Abstract
Background
The worldwide distributed hematophagous poultry red mite Dermanyssus gallinae (De Geer, 1778) is one of the most important pests of poultry. Even though 35 acaricide compounds are available, control of D. gallinae remains difficult due to acaricide resistances as well as food safety regulations. The current study was carried out to identify putative excretory/secretory (pES) proteins of D. gallinae since these proteins play an important role in the host-parasite interaction and therefore represent potential targets for the development of novel intervention strategies. Additionally, putative transmembrane proteins (pTM) of D. gallinae were analyzed as representatives of this protein group also serve as promising targets for new control strategies.
Methods
D. gallinae pES and pTM protein prediction was based on putative protein sequences of whole transcriptome data which was parsed to different bioinformatical servers (SignalP, SecretomeP, TMHMM and TargetP). Subsequently, pES and pTM protein sequences were functionally annotated by different computational tools.
Results
Computational analysis of the D. gallinae proteins identified 3,091 pES (5.6%) and 7,361 pTM proteins (13.4%). A significant proportion of pES proteins are considered to be involved in blood feeding and digestion such as salivary proteins, proteases, lipases and carbohydrases. The cysteine proteases cathepsin D and L as well as legumain, enzymes that cleave hemoglobin during blood digestion of the near related ticks, represented 6 of the top-30 BLASTP matches of the poultry red mite’s secretome. Identified pTM proteins may be involved in many important biological processes including cell signaling, transport of membrane-impermeable molecules and cell recognition. Ninjurin-like proteins, whose functions in mites are still unknown, represent the most frequently occurring pTM.
Conclusion
The current study is the first providing a mite’s secretome as well as transmembranome and provides valuable insights into D. gallinae pES and pTM proteins operating in different metabolic pathways. Identifying a variety of molecules putatively involved in blood feeding may significantly contribute to the development of new therapeutic targets or vaccines against this poultry pest.
Keywords: Next generation sequencing, In silico, Excretory/secretory proteins, Secreted proteins, Transmembrane proteins, Acari, Blood digestion, Embryogenesis, Ninjurin
Background
The poultry red mite Dermanyssus gallinae (De Geer, 1778) is a worldwide distributed parasitic mite of poultry. It affects its hosts by blood feeding, causing skin irritations, weight loss, restlessness, feather pecking, and an increased incidence of cannibalism [1,2]. Furthermore, in cases with a high infestation rate it may even cause death due to anemia. As a consequence, the parasite leads to high economic losses in poultry farming with estimated annual costs of €130 million throughout the European Union alone. Therefore, the poultry red mite is the major pest for poultry farming [2,3]. The prevalence of D. gallinae depends on flock systems: infestation rates were 4% in cage systems but 33% in alternative systems and 67% of backyard flocks [3,4]. In different countries, D. gallinae prevalence rates can reach up to 80-90% as shown for the United Kingdom, The Netherlands, Italy, Serbia, Montenegro, Morocco and Japan [3]. Control of the poultry red mite is extremely difficult even though 35 effective compounds of different acaricide groups such as pyrethroids or carbamates are available [2]. However, repeated or long-term chemical control may often lead to acaricide resistance of D. gallinae, as shown for carbaryl, permethrin, or DTT [5-8]. Increasing resistance combined with a lack of newly discovered acaricide substance groups show the importance of new development and intervention strategies to ensure animal welfare and to reduce economic losses in poultry farming. Such strategies might be targeted drug development by identifying drug targets and substance libraries screening. Alternatively, future control strategies could rely on vaccine development, which seems a feasible way to combat this hematophagous parasite. Homologous immunization of laying hens with soluble proteins extracted from D. gallinae achieved 50.6% mite mortality [9]. Heterologous immunization of poultry with recombinant Rhipicephalus microplus (formerly Boophilus microplus) Bm86, a membrane-bound midgut surface protein which is used as a vaccine antigen against the mentioned cattle tick, increased D. gallinae mortality by 23% (not significant) compared to the control group, whereas heterologous poultry immunization with recombinant subolesin originating from the mosquito Aedes albopictus increased D. gallinae mortality by 35.1% (p = 0.009) [10]. However, to date, no vaccine candidate with appropriate potential of mite control is available.
Excretory/secretory (ES) proteins play an important role in the host-parasite interface while acting as virulence factors or immune regulators to host immune recognition. Thus, they are crucial for survival of the parasite inside and outside the host organism [11,12]. As ES proteins are supposed to be involved in causing clinical infections in the host organism, they represent a favored group of antigens for the development of new therapeutical solutions e.g. as vaccine candidates or drug targets [12-14]. The current study was conducted to identify and functionally annotate putative ES (pES) and transmembrane (pTM) proteins of D. gallinae by in silico analysis of 454 pyrosequencing generated transcriptome data, which include all developmental stages of starved as well as fed mites [15]. These first analyses of the secretome as well as transmembranome of an acarid species provide potential D. gallinae drug targets or vaccine candidates against this major poultry pest.
Methods
Identification of D. gallinae pES and pTM proteins
D. gallinae pES and pTM protein identification was based on putative protein sequences of whole transcriptome data recently made available by Schicht et al.[15]. Those transcriptome data were generated by two 454-pyrosequencing runs of a pooled cDNA sample of all developmental stages (from egg to the adult stage) and sex of starved as well as freshly blood fed D. gallinae mites. Conceptual translation of the resulting 267,464 D. gallinae nucleotide sequences produced 55,129 (20.6%) coding regions derived from 17,860 isotigs, 24 contigs and 37,245 singletons.
In silico prediction of pES and pTM protein was carried out according to the protocol of Garg and Ranganathan [12], who conducted pES protein prediction by combining the computational tools SignalP [16], SecretomeP [17], TargetP [18,19] and TMHMM [20,21]. The SignalP software package (version 4.1, http://www.cbs.dtu.dk/services/SignalP/) was used for identifying classical secretory proteins. All putative D. gallinae proteins which were not classified to contain signal peptide cleavage sites were further analyzed with SecretomeP (version 2.0, http://www.cbs.dtu.dk/services/SecretomeP/) for predicting non-classical secreted proteins. To limit false positive results the neural network (NN) score of ≥0.9 was set as described by Garg and Ranganathan [12]. To include only truly secreted D. gallinae proteins in subsequent analyses, proteins predicted to be secreted by either of the above mentioned software analyses were subsequently scanned for the presence of mitochondrial sequences by TargetP (version 1.1, http://www.cbs.dtu.dk/services/TargetP/) and transmembrane helices by TMHMM (version 2.0, http://www.cbs.dtu.dk/services/TMHMM/). Protein sequences identified to be of mitochondrial origin or exhibiting transmembrane helices were excluded from the “secreted” data set. Prediction of D. gallinae pTM proteins was carried out separately by scanning the putative D. gallinae protein sequences with TMHMM.
Identification of protein homologs
For identifying homologous proteins, pES and pTM proteins were BLASTed (BLASTP) against the non-redundant (nr) database using the Blast2Go (b2g) software suite [22,23]. E-value cut-off was set at 1.0E-6.
Functional annotation
Supported by b2g, D. gallinae pES and pTM proteins were functionally mapped to Gene Ontology terms [24,25] and annotated by setting default parameters (E-Value-Hit-Filter: 1.0E-6; Annotation cut-off: 55; GO weight: 5; Hsp-Hit Coverage cut-off: 0). Additionally, pES and pTM proteins were associated to protein families, domains and functional sites through InterProScan [26]. InterProScan integrated the following protein signature data bases: BlastProDom, FPrintScan, HMM-PIR, HMM-Pfam, HMM-Smart, HMM-Tigr, ProfileScan, Pattern Scan, Superfamily, Gene3D and HMM-Panther. pES and pTM proteins were subsequently passed to KOBAS2.0 [27] to identify statistically enriched related KO (KEGG Orthology) terms and KEGG pathways [28]. KOBAS was also used for KEGG gene mapping to identify pathways which are shared with the black-legged (deer) tick Ixodes scapularis as an example of a species belonging to the super order Parasitiformes. I. scapularis was chosen since its genome data are available [The Ixodes scapularis Genome project, http://extension.entm.purdue.edu/igp/; [29]].
Results
Predicted D. gallinae secretome and transmembranome size
Of the 55,129 putative D. gallinae protein sequences [15], a number of 2,935 sequences (5.3%) were predicted to contain a signal peptide cleavage site by SignalP. Of the remaining sequences, SecretomeP classified 1,450 protein sequences (2.6%) as non-classical secreted proteins. The putative 4,385 secreted proteins were parsed to TargetP and TMHMM resulting in 341 protein sequences (7.8%) predicted to be of mitochondrial origin and 953 protein sequences (21.7%) predicted to contain transmembrane helices. Potential mitochondrial and transmembrane proteins were excluded from the data set resulting in 3,091 D. gallinae pES protein sequences, representing 5.6% of the putative protein dataset. pTM protein prediction by TMHMM resulted in 7,361 (13.4%) sequences out of the 55,129 D. gallinae putative protein sequences.
pES and pTM protein identification
Of the 3,091 D. gallinae pES protein sequences, a total of 1,083 (35.0%) showed significant BLASTP matches with proteins deposited in the GenBank database. A percentage of 88.7% (961 sequences) of D. gallinae pES proteins matched with proteins of the phytoseiid predatory mite Metaseiulus occidentalis followed with a considerably lower number of sequences by the tick species I. scapularis and Rh. pulchellus (Figure 1). D. gallinae pES proteins were identified as proteases [e.g. cysteine proteases such as cathepsins (45 sequences) or legumains (11 sequences)], cuticle proteins (27 sequences), salivary proteins (24 sequences), mucins (5 sequences), vitellogenins (4 sequences) and many more. About 20% of pES proteins represented uncharacterized or hypothetical/predicted protein homologs. An overview of BLASTP top-hits is given in Table 1, whereas detailed BLASTP results are listed in Additional file 1.
Figure 1.
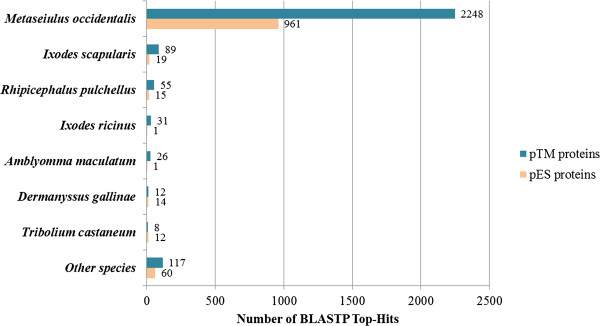
BLASTP top-hit species distribution of D. gallinae pES and pTM protein sequences.
Table 1.
Top 30 BLASTP matches of D. gallinae pES proteins
| Hit description | Species | Hit accession | No. of sequences |
|---|---|---|---|
| Predicted: uncharacterized protein LOC100904214 |
M. occidentalis |
XP_003739349 |
15 |
| Predicted: uncharacterized protein LOC100908413 |
M. occidentalis |
XP_003743523 |
11 |
| Legumain-like protease precursor |
M. occidentalis |
XP_003746513 |
10 |
| Cathepsin l-like |
M. occidentalis |
XP_003747496 |
10 |
| Cathepsin l-1 |
D. gallinae |
CCC33064 |
9 |
| TBC1 domain family member 10A-like |
M. occidentalis |
XP_003737545 |
8 |
| Predicted: uncharacterized protein LOC100898836 |
M. occidentalis |
XP_003742669 |
8 |
| Predicted: uncharacterized protein LOC100906884 |
M. occidentalis |
XP_003743278 |
8 |
| Predicted: cathepsin L-like |
M. occidentalis |
XP_003743816 |
8 |
| Predicted: cuticle protein 10.9-like |
M. occidentalis |
XP_003744493 |
8 |
| Predicted: uncharacterized protein LOC100900702 |
M. occidentalis |
XP_003746592 |
8 |
| Predicted: uncharacterized protein LOC100900705 |
M. occidentalis |
XP_003747300 |
8 |
| Predicted: uncharacterized protein LOC100905853 |
M. occidentalis |
XP_003739512 |
7 |
| Predicted: phospholipase A2-like |
M. occidentalis |
XP_003747592 |
7 |
| Predicted: uncharacterized protein LOC100900008 |
M. occidentalis |
XP_003737797 |
6 |
| Chymotrypsin-like elastase family member 2A-like |
M. occidentalis |
XP_003740761 |
6 |
| Predicted: soluble calcium-activated nucleotidase 1-like |
M. occidentalis |
XP_003741943 |
6 |
| Predicted: cathepsin L-like |
M. occidentalis |
XP_003738729 |
5 |
| Predicted: peroxidasin-like |
M. occidentalis |
XP_003743011 |
5 |
| Predicted: peroxiredoxin-4-like |
M. occidentalis |
XP_003743456 |
5 |
| Predicted: probable serine carboxypeptidase CPVL-like isoform 1 |
M. occidentalis |
XP_003748333 |
5 |
| Predicted: probable serine carboxypeptidase CPVL-like isoform 2 |
M. occidentalis |
XP_003748334 |
5 |
| Secreted salivary gland |
I. persulcatus |
BAH09304 |
4 |
| Cathepsin D-1 |
D. gallinae |
CCC33063 |
4 |
| Predicted: uncharacterized protein LOC100903770 |
M. occidentalis |
XP_003737317 |
4 |
| Predicted: ferritin, liver middle subunit-like isoform 1 |
M. occidentalis |
XP_003737398 |
4 |
| Predicted: cuticle protein 10.9-like |
M. occidentalis |
XP_003739044 |
4 |
| Predicted: frizzled-4-like |
M. occidentalis |
XP_003740054 |
4 |
| Predicted: epidermal growth factor receptor-like |
M. occidentalis |
XP_003740683 |
4 |
| Predicted: uncharacterized protein LOC100905037 | M. occidentalis | XP_003740704 | 4 |
BLASTP homology search of the 7,361 pTM protein sequences revealed 2,586 (35.1%) matches with published protein sequences. Of these, 2,248 (86.9%) were homologous to M. occidentalis (cf. Figure 1). Besides hypothetical and uncharacterized proteins, representing about 15% of sequence homologs, ion channels, receptors and several transporters [e.g. ABC transporter (12 sequences), glucose (12 sequences), amino acid (44 sequences), protein (12 sequences), zinc (24 sequences) or ion transporter (39 sequences)] could be identified. A large part of D. gallinae pTM protein sequences were assigned to ninjurin-like proteins (96 sequences). Furthermore, proteins of the mite’s neuronal network such as GABA (2 sequences), glutamate (31 sequences) or acetylcholine (13 sequences) receptors and transporter (GABA: 11 sequences, glutamate: 4 sequences, acetylcholine: 2 sequences) were identified (Table 2 and Additional file 2).
Table 2.
Top 30 BLASTP matches of D. gallinae pTM proteins
| Hit description | Species | Hit accession | No. of sequences |
|---|---|---|---|
| Predicted: ninjurin-2-like |
M. occidentalis |
XP_003737707 |
96 |
| Predicted: elongation of very long chain fatty acids protein 7-like |
M. occidentalis |
XP_003739733 |
50 |
| Predicted: nose resistant to fluoxetine protein 6-like |
M. occidentalis |
XP_003740562 |
38 |
| Hypothetical protein |
Rh. pulchellus |
JAA57419 |
28 |
| Hypothetical protein |
Rh. pulchellus |
JAA54392 |
21 |
| Predicted: steroid 17-alpha-hydroxylase/17,20 lyase-like |
M. occidentalis |
XP_003742972 |
18 |
| Predicted: adipocyte plasma membrane-associated protein-like |
M. occidentalis |
XP_003748317 |
16 |
| Predicted: cytochrome b561-like |
M. occidentalis |
XP_003737194 |
16 |
| Predicted: voltage-dependent calcium channel type A subunit alpha-1-like |
M. occidentalis |
XP_003743405 |
14 |
| Predicted: uncharacterized protein LOC100906895 |
M. occidentalis |
XP_003747421 |
13 |
| Predicted: solute carrier family 22 member 6-like |
M. occidentalis |
XP_003744619 |
13 |
| hypothetical protein |
A. maculatum |
AEO35694 |
12 |
| Predicted: canalicular multispecific organic anion transporter 1, partial |
M. occidentalis |
XP_003738712 |
12 |
| Predicted: uncharacterized protein LOC100907708 |
M. occidentalis |
XP_003742569 |
12 |
| integral membrane protein, putative |
I. scapularis |
XP_002412964 |
12 |
| Predicted: excitatory amino acid transporter 4-like |
M. occidentalis |
XP_003737125 |
11 |
| Predicted: uncharacterized protein LOC100904062 |
M. occidentalis |
XP_003737318 |
11 |
| Predicted: lysophospholipid acyltransferase 7-like |
M. occidentalis |
XP_003746004 |
10 |
| Predicted: PRA1 family protein 3-like |
M. occidentalis |
XP_003739732 |
10 |
| Predicted: uncharacterized protein LOC100909236 |
M. occidentalis |
XP_003746970 |
10 |
| Predicted: protein transport protein Sec61 subunit gamma-like |
M. occidentalis |
XP_003742336 |
10 |
| Putative lipid exporter abca1, partial |
Rh. pulchellus |
JAA63751 |
9 |
| Predicted: putative sodium-coupled neutral amino acid transporter 7-like |
M. occidentalis |
XP_003744784 |
9 |
| Predicted: vacuolar ATPase assembly integral membrane protein VMA21-like |
M. occidentalis |
XP_003744117 |
9 |
| Predicted: aquaporin-10-like |
M. occidentalis |
XP_003745025 |
8 |
| Predicted: aldehyde dehydrogenase, dimeric NADP-preferring-like |
M. occidentalis |
XP_003746957 |
8 |
| Putative caax prenyl protease 1 log danio rerio zinc metallopeptidase ste24 |
I. ricinus |
JAA68353 |
8 |
| Predicted: membrane-bound O-acyltransferase domain-containing protein 2-like |
M. occidentalis |
XP_003747942 |
8 |
| Predicted: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial-like |
M. occidentalis |
XP_003738022 |
8 |
| Predicted: patched domain-containing protein 3-like | M. occidentalis | XP_003747353 | 8 |
Functional annotation of D. gallinae pES and pTM proteins
Functional annotation of D. gallinae pES and pTM proteins was based on Gene Ontology terms and assignment of protein families, protein domains and functional sites. Of the 3,091 D. gallinae pES proteins, 448 were assigned to 1,946 GO terms divided into 870 GO terms originating from the GO domain Biological Process, 383 GO terms from the Cellular Component domain and 693 GO terms from the Molecular Function domain (Additional file 3). Of the latter, 574 GO terms could be assigned to a third level subcategory (Figure 2), whereby the term hydrolase activity represented with 140 annotations - e.g. cathepsin L, cathepsin K, legumain or secreted mucin - nearly one fourth of assigned GO terms. This term originates together with the terms isomerase activity (9 annotations), ligase activity (9), lyase activity (2), oxidoreductase activity (37), small proteins activating enzyme activity (1) and transferase activity (43) from the parental GO term catalytic activity, which included 42.0% of all Molecular Function Ontology terms assigned to a third level subcategory. The second largest share was contributed to by the parental term binding (34.2%), represented amongst others by the third level subcategory terms protein binding (75 annotations), peptide binding (3), nucleic acid binding (34), carbohydrate binding (6) and lipid binding (3).
Figure 2.
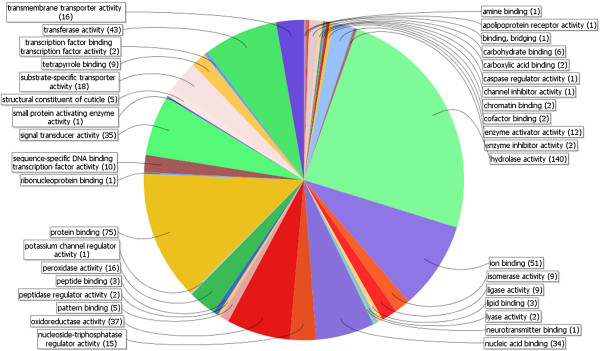
Molecular function ontology distribution of D. gallinae pES proteins on third level subcategory.
Of the total 7,361 D. gallinae pTM proteins, 1,253 were mapped to 6,610 GO terms originating from 3,097 Biological Processes terms, 1,720 Molecular Function terms and 1,793 Cellular Component terms (Additional file 4). On a third level subcategory, 1,637 GO terms were assigned to the Molecular Function domain (Figure 3), the largest part of which (325 annotations) being mapped to transmembrane transporter activity, followed by substrate-specific transporter activity (287) and hydrolase activity (196).
Figure 3.
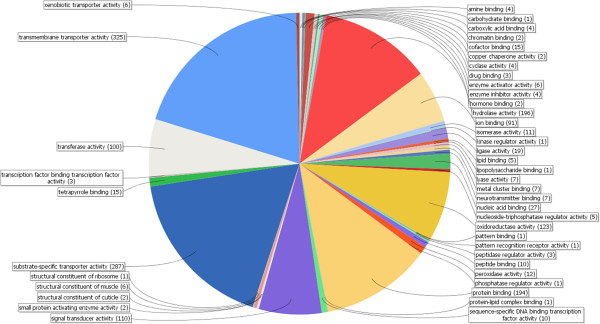
Molecular function ontology distribution of D. gallinae pTM proteins on third level subcategory.
InterPro annotation of pES protein sequences resulted in 531 different assigned protein domains and families. A major part of D. gallinae pES proteins represented proteases assumed to be involved in proteolytic digestion. Another significant share was Immunoglobin-like proteins or subtypes involved in many functions such as cell-cell recognition, cell-surface receptors, muscle structure and immune functions. InterProScan of pTM protein sequences revealed 859 protein domains and families of which ninjurin was the most frequently occurring domain. A further large share of pTM protein sequences were assigned to different transporter systems such as ion channels, ABC transporter or symporter, and cytochrome P450 domains. The most frequently occurring protein domains and families of pES and pTM proteins of D. gallinae are shown in Table 3.
Table 3.
Top 25 protein domains and families of D. gallinae pES and pTM proteins
|
pES proteins |
pTM proteins |
||
|---|---|---|---|
| InterPro code and description | No. of pES proteins (%) | InterPro code and description | No. of pTM proteins (%) |
| IPR013128 Peptidase C1A, papain |
42 (1.36) |
IPR007007 Ninjurin |
97 (1.32) |
| IPR013201 Proteinase inhibitor I29, cathepsin propeptide |
41 (1.33) |
IPR016196 Major facilitator superfamily domain, general substrate transporter |
56 (0.76) |
| IPR013783 Immunoglobulin-like fold |
34 (1.10) |
IPR002076 GNS1/SUR4 membrane protein |
53 (0.72) |
| IPR009003 Peptidase cysteine/serine, trypsin-like |
32 (1.04) |
IPR016040 NAD(P)-binding domain |
49 (0.67) |
| IPR001254 Peptidase S1/S6, chymotrypsin/Hap |
31 (1.00) |
IPR001128 Cytochrome P450 |
46 (0.62) |
| IPR000618 Insect cuticle protein |
27 (0.87) |
IPR005821 Ion transport domain |
44 (0.60) |
| IPR007110 Immunoglobulin-like |
25 (0.81) |
IPR002401 Cytochrome P450, E-class, group I |
34 (0.46) |
| IPR000668 Peptidase C1A, papain C-terminal |
24 (0.78) |
IPR017452 GPCR, rhodopsin-like, 7TM |
30 (0.41) |
| IPR001314 Peptidase S1A, chymotrypsin-type |
24 (0.78) |
IPR000175 Sodium:neurotransmitter symporter |
29 (0.39) |
| IPR018114 Peptidase S1/S6, chymotrypsin/Hap, active site |
23 (0.74) |
IPR002198 Short-chain dehydrogenase/reductase SDR |
29 (0.39) |
| IPR000169 Cysteine peptidase, cysteine active site |
22 (0.71) |
IPR013783 Immunoglobulin-like fold |
29 (0.39) |
| IPR012336 Thioredoxin-like fold |
18 (0.58) |
IPR000276 GPCR, rhodopsin-like |
27 (0.37) |
| IPR025661 Cysteine peptidase, asparagine active site |
18 (0.58) |
IPR002347 Glucose/ribitol dehydrogenase |
27 (0.37) |
| IPR013098 Immunoglobulin I-set |
14 (0.45) |
IPR001140 ABC transporter, transmembrane domain |
24 (0.33) |
| IPR013781 Glycoside hydrolase, catalytic domain |
14 (0.45) |
IPR017940 ABC transporter, integral membrane type 1 |
24 (0.33) |
| IPR017853 Glycoside hydrolase, superfamily |
14 (0.45) |
IPR020846 Major facilitator superfamily domain |
24 (0.33) |
| IPR002007 Heme peroxidase, animal |
13 (0.42) |
IPR017972 Cytochrome P450, conserved site |
23 (0.31) |
| IPR010255 Heme peroxidase |
13 (0.42) |
IPR011527 ABC transporter, transmembrane domain, type 1 |
22 (0.30) |
| IPR003598 Immunoglobulin subtype 2 |
12 (0.39) |
IPR011042 Six-bladed beta-propeller, TolB-like |
21 (0.29) |
| IPR003599 Immunoglobulin subtype |
12 (0.39) |
IPR013099 Ion transport 2 |
21 (0.29) |
| IPR001096 Peptidase C13, legumain |
11 (0.36) |
IPR006201 Neurotransmitter-gated ion-channel |
20 (0.27) |
| IPR001563 Peptidase S10, serine carboxypeptidase |
11 (0.36) |
IPR004299 Membrane bound O-acyl transferase, MBOAT |
19 (0.26) |
| IPR013083 Zinc finger, RING/FYVE/PHD-type |
11 (0.36) |
IPR011701 Major facilitator superfamily |
18 (0.24) |
| IPR002018 Carboxylesterase, type B |
10 (0.32) |
IPR005828 General substrate transporter |
17 (0.23) |
| IPR011009 Protein kinase-like domain | 10 (0.32) | IPR018108 Mitochondrial substrate/solute carrier | 17 (0.23) |
KEGG pathway mapping revealed 252 pES proteins to be involved in 180 pathways. The most frequently assigned pathway was lysosome, followed by antigen processing and presentation. Phagosome was the third most frequent pathway. Mapping of D. gallinae pES protein sequences against I. scapularis KEGG GENES resulted in assigning 133 pES proteins to 60 pathways. The analysis revealed lysosome as the most frequent and phagosome as the third most frequent pathway as well, whereas the second most frequent pathway was represented by metabolic pathways. The top 15 KEGG pathways as well as I. scapularis pathways of pES protein sequences are listed in Table 4. A total of 611 pTM protein sequences were found to be involved in 210 KEGG pathways with protein processing in endoplasmic reticulum being the most frequently occurring pathway, followed by lysosome and Huntington’s disease (cf. Table 5). Furthermore, pTM protein sequences were mapped to neuronal processes such as neuroactive ligand-receptor interaction or glutamatergic synapse. KEGG Gene mapping of D. gallinae pTM protein sequences to I. scapularis revealed 331 pTM protein sequences to be involved in 60 pathways. The most frequently shared assigned pathway were metabolic pathways followed by protein processing in endoplasmic reticulum and lysosome.
Table 4.
Top 15 KEGG pathways of D. gallinae pES proteins
| KEGG pathway | No. of sequences (%) | No. of enzymes | I. sapularis KEGG pathway | No. of sequences (%) | No. of NCBI Gene ID’s |
|---|---|---|---|---|---|
| ko04142 Lysosome |
58 (1.88) |
17 |
isc04142 Lysosome |
41 (1.33) |
12 |
| ko04612 Antigen processing and presentation |
37 (1.20) |
5 |
isc01100 Metabolic pathways |
26 (0.84) |
21 |
| ko04145 Phagosome |
29 (0.94) |
7 |
isc04145 Phagosome |
18 (0.58) |
6 |
| ko05323 Rheumatoid arthritis |
22 (0.71) |
4 |
isc04141 Protein processing in endoplasmic reticulum |
17 (0.55) |
12 |
| ko04141 Protein processing in endoplasmic reticulum |
19 (0.61) |
14 |
isc04310 Wnt signaling pathway |
9 (0.2) |
6 |
| ko00230 Purine metabolism |
16 (0.52) |
7 |
isc00230 Purine metabolism |
8 (0.26) |
5 |
| ko00240 Pyrimidine metabolism |
16 (0.52) |
7 |
isc00240 Pyrimidine metabolism |
8 (0.26) |
5 |
| ko05166 HTLV-I infection |
14 (0.45) |
8 |
isc00511 Other glycan degradation |
6 (0.19) |
3 |
| ko04310 Wnt signaling pathway |
12 (0.39) |
7 |
isc03008 Ribosome biogenesis in eukaryotes |
6 (0.19) |
3 |
| ko05200 Pathways in cancer |
12 (0.39) |
6 |
isc03018 RNA degradation |
5 (0.16) |
2 |
| ko04510 Focal adhesion |
11 (0.36) |
7 |
isc04120 Ubiquitin mediated proteolysis |
4 (0.13) |
4 |
| ko00860 Porphyrin and chlorophyll metabolism |
9 (0.29) |
3 |
isc00600 Sphingolipid metabolism |
4 (0.13) |
3 |
| ko04010 MAPK signaling pathway |
9 (0.29) |
6 |
isc03013 RNA transport |
4 (0.13) |
3 |
| ko04972 Pancreatic secretion |
9 (0.29) |
7 |
isc04070 Phosphatidylinositol signaling system |
4 (0.13) |
3 |
| ko04020 Calcium signaling pathway | 8 (0.26) | 5 | isc03015 mRNA surveillance pathway | 4 (0.13) | 2 |
Table 5.
Top 15 KEGG pathways of D. gallinae pTM proteins
| KEGG pathway | No. of sequences (%) | No. of enzymes | I. sapularis KEGG pathway | No. of sequences (%) | No of NCBI Gene ID’s |
|---|---|---|---|---|---|
| ko04141 Protein processing in endoplasmic reticulum |
60 (0.81) |
25 |
isc01100 Metabolic pathways |
104 (1.43) |
52 |
| ko04142 Lysosome |
52 (0.71) |
15 |
isc04141 Protein processing in endoplasmic reticulum |
54 (0.73) |
21 |
| ko05016 Huntington’s disease |
52 (0.71) |
21 |
isc04142 Lysosome |
50 (0.68) |
13 |
| ko00190 Oxidative phosphorylation |
48 (0.65) |
24 |
isc00190 Oxidative phosphorylation |
39 (0.53) |
16 |
| ko04020 Calcium signaling pathway |
44 (0.6) |
18 |
isc04145 Phagosome |
23 (0.31) |
8 |
| ko05010 Alzheimer’s disease |
43 (0.58) |
21 |
isc04080 Neuroactive ligand-receptor interaction |
22 (0.30) |
11 |
| ko04080 Neuroactive ligand-receptor interaction |
42 (0.57) |
24 |
isc03060 Protein export |
20 (0.27) |
7 |
| ko04724 Glutamatergic synapse |
42 (0.57) |
16 |
isc00564 Glycerophospholipid metabolism |
19 (0.26) |
7 |
| ko05012 Parkinson’s disease |
41 (0.56) |
16 |
isc00510 N-Glycan biosynthesis |
18 (0.24) |
10 |
| ko02010 ABC transporters |
34 (0.46) |
15 |
isc04146 Peroxisome |
16 (0.22) |
6 |
| ko04972 Pancreatic secretion |
34 (0.46) |
14 |
isc04310 Wnt signaling pathway |
13 (0.18) |
6 |
| ko00564 Glycerophospholipid metabolism |
32 (0.43) |
13 |
isc00600 Sphingolipid metabolism |
11 (0.15) |
6 |
| ko04970 Salivary secretion |
30 (0.41) |
13 |
isc04120 Ubiquitin mediated proteolysis |
11 (0.15) |
6 |
| ko04145 Phagosome |
27 (0.37) |
12 |
isc01040 Biosynthesis of unsaturated fatty acids |
10 (0.14) |
4 |
| ko04723 Retrograde endocannabinoid | 28 (0.38) | 14 | isc00020 Citrate cycle (TCA cycle) | 9 (0.12) | 4 |
Discussion
The secretome is a part of the proteome of an organism and includes all proteins secreted by the cell including those of the extracellular matrix, proteins shed from the cell membrane and vesicle proteins like microsomal vesicles [12,30-32]. Transmembrane proteins are a group of membrane proteins containing subunits exposed on both sides of a cell membrane. They compose approximately 30% of a typical genome and are involved in many important biological processes including cell signaling, transport of membrane-impermeable molecules and cell recognition [33]. As the present D. gallinae secretome as well as transmembranome predictions and analyses were based on putative proteins of transcriptome data including all developmental stages of starved as well as fed whole body poultry red mites [15], a broad spectrum of pES and pTM proteins originating from different metabolic pathways was expected. About one fifth (19%) of D. gallinae putative proteins was assigned to the mite’s secretome or transmembranome, divided into 5.6% (3,091) secreted and 13.4% (7,361) transmembrane proteins, respectively. Of these predicted proteins, ~35.0% showed significant matches with known protein sequences, with the predatory phytoseiid mite M. occidentalis being the top-1 hit of 89% D. gallinae pES proteins and 87% pTM proteins. This was not unexpected since both mite species share a close relationship and furthermore, the genome as well as the transcriptome of M. occidentalis is sequenced and available in the common data bases [34,35]. The share of 65% pES and pTM proteins which could not be identified via BLAST and thus were categorized as “novel” are either parasite specific proteins or even unique for the poultry red mite, underlining the importance of further research on protein characterization.
The major part of pES proteins were identified as hydrolases and of these, a large share was cysteine proteases. Cysteine proteases are important in different biochemical and physiological processes of arthropods like embryogenesis [36-42]. The arthropod embryo needs a lot of nutrients during its development. It obtains its nutrition from egg reserve material consisting of amino acids, carbohydrates and lipids stored in yolk granules. To make these nutrients available, enzymatic machinery is needed [39]. Degradation of the yolk protein vitellin is triggered by acidification of the yolk granules, activating as a consequence cysteine proteinases like cathepsin L and B [37-39,43] and aspartic proteinases like cathepsin D [44]. Besides embryogenesis, these cathepsins may be essential in the proteolytic digestion of the mite’s blood meal. This might be extrapolated from the well-studied blood digestion in the closely related ticks [45-49] since both, ticks and D. gallinae share anatomic similarity of the intestinal tract belonging to the Parasitiformes type. As summarized by Horn et al.[45], the primary cleavage of hemoglobin in the hard tick I. ricinus is accomplished by the endopeptidase cathepsin D, which is supported by the catalytic activity of cathepsin L and legumain. These three proteases represent 6 of the top-30 BLASTP matches of the poultry red mite’s secretome (cf. Table 1). The protein family and domain analysis of D. gallinae pES proteins revealed a high proportion of cysteine peptidases C1A, papain (IPR013128: 42 pES sequences; IPR000668: 22 pES sequences) and C13, legumain (IPR001096: 11 pES sequences). Santamaria et al.[50] compared the number of different protein families of the phytophageous mite Tetranychus urticae to different arthropod species (genome data of 10 insects, 1 crustacean and 1 tick) and found that C1A papain-like peptidases are common in all species, whereas a high number of C13 legumain-like peptidases (19 genes) was found in T. urticae mites compared to the other arthropods. The eleven predicted legumain-like peptidases in D. gallinae suggest that extensive expansion of this protein family amongst mites might not be unusual. KEGG pathway mapping predicted pES proteins to be most frequently located in the lysosome in both pathways of all organisms and the tick I. scapularis alone. This might result from the large number of D. gallinae pES proteins identified as proteases like cathepsins, which may act as endopeptidases in the lysosome [51]. As in mites, digestive processes of ticks take place in the acid endosomal/ lysosomal vesicles of gut epithelia cells, contrary to other blood feeding arthropods [52,53].
With the identification and analyses of different blood feeding-induced molecules of ticks, an antigen against the cattle tick Rhipicephalus microplus (formerly Boophilus microplus) was found [54] leading to successful vaccine development against this tick (TickGARD plus™/Gavac™). The antigen Bm86 is a membrane-bound glycoprotein of the tick’s intestinal tract [54,55]. As a gut protein, Bm86 is not part of the normal host-tick interaction and therefore does not stimulate an immunological response under normal circumstances. However, vaccination with this “concealed” or “hidden” antigen induced an immunological response of the cattle host consequently damaging the blood feeding tick [56,57]. When considering potential feeding-induced predicted D. gallinae pTM and pES molecules as concealed antigens, a broad range of new vaccine candidates against the poultry red mite is provided by the present study, e.g. legumains, chymotrypsins or cathepsins. Of these, cathepsin D and L were suggested by Bartley et al.[58] as part of a multi-component vaccine against the poultry red mite due to their efficiency in an in vitro feeding assay. Notably, BLASTP search of predicted D. gallinae pTM proteins did not result in a Rh. microplus Bm86 hit amongst the top-20 BLAST Hits of each D. gallinae pTM protein sequence. This may indicate that no Bm86 homologue is present in the poultry red mite, although as this search was based on transcriptomic data it could be present albeit at low abundance or alternatively it may not be expressed. Specific analysis of the mite’s gut transcripts could test this hypothesis. However, in contrast to ticks, dissection of D. gallinae’s gut is not possible [59] due to its small size. This is, depending on the developmental stage, about 0.39-1 mm in length and 0.26-0.64 mm in width [60]. Thus, estimating gut-derived pTM proteins remains complicated as proteases and other molecules suggested to be involved in blood digestion may also play a role in other processes. Besides “hidden” antigens “exposed” antigens like salivary proteins are also discussed as vaccine candidates against ticks [57,61] as they may act as immunomodulators [62,63]. Therefore, further research on the 24 predicted salivary proteins of D. gallinae could also promote vaccine development.
The BLASTP top-hit of the D. gallinae transmembranome is represented by a ninjurin-2-like protein. In mammals, ninjurin acts as an adhesion molecule which is induced by nerve injury and promotes axonal growth but is also expressed in a number of other tissues, predominantly in epithelial cells [64]. Correlation of ninjurin upregulation with wounding incidents was also shown in adult Drosophila melanogaster flies [65] and in the mosquito Anopheles gambiae ninjurin is suggested to play an important role in innate immune response, probably in signaling and cell communication [66]. Whether the abundance of ninjurin-like proteins amongst the D. gallinae pTM proteins results from the preparation steps (immobilized living mites were collected via forceps which might have injured the mites), from immune functions or from completely different, hitherto unknown functions in mites remains unclear. Another interesting and unexpected pTM protein homolog was the third most common BLASTP top-hit, which is predicted as being nose resistant to fluoxetine protein 6-like (nrf-6). This transmembrane protein, which acts in the intestine of Caenorhabditis elegans and confers sensitivity to the antidepressant fluoxetine (Prozak), is suggested to play a role in drug or yolk transport across membranes [67,68]. The D. melanogaster beltless (blt) gene shares homologies with the C. elegans nrf-6 and is crucial during oogenesis and embryogenesis, but also expressed in the adult nervous system and thus suggested to be important for neuronal functioning [69]. In mites, function and localization of genes involved in neurological processes is largely unknown. Research work is needed to characterize D. gallinae pTM proteins involved in neurological processes, since - if one extrapolates from the mode of action of most acaricides - they could represent promising candidates for identifying new drug targets against poultry red mites.
Conclusion
The present study is the first of its kind analyzing an acarid secretome as well as transmembranome in silico. It provides valuable insights into a pool of D. gallinae proteins, which might be relatively easily accessible candidates for drugs or immunological components like antibodies and thus represent potential drug targets or vaccine candidates. In particular, pES or pTM proteins suggested to be involved in blood feeding and digestion or neurological processes provide a promising basis for further research on new intervention strategies against D. gallinae, which is considered one of the most serious pests of poultry. Nevertheless, future studies on the proteomic level are highly desirable to confirm the predicted secretome and transmembranome of the red poultry mite as in silico analyses rely on the use of algorithms to identify sequence features (i.e. signal peptide motifs) and thus do not necessarily accurately reflect the entire biological truth.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CS conceived and designed the research plan, coordinated the study and participated in writing of the manuscript. SS and WQ performed bioinformatic analyses. SS, WQ, LP and CS analyzed and interpreted the data. SS drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
BLASTP top-hits of D. gallinae pES proteins.
BLASTP top-hits of D. gallinae pTM proteins.
Gene Ontology term distribution of D. gallinae pES proteins.
Gene Ontology term distribution of D. gallinae pTM proteins.
Contributor Information
Sabine Schicht, Email: sabine.schicht@tiho-hannover.de.
Weihong Qi, Email: weihong.qi@fgcz.ethz.ch.
Lucy Poveda, Email: lucy.poveda@fgcz.uzh.ch.
Christina Strube, Email: christina.strube@tiho-hannover.de.
References
- Kirkwood AC. Anaemia in poultry infested with the red mite Dermanyssus gallinae. Vet Rec. 1967;80:514–516. doi: 10.1136/vr.80.17.514. [DOI] [PubMed] [Google Scholar]
- Chauve C. The poultry red mite Dermanyssus gallinae (De Geer, 1778): current situation and future prospects for control. Vet Parasitol. 1998;79:239–245. doi: 10.1016/S0304-4017(98)00167-8. [DOI] [PubMed] [Google Scholar]
- Sparagano O, Pavlicevic A, Murano T, Camarda A, Sahibi H, Kilpinen O, Mul M, van Emous R, le Bouquin S, Hoel K, Cafiero MA. Prevalence and key figures for the poultry red mite Dermanyssus gallinae infections in poultry farm systems. Exp Appl Acarol. 2009;48:3–10. doi: 10.1007/s10493-008-9233-z. [DOI] [PubMed] [Google Scholar]
- Hoglund J, Nordenfors H, Uggla A. Prevalence of the poultry red mite, Dermanyssus gallinae, in different types of production systems for egg layers in Sweden. Poult Sci. 1995;74:1793–1798. doi: 10.3382/ps.0741793. [DOI] [PubMed] [Google Scholar]
- Beugnet F, Chauve C, Gauthey M, Beert L. Resistance of the red poultry mite to pyrethroids in France. Vet Rec. 1997;140:577–579. doi: 10.1136/vr.140.22.577. [DOI] [PubMed] [Google Scholar]
- Marangi M, Cafiero MA, Capelli G, Camarda A, Sparagano OA, Giangaspero A. Evaluation of the poultry red mite, Dermanyssus gallinae (Acari: Dermanyssidae) susceptibility to some acaricides in field populations from Italy. Exp Appl Acarol. 2009;48:11–18. doi: 10.1007/s10493-008-9224-0. [DOI] [PubMed] [Google Scholar]
- Zeman P, Zelezny J. The susceptibility of the poultry red mite, Dermanyssus gallinae (De Geer, 1778), to some acaricides under laboratory conditions. Exp Appl Acarol. 1985;1:17–22. doi: 10.1007/BF01262196. [DOI] [PubMed] [Google Scholar]
- Zeman P. Encounter the poultry red mite resistance to acaricides in Czechoslovak poultry-farming. Folia Parasitol (Praha) 1987;34:369–373. [PubMed] [Google Scholar]
- Harrington D, Din HM, Guy J, Robinson K, Sparagano O. Characterization of the immune response of domestic fowl following immunization with proteins extracted from Dermanyssus gallinae. Vet Parasitol. 2009;160:285–294. doi: 10.1016/j.vetpar.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Harrington D, Canales M, de la Fuente J, de Luna C, Robinson K, Guy J, Sparagano O. Immunisation with recombinant proteins subolesin and Bm86 for the control of Dermanyssus gallinae in poultry. Vaccine. 2009;27:4056–4063. doi: 10.1016/j.vaccine.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G, Ranganathan S. In silico secretome analysis approach for next generation sequencing transcriptomic data. BMC Genomics. 2011;12(Suppl 3):S14. doi: 10.1186/1471-2164-12-S3-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez PJ, Zhan B, Bethony JM, Loukas A, Williamson A, Goud GN, Hawdon JM, Dobardzic A, Dobardzic R, Ghosh K. et al. Progress in the development of a recombinant vaccine for human hookworm disease: the human hookworm vaccine initiative. Int J Parasitol. 2003;33:1245–1258. doi: 10.1016/S0020-7519(03)00158-9. [DOI] [PubMed] [Google Scholar]
- Bonin-Debs AL, Boche I, Gille H, Brinkmann U. Development of secreted proteins as bio therapeutic agents. Expert Opin Biol Ther. 2004;4:551–558. doi: 10.1517/14712598.4.4.551. [DOI] [PubMed] [Google Scholar]
- Schicht S, Qi W, Poveda L, Strube C. Whole transcriptome analysis of the poultry red mite Dermanyssus gallinae (De Geer, 1778) Parasitology. in press. [DOI] [PubMed]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. The gene ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2011;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Wikel SK. The Ixodes scapularis genome project: an opportunity for advancing tick research. Trends Parasitol. 2005;21:151–153. doi: 10.1016/j.pt.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Caccia D, Dugo M, Callari M, Ongarzone I. Bioinformatics tools for secretome analysis. Biochim Biophys Acta. 2013. in press. [DOI] [PubMed]
- Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. A proteomic view on genome-based signal peptide predictions. Genome Res. 2001;11:1484–1502. doi: 10.1101/gr.182801. [DOI] [PubMed] [Google Scholar]
- Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev. 2000;64:515–547. doi: 10.1128/MMBR.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinforma. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy MA. The predatory mite Metaseiulus occidentalis: mitey small and mitey large genomes. Bioessays. 2009;31:581–590. doi: 10.1002/bies.200800175. [DOI] [PubMed] [Google Scholar]
- Hoy MA, Yu F, Meyer JM, Tarazona OA, Jeyaprakash A, Wu K. Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): a cautionary tale about possible contamination by prey sequences. Exp Appl Acarol. 2012;59:283–296. doi: 10.1007/s10493-012-9603-4. [DOI] [PubMed] [Google Scholar]
- Sojka D, Francischetti IM, Calvo E, Kotsyfakis M. Cysteine proteases from blood feeding arthropod ectoparasites. Adv Exp Med Biol. 2011;712:177–191. doi: 10.1007/978-1-4419-8414-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F. Yolk degradation in tick eggs: II. Evidence that cathepsin L-like proteinase is stored as a latent, acid-activable proenzyme. Arch Insect Biochem Physiol. 1990;14:237–252. doi: 10.1002/arch.940140404. [DOI] [PubMed] [Google Scholar]
- Fagotto F. Yolk degradation in tick eggs: I. Occurrence of a cathepsin L-like acid proteinase in yolk spheres. Arch Insect Biochem Physiol. 1990;14:217–235. doi: 10.1002/arch.940140403. [DOI] [PubMed] [Google Scholar]
- Seixas A, Dos Santos PC, Velloso FF, Da Silva Vaz I Jr, Masuda A, Horn F, Termignoni C. A Boophilus microplus vitellin-degrading cysteine endopeptidase. Parasitology. 2003;126:155–163. doi: 10.1017/S0031182002002731. [DOI] [PubMed] [Google Scholar]
- Estrela A, Seixas A, Termignoni C. A cysteine endopeptidase from tick (Rhipicephalus (Boophilus) microplus) larvae with vitellin digestion activity. Comp Biochem Physiol B Biochem Mol Biol. 2007;148:410–416. doi: 10.1016/j.cbpb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Uchida K, Ohmori D, Ueno T, Nishizuka M, Eshita Y, Fukunaga A, Kominami E. Preoviposition activation of Cathepsin-like proteinases in degenerating ovarian follicles of the mosquito Culex pipiens pallens. Dev Biol. 2001;237:68–78. doi: 10.1006/dbio.2001.0357. [DOI] [PubMed] [Google Scholar]
- Cho WL, Tsao SM, Hays AR, Walter R, Chen JS, Snigirevskaya ES, Raikhel AS. Mosquito Cathepsin B-like protease involved in embryonic degradation of vitellin is produced as a latent extra ovarian precursor. J Biol Chem. 1999;274:13311–13321. doi: 10.1074/jbc.274.19.13311. [DOI] [PubMed] [Google Scholar]
- Medina M, Leon P, Vallejo CG. Drosophila Cathepsin B-like proteinase: a suggested role in yolk degradation. Arch Biochem Biophys. 1988;263:355–363. doi: 10.1016/0003-9861(88)90646-7. [DOI] [PubMed] [Google Scholar]
- Logullo C, Vaz Ida S, Sorgine MH, Paiva-Silva GO, Faria FS, Zingali RB, De Lima MF, Abreu L, Oliveira EF, Alves EW. et al. Isolation of an aspartic proteinase precursor from the egg of a hard tick, Boophilus microplus. Parasitology. 1998;116(Pt 6):525–532. doi: 10.1017/s0031182098002698. [DOI] [PubMed] [Google Scholar]
- Horn M, Nussbaumerova M, Sanda M, Kovarova Z, Srba J, Franta Z, Sojka D, Bogyo M, Caffrey CR, Kopacek P, Mares M. Hemoglobin digestion in blood-feeding ticks: mapping a multipeptidase pathway by functional proteomics. Chem Biol. 2009;16:1053–1063. doi: 10.1016/j.chembiol.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM, Labruna MB, Mans BJ, Maruyama SR, Francischetti IM, Barizon GC, dMS IK. The sialotranscriptome of Antricola delacruzi female ticks is compatible with non-hematophagous behavior and an alternative source of food. Insect Biochem Mol Biol. 2012;42:332–342. doi: 10.1016/j.ibmb.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alim MA, Tsuji N, Miyoshi T, Islam MK, Hatta T, Yamaji K, Fujisaki K. Developmental stage- and organ-specific expression profiles of asparaginyl endopeptidases/legumains in the ixodid tick Haemaphysalis longicornis. J Vet Med Sci. 2008;70:1363–1366. doi: 10.1292/jvms.70.1363. [DOI] [PubMed] [Google Scholar]
- Franta Z, Frantova H, Konvickova J, Horn M, Sojka D, Mares M, Kopacek P. Dynamics of digestive proteolytic system during blood feeding of the hard tick Ixodes ricinus. Parasit Vectors. 2010;3:119. doi: 10.1186/1756-3305-3-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Sonenshine DE, Valenzuela JG. Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae) BMC Genomics. 2008;9:552. doi: 10.1186/1471-2164-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria ME, Hernandez-Crespo P, Ortego F, Grbic V, Grbic M, Diaz I, Martinez M. Cysteine peptidases and their inhibitors in Tetranychus urticae: a comparative genomic approach. BMC Genomics. 2012;13:307. doi: 10.1186/1471-2164-13-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohley P, Seglen PO. Proteases and proteolysis in the lysosome. Experientia. 1992;48:151–157. doi: 10.1007/BF01923508. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of the tick. New York: Oxford University Press; 1991. [Google Scholar]
- Coons LB, Alberti G. In: Microscopic anatomy of invertebrates. Volume 8B. Harrison FW, Foelix RF, editor. New York: Wiley-Liss; 1999. The acari-ticks; pp. 267–514. [Google Scholar]
- Willadsen P, Riding GA, McKenna RV, Kemp DH, Tellam RL, Nielsen JN, Lahnstein J, Cobon GS, Gough JM. Immunologic control of a parasitic arthropod. Identification of a protective antigen from Boophilus microplus. J Immunol. 1989;143:1346–1351. [PubMed] [Google Scholar]
- Gough JM, Kemp DH. Localization of a low abundance membrane protein (Bm86) on the gut cells of the cattle tick Boophilus microplus by immunogold labeling. J Parasitol. 1993;79:900–907. doi: 10.2307/3283728. [DOI] [PubMed] [Google Scholar]
- Willadsen P. Anti-tick vaccines. Parasitology. 2004;129(Suppl):S367–S387. doi: 10.1017/s0031182003004657. [DOI] [PubMed] [Google Scholar]
- Nuttall PA, Trimnell AR, Kazimirova M, Labuda M. Exposed and concealed antigens as vaccine targets for controlling ticks and tick-borne diseases. Parasite Immunol. 2006;28:155–163. doi: 10.1111/j.1365-3024.2006.00806.x. [DOI] [PubMed] [Google Scholar]
- Bartley K, Huntley JF, Wright HW, Nath M, Nisbet AJ. Assessment of Cathepsin D and L-like proteinases of poultry red mite, Dermanyssus gallinae (De Geer), as potential vaccine antigens. Parasitology. 2012;139:755–765. doi: 10.1017/S0031182011002356. [DOI] [PubMed] [Google Scholar]
- Nisbet AJ, Billingsley PF. A comparative survey of the hydrolytic enzymes of ectoparasitic and free-living mites. Int J Parasitol. 2000;30:19–27. doi: 10.1016/S0020-7519(99)00169-1. [DOI] [PubMed] [Google Scholar]
- Sikes RK, Chamberlain RW. Laboratory observations on three species of bird mites. J Parasitol. 1954;40:691–697. doi: 10.2307/3273713. [DOI] [PubMed] [Google Scholar]
- Shahein YE, Abouelella AM, Hussein NA, Hamed RR, El-Hakim AE, Abdel-Shafy S, Tork SE. Identification of four novel Rhipicephalus annulatus upregulated salivary gland proteins as candidate vaccines. Protein J. 2013;32:392–398. doi: 10.1007/s10930-013-9498-x. [DOI] [PubMed] [Google Scholar]
- Alarcon-Chaidez FJ, Sun J, Wikel SK. Transcriptome analysis of the salivary glands of Dermacentor andersoni Stiles (Acari: Ixodidae) Insect Biochem Mol Biol. 2007;37:48–71. doi: 10.1016/j.ibmb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG. High-throughput approaches to study salivary proteins and genes from vectors of disease. Insect Biochem Mol Biol. 2002;32:1199–1209. doi: 10.1016/S0965-1748(02)00083-8. [DOI] [PubMed] [Google Scholar]
- Araki T, Milbrandt J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron. 1996;17:353–361. doi: 10.1016/S0896-6273(00)80166-X. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo F, Ghani Y, Kafatos FC, Christophides GK. Comprehensive genetic dissection of the hemocyte immune response in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9:e1003145. doi: 10.1371/journal.ppat.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy RK, Thomas JH. Fluoxetine-resistant mutants in C. elegans define a novel family of transmembrane proteins. Mol Cell. 1999;4:143–152. doi: 10.1016/s1097-2765(00)80362-7. [DOI] [PubMed] [Google Scholar]
- Choy RK, Kemner JM, Thomas JH. Fluoxetine-resistance genes in Caenorhabditis elegans function in the intestine and may act in drug transport. Genetics. 2006;172:885–892. doi: 10.1534/genetics.103.024869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Dimitrijevic N, Manev H. Identification of a novel Drosophila gene, beltless, using injectable embryonic and adult RNA interference (RNAi) BMC Genomics. 2003;4:33. doi: 10.1186/1471-2164-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BLASTP top-hits of D. gallinae pES proteins.
BLASTP top-hits of D. gallinae pTM proteins.
Gene Ontology term distribution of D. gallinae pES proteins.
Gene Ontology term distribution of D. gallinae pTM proteins.


