse-labeling with EdU. The pattern of the cell cycle and DDS response to EdU incorporation varies depending on the cell type and the status of p53.
Acknowledgments
Grant sponsors: NCI, Grant number: RO1 28 704
Grant sponsors: Robert Andrew Welke Foundation for Cancer Research and Jagiellonian University
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Muller WU, Streffer C, Molls M, Gluck L. Radiotoxicities of [3H] thymidine and [3H] arginine compared in mouse embryos in vitro. Radiat Res. 1987;110:192–198. [PubMed] [Google Scholar]
- 2.Staiano-Coico L, Darzynkiewicz Z, Hefton JM, Dutkowski R, Darlington GJ, Weksler M. Increased sensitivity of lymphocytes from old humans to cell cycle arrest and chromosomal damage induced by 3H-TdR. Science. 1983;219:1335–1337. doi: 10.1126/science.6828861. [DOI] [PubMed] [Google Scholar]
- 3.Jorgenson TJ, Olive PL, Durand RE. DNA strand breaks in Chinese hamster V79 cells caused by low levels of incorporated [3H] and [14C] thymidine. Int J Rad Biol Relat Stud Phys Chem Med. 1987;51:673–680. doi: 10.1080/09553008414552201. [DOI] [PubMed] [Google Scholar]
- 4.Muller WU, Heckeley N, Streffer C. Effects of cell cycle exposure to 3H- thymidine or 3H-arginine on development and cell proliferation of mouse embryos. Radiat Environ Biophys. 1996;35:267–271. doi: 10.1007/s004110050039. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman ER, Davidson RL. Biological and biochemical effects of bromodeoxyuri-dine and deoxycytidine on Syrian hamster melanoma cells. Somatic Cell Genet. 1978;4:587–601. doi: 10.1007/BF01542928. [DOI] [PubMed] [Google Scholar]
- 6.Diermeier S, Schmidt-Bruecken E, Kubbies M, Kunz-Schughart LA, Brockhoff G. Exposure to continuous bromodeoxyuridine (BrdU) differentially affects cell cycle progression of human breast and bladder cancer cell lines. Cell Prolif. 2004;37:195–206. doi: 10.1111/j.1365-2184.2004.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehner B, Sandner B, Marschallinger J, Lehner C, Furtner T, Coullard-Despres S, Rivera FJ, Brockhoff G, Bauer HC, Weidner N, et al. The dark side of BrdU in neural stem cell biology: Detrimental effects on cell cycle. Differentiation and survival. Cell Tissue Res. 2011;345:313–328. doi: 10.1007/s00441-011-1213-7. [DOI] [PubMed] [Google Scholar]
- 8.Levkoff LH, Marshall GP, II, Ross HH, Caldeira M, Reynolds BA, Cakiroglu M, Mariani CL, Streit WJ, Laywell ED. Bromodeoxyuridine inhibits cancer cell proliferation in vitro and in vivo. Neoplasia. 2008;10:804–816. doi: 10.1593/neo.08382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell MA, He X, Svendsen CN. 5-Bromo-2′-deoxyuridine is selectively toxic to neuronal precursors in vitro. Eur J Neurosci. 2005;22:2965–2970. doi: 10.1111/j.1460-9568.2005.04504.x. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Traganos F, Melamed MR, Darzynkiewicz Z. Detection of 5-bromo-2-deoxyuridine incorporated into DNA by labeling strand breaks induced by photolysis (SBIP) Int J Oncol. 1994;4:1157–1161. doi: 10.3892/ijo.4.6.1157. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, King MA, Halicka HD, Traganos F, Okafuji M, Darzynkiewicz Z. Histone H2AX phosphorylation induced by selective photolysis of BrdU-labeled DNA with UV light: Relation to cell cycle phase. Cytometry A. 2004;62A:1–7. doi: 10.1002/cyto.a.20086. [DOI] [PubMed] [Google Scholar]
- 12.Turner DP, Cortellino S, Schupp JE, Caretti E, Loh T, Kinsella TJ, Bellacosa A. The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res. 2006;66:7686–7693. doi: 10.1158/0008-5472.CAN-05-4488. [DOI] [PubMed] [Google Scholar]
- 13.Taverna P, Hwang HS, Schupp JE, Radivoevitch T, Session NN, Reddy G, Zarling DA, Kinsella TJ. Inhibition of base excision repair potentiates iododeoxyuridine-induced cytotoxicity and radiosensitization. Cancer Res. 2003;63:838–846. [PubMed] [Google Scholar]
- 14.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Arora A, Min W, Roifman CM, Grunebaum E. EdU uptake is an alternative non-radioactive assay to 3H thymidine uptake for in vivo measurement of mice T-cell proliferations. J Immunol Methods. 2009;350:29–35. doi: 10.1016/j.jim.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Darzynkiewicz Z, Zhao H, Halicka HD, Li J. Cytometry of DNA replication and RNA synthesis: Historical perspective and recent advances based on “click chemistry”. Cytometry A. 2011;79A:328–337. doi: 10.1002/cyto.a.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamelik RM, Krishan A. Click-iT assay with improved DNA distribution histograms. Cytometry A. 2009;75A:862–865. doi: 10.1002/cyto.a.20780. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Sun Y, Lin G, Zhang R, Zhang K, Xie J, Wang L, Li J. Multicolor flow cytome-try analysis of the proliferations of T-lymphocyte subsets in vitro by EdU incorporation. Cytometry A. 2012;81A:901–909. doi: 10.1002/cyto.a.22113. [DOI] [PubMed] [Google Scholar]
- 19.Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2-deoxyuridine incorporation with click chemistry an alternative to using 5-bromo-2-deoxyuridine antibodies. Biotechniques. 2008;44:927–929. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- 20.Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A. 2009;75A:535–546. doi: 10.1002/cyto.a.20712. [DOI] [PubMed] [Google Scholar]
- 21.Ross HH, Rahman M, Levkoff LH, Millete S, Martin-Carreras T, Dunbar EM, Reynolds BA, Laywell ED. Ethynyldeoxyuridine (EdU) suppresses in vitro population expansion and in vivo tumor progression of human glioblastoma cells. J Neurooncol. 2011;105:485–498. doi: 10.1007/s11060-011-0621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohlmeier F, Maya-Mendoza A, Jackson DA. EdU induces DNA damage response and cell death in mESC culture. Chromosome Res. 2013;21:87–100. doi: 10.1007/s10577-013-9340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao H, Traganos F, Dobrucki J, Wlodkowic D, Darzynkiewicz Z. Induction of DNA damage response by the supravital probes of nucleic acids. Cytometry A. 2009;75A:510–519. doi: 10.1002/cyto.a.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka T, Huang X, Halicka HD, Zhao H, Traganos F, Albino AP, Dai W, Darzynkiewicz Z. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry A. 2007;71A:648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darzynkiewicz Z, Zhao H, Halicka HD, Rybak P, Dobrucki J, Wlodkowic D. DNA damage signaling assessed in individual cells in relation to the cell cycle phase and induction of apoptosis. Crit Rev Clin Lab Sci. 2012;49:199–217. doi: 10.3109/10408363.2012.738808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 27.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H, Dobrucki J, Rybak P, Traganos F, Halicka HD, Darzynkiewicz Z. Induction of DNA damage signaling by oxidative stress in relation to DNA replication as detected using the “Click Chemistry”. Cytometry A. 2011;79A:897–902. doi: 10.1002/cyto.a.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao H, Dobrucki J, Rybak P, Traganos F, Darzynkiewicz Z. Relationship of DNA damage signaling induced by DNA topoisomerase inhibitors camptothecin/topote-can, mitoxantrone or etoposide and DNA replication. Cytometry A. 2012;81A:45–51. doi: 10.1002/cyto.a.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz JL, Jordan E, Evans HH, Lenarczyk M, Liber H. The TP53 dependence of radiation–induced chromosome instability in human lymphoblastoid lines. Radiat Res. 2003;159:730–736. doi: 10.1667/rr3005. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka T, Kurose A, Huang X, Traganos F, Dai W, Darzynkiewicz Z. Extent of constitutive histone H2AX phosphorylation on Ser-139 varies in cells with different TP53 status. Cell Prolif. 2006;39:313–323. doi: 10.1111/j.1365-2184.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halicka HD, Zhao H, Li J, Lee YS, Hsieh TC, Wu JM, Darzynkiewicz Z. Potential anti-aging agents suppress the level of constitutive DNA mage and mTOR- signaling. Aging. 2012;4:952–965. doi: 10.18632/aging.100521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pozarowski P, Holden E, Darzynkiewicz Z. Laser scanning cytometry: Principles and applications. An update Methods Mol Biol. 2013;913:187–212. doi: 10.1007/978-1-62703-056-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka T, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5:1940–1945. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, Kajstura M, Halicka HD, Traganos F, Darzynkiewicz Z. Constitutive his-tone H2AX phosphorylation and ATM activation are strongly amplified during mitogenic stimulation of lymphocytes. Cell Prolif. 2007;40:1–13. doi: 10.1111/j.1365-2184.2007.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Tanaka T, Halicka HD, Traganos F, Zarebski M, Dobrucki J, Darzynkiewicz Z. Cytometric assessment of DNA damage by exogenous and endogenous oxidants reports the aging-related processes. Cytometry A. 2007;71A:905–914. doi: 10.1002/cyto.a.20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wlodkowic D, Skommer J, Darzynkiewicz Z. Cytometry in cell necrobiology revisited Recent advances and new vistas. Cytometry A. 2010;77A:591–606. doi: 10.1002/cyto.a.20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wlodkowic D, Telford W, Skommer J, Darzynkiewicz Z. Apoptosis and beyond: Cytometry in studies of programmed cell death. Methods Cell Biol. 2011;103:55–98. doi: 10.1016/B978-0-12-385493-3.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn J, Urist M, Prives C. The Chk2 protein kinase. DNA Repair. 2004;3:1039–1047. doi: 10.1016/j.dnarep.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 40.Darzynkiewicz Z, Zhao H, Halicka HD, Rybak P, Dobrucki J, Wlodkowic D. DNA damage signaling assessed in individual cells in relation to the cell cycle phase and induction of apoptosis. Crit Rev Clin Lab Sci. 2012;49:199–217. doi: 10.3109/10408363.2012.738808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luther E, Kamentsky LA. Resolution of mitotic cells using laser scanning cytometry. Cytometry. 1996;23:272–278. doi: 10.1002/(SICI)1097-0320(19960401)23:4<272::AID-CYTO2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 42.Juan G, Traganos F, James WM, Ray JM, Roberge M, Sauve DM, Anderson H, Darzynkiewicz Z. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry. 1998;32:71–77. doi: 10.1002/(sici)1097-0320(19980601)32:2<71::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 43.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key-players? Good targets? Nat Rev Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph J. Cdc25 phosphatases: Structure, specificity and mechanism. Biochemistry. 2007;46:3595–3604. doi: 10.1021/bi700026j. [DOI] [PubMed] [Google Scholar]
- 45.Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami M, Traganos F. Cytometry in cell necrobiology Analysis of apoptosis and accidental cell death (necrosis) Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- 46.Djuzenova CS, Elsner I, Katzer A, Worschech E, Distel LV, Flentje M, Polat B. Radio-sensitivity in breast cancer assessed by the histone γ-H2AX and 53BP1 foci. Radiat Oncol. 2013;8:98. doi: 10.1186/1748-717X-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng CKM, Wong MYP, Lam RKK, Ho JPY, Chiu SK, Yu KN. p53 binding protein 1 foci as a biomarker of DNA double strand breaks induced by ionizing radiation. Nucl Instrum Methods Phys Res A. 2011;660:116–120. [Google Scholar]
- 48.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 49.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Melamed MR, Darzynkiewicz Z. Detection of apoptosis and DNA replication by differential labeling of DNA strand breaks with fluorochromes of different color. Exp Cell Res. 1996;222:28–37. doi: 10.1006/excr.1996.0004. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Traganos F, Melamed MR, Darzynkiewicz Z. Single step procedure for DNA strand break labeling Detection of apoptosis and DNA replication. Cytometry. 1995;20:172–180. doi: 10.1002/cyto.990200210. [DOI] [PubMed] [Google Scholar]


