Abstract
Background
Sulfide is a common toxin to animals and is abundant in coastal and aquatic sediments. Sulfur dioxygenase (SDO) is thought to be the key enzyme involved in sulfide oxidation in some organisms. The echiuran worm, Urechis unicinctus, inhabits coastal sediment and tolerates high concentrations of sulfide. The SDO is presumably important for sulfide tolerance in U. unicinctus.
Results
The full-length cDNA of SDO from the echiuran worm U. unicinctus, proven to be located in the mitochondria, was cloned and the analysis of its sequence suggests that it belongs to the metallo-β-lactamase superfamily. The enzyme was produced using an E. coli expression system and the measured activity is approximately 0.80 U mg protein−1. Furthermore, the expression of four sub-segments of the U. unicinctus SDO was accomplished leading to preliminary identification of functional domains of the enzyme. The identification of the conserved metal I (H113, H115, H169 and D188), metal II (D117, H118, H169 and H229) as well as the potential glutathione (GSH) (R197, Y231, M279 and I283) binding sites was determined by enzyme activity and GSH affinity measurements. The key residues responsible for SDO activity were identified by analysis of simultaneous mutations of residues D117 and H118 located close to the metal II binding site.
Conclusion
The recombinant SDO from U. unicinctus was produced, purified and characterized. The metal binding sites in the SDO were identified and Y231 recognized as the mostly important amino acid residue for GSH binding. Our results show that SDO is located in the mitochondria where it plays an important role in sulfide detoxification of U. unicinctus.
Introduction
Sulfide, a common toxin, may be harmful for organisms by reducing the affinity of hemoglobin to oxygen [1], inhibiting the activity of cytochrome c oxidase and succinate oxidase complexes [2], [3], depolarizing mitochondria [4], inducing apoptosis [5], and causing oxidative damage to RNA and DNA [6]. In marine sediments sulfide accumulates because of the existence of anaerobic sulfate-reducing bacteria [7]. Animals in permanent burrows are frequently exposed to sulfide during low tides; for example, sulfide could reach 66 µM in the burrow water where the echiuran worm Urechis caupo lives [8] and variety of defensive responses are adopted by animals living in sediments. Mitochondrial oxidation is considered the primary pathway used to detoxify sulfide in the worms living in sediments where prolonged exposure to toxic sulfide occurs [9].
In mitochondria, an enzymatic system involving three enzymes, sulfide: quinine oxidoreductase (SQR), sulfur dioxygenase (SDO) and sulfur transferase (ST), are involved in oxidative sulfide detoxification. Two models for sulfide oxidation [10], [11] have been proposed as shown in Figure 1. SDO plays an essential role in both by oxidizing sulfane sulfur of glutathione persulfide (GSS−) to sulfite by using O2. SDO in humans was initially recognized as ETHE1 protein (ethylmalonic encephalopathy 1) since it was recognized that ETHE1 gene mutation leads to ethylmalonic encephalopathy (EE) [12]. Recently, Tiranti et al. [13] suggested that ETHE1 possesses SDO activity and is involved in the oxidation of sulfide since SDO activity 1) is absent in EE patients and ETHE1 −/− mice, and 2) increases when human ETHE1 is overexpressed in Hela or E. coli cells. Moreover, in Arabidopsis thaliana, ETHE1 also catalyzes the GSS−-dependent activity with consumption of oxygen at a rate of 7.95±0.71 µmol O2 min−1 mg−1 [14]. Thus, at present, the function of ETHE1 is mostly focused in its SDO activity and the biochemical characterization and kinetic properties of the human enzyme shows a Michaelis constant (KM) for GSSH of 0.34±0.03 mM and a Vmax of 113±4 µmol min −1 mg protein −1 [15]. To date, most of the research concerning the SDO enzyme has been restricted to mammal and plant sources and no information in invertebrates, in particular those that have sulfide tolerance, was reported.
Figure 1. The two proposed sulfide oxidation models.
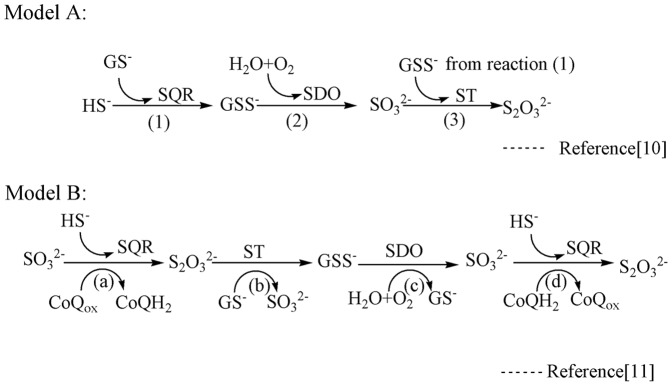
Abbreviations: SQR, sulfide: quinine oxidoreductase; ST, sulfur transferase; SDO, sulfur dioxygenase; GS−, glutathione; GSS−, glutathione persulfide. Model A: The sulfide can be converted to GSS− by SQR, and the sulfane sulfur of GSS− is oxidized to sulfite using O2 by SDO in the mitochondrial-matrix; finally, the generated sulfite is converted by ST catalysis to thiosulfate, which is less toxic to the organism. Model B: Thiosulfate biosynthesis occurs in the first step of sulfide oxidation catalyzed by SQR with sulfite as the acceptor of the sulfane sulfur, then thiosulfate can act as the ST substrate to produce GSS− and regenerate the sulfite; finally, the sulfane sulfur of GSS− is oxidized to sulfite using O2 by SDO. The newly generated sulfite could then enter the cycle again.
The echiuran worm Urechis unicinctus is mainly distributed in China, Korea, Russia and Japan, and inhabits marine sediments, especially intertidal and subtidal mudflats [16], [17]. It has been reported that U. unicinctus can tolerate, use and metabolize environmental sulfide [18], [19], [20]. Furthermore, the presence of the U. unicinctus SQR was revealed in different tissues and upon exposure to different sulfide concentrations at the mRNA, protein and enzyme activity levels [21], [22]. This study aims at increasing our understanding of sulfide metabolic adaptation as well as exploring the function and catalytic mechanism of SDO in U. unicinctus. The full length SDO cDNA as well as four sub-segmental sequences were cloned and expressed in E. coli allowing for the elucidation of domains responsible for enzyme activity and the catalytic mechanism of U. unicinctus SDO.
Materials and Methods
Cloning of target full length cDNA in U. unicinctus
The nested degenerate primers for cloning SDO cDNA fragment were designed according to the evolutionary conserved domains of SDO cDNA in other species obtained from the National Center for Biotechnology Information (NCBI). The primary PCR was conducted using the forward (5′-CAYGCNGAYCAYATHACNGG-3′) and reverse degenerate primers (5′-GTARTCRTGNGCNGGRTANA-3′), and the body wall cDNA of U. unicinctus as a template. Semi-nested PCR was conducted using 2000× diluted primary PCR product as the template and the semi-nested reverse degenerate primer was changed to 5′-TGGAARTCNGTNCKNCCRCA-3′. The PCR product was purified, subcloned into pMD18-T vector (Takara, Otsu, Japan), and then transformed into E. coli-DH5α competent cells (Takara). The obtained fragments were sequenced using an ABI PRISM 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA) and the sequence alignment was performed using Blastx from the NCBI, and was preliminarily confirmed as the SDO sequence fragment.
The 3′ and 5′ RACE ready first-strand cDNA was synthesized using the SMARTer™ RACE kit according to the manufacturer's instructions (Clontech, Mountain View, CA, USA). The specific primers, GSP-5′ (5′-GCCATCCCTTTCTCATGCCAAACAT-3′) and GSP-3′ (5′-TGTGATTGCCGAATGCAGTCAAGCT-3′), were designed to clone the 5′ and 3′ regions of SDO, respectively. The RACE PCR reactions were performed using the Advantage II Polymerase Mix (Clontech). The 3′ and 5′-RACE products were gel-purified, subcloned, sequenced, and assembled with SeqMan Pro (DNA STAR, Madison, WI, USA).
Sequence analysis
The sequence similarity of SDO with those from other species was analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the protein structure and biochemical properties were predicted using the ExPASy proteomics server (http://www.expasy.org/tools/). The three-dimensional sequence model was predicted with the alignment mode in Swiss Model using the SDO protein from A. thaliana (PDB ID: 2GCU) as the template [23], [24], [25]. The multiple alignments of SDO were generated with ClustalX 2.1 and the phylogenetic tree was constructed using the MEGA program 5.0 by the Neighbor-Joining method using the Poisson correction amino acid substitution model and the complete deletion gaps option. Bootstrap values from 1000 replicates were calculated and indicated at branch points on the neighbor-joining tree.
Prokaryotic expression, purification and refolding of U. unicinctus SDO
Based in protein structural predictions, four sub-segments (Del-1-Del-4) of the U. unicinctus SDO open reading frame (ORF) and the complete ORF sequence were chosen, as shown in Figure 2, and amplified using the primers in Table 1. The obtained cDNA sequences were cloned into the pET28a plasmid, sequenced and transformed into E. coli BL21 (DE3). These expressed proteins contained 6-His tags at both C-terminal and N-terminal for purification purposes on a nickel affinity column. An overnight culture of E. coli BL21 (DE3) was grown in 1 mL Luria Bertani (LB) with kanamycin (30 µg/mL) at 37°C and then inoculated into 100 mL of LB media also supplemented with kanamycin. When the OD600 reached 0.5 expression was induced by adding 1 mM IPTG (isopropyl- β-D-1-thiogalactopyranoside) and the culture was grown for an additional 5 h period. The cells were then harvested by centrifugation at 10,000 g for 10 min at 4°C, suspended in 50 mM phosphate buffered solution (PBS, pH 7.4), and broken by ultrasonication on ice. The target recombinant proteins were found primarily in inclusion bodies. The five proteins were dissolved in 4 mL of buffer composed of 8 M urea, 10 mM Tris-HCl (pH 8.0) and 0.1 M trisodium phosphate. The recombinant proteins were purified by Ni-NTA affinity chromatography according to the manufacturer's protocol (Novagen, Darmstadt, Germany), and the purity of the eluted samples was analyzed by SDS-PAGE. The recombinant proteins were refolded by adding the refolding buffer (50 mM glycine, 100 mM phosphate buffer, 200 mM NaCl, 5 mM EDTA, 5 mM FeCl2, 10 mM DTT) with stirring to dilute the 8 M urea to 1 M, and then allowing the solution to stir at 4°C for a further 12 h. The urea was removed by dialysis; the soluble and aggregated fractions of the renaturation mixture were separated by centrifugation (10,000 g, 4°C, and 10 min) and analyzed by SDS-PAGE to confirm that the soluble fraction was successfully refolded.
Figure 2. Schematic diagram of SDO sub-segment expression.
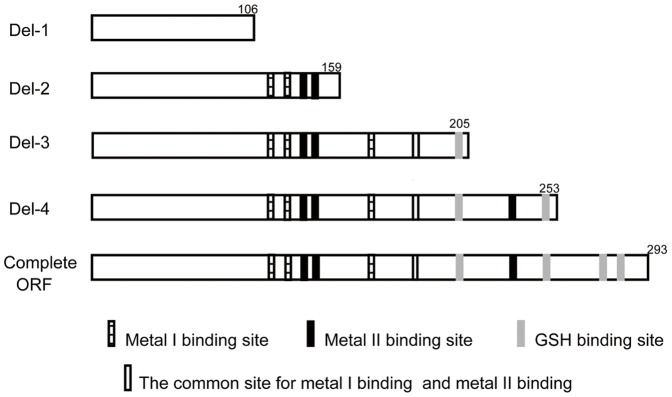
Del-1: no structural amino acid site; Del-2: two metal I binding site residues, two metal II binding site residues and no GSH binding site; Del-3: complete metal I binding site, three metal II binding site residues and one GSH binding site; Del-4: complete metal I binding site and metal II binding site as well as three GSH binding sites; complete ORF: complete metal I binding sites and metal II binding sites as well as GSH binding sites. The numbers indicated the expressed sub-segment amino acid length.
Table 1. Primers for the five cDNA sequences of U. unicinctus SDO.
| Forward primer | Reserved primer | Size (bp) | |
| Del-1 | GAGCTCATGCTTTCGTCCGTATGTGG | CTCGAGTAGATTGAGCCCGAGTTCTTT | 318 |
| Del-2 | GAGCTCATGCTTTCGTCCGTATGTGG | CTCGAGGAATTGACCAAACTCGATACC | 477 |
| Del-3 | GAGCTCATGCTTTCGTCCGTATGTGG | CTCGAGTGAGCTTCCTTGTTGGAAATC | 615 |
| Del-4 | GAGCTCATGCTTTCGTCCGTATGTGG | CTCGAGAATTGGTTTAGTGAGGCGGG | 759 |
| Complete ORF | GAGCTCATGCTTTCGTCCGTATGTGG | CTCGAGGGACTTGCTTGGGGGTGATT | 879 |
Double mutant of U. unicinctus SDO
Double mutagenesis was conducted using a fast mutagenesis system kit (Transgen, Beijing, China) and the mutations were generated using the primers, forward: ACACACATGTGCACGCTGAGGCTGTGACTGGTA and reverse: GCC TCAGCGTGCACATGTGTGTTGACTGCATA (the bold and italic letters show the bases deviating from the original sequence) to obtain the double mutant, D117E/H118A. The resulting sequence was analyzed by DNA sequencing and the refolded mutated SDO was attained as described above.
Enzyme activity assays
SDO activity (1U = 1 µmol GSS− min−1) was measured by the consumption of the substrate GSS− according to the method of Hildebrandt and Grieshaber [10]. The reaction mixture (2 mL) consisted of 0.1 M potassium phosphate buffer (pH 7.4), 1 mM GSH and 3 µg L−1 of the refolded SDOs. The reaction was started by adding 30 µL of a saturated acetonic sulfur solution. Samples and 250 µL aliquots were taken from the reaction mixture at various time intervals and incubated with 375 µL HCl (10 M) and 375 µL methylene blue (75 µM) at room temperature for 30 min. The absorption of oxidized methylene blue was measured at 670 nm, and subtracted from that of the blanks containing buffer instead of the sample. GSS− consumption was determined considering the amount of methylene blue reduced.
Enzyme kinetics analysis
To explore the kinetic characterization of the recombinant SDOs, the substrate GSS− was prepared by anaerobically mixing GSH (10 mM in 200 mM sodium phosphate, pH 7.4) with saturated acetonic sulfur solution. The GSS− concentration was determined as described above. The reaction mixture (2 ml) contained 0.1 M potassium phosphate buffer (pH 7.4), GSS− (gradient concentrations) and 3 µg L−1 of the refolded SDOs. The reactions were started by adding enzyme SDO preparations, and oxygen consumption was recorded using the Oxytherm liquid-phase oxygen measurement system (Hansatech, Pentney, U.K.) at 25°C. Kinetic parameters were calculated by a non-linear least square analysis of the data fitted to the Michaelis–Menten equation using the enzyme kinetics module of Sigmaplot version 12.0 (Systat Software, Erkrath, Germany).
GSH-affinity determination
A solution containing 3 µg L−1 refolded SDO in 100 mM phosphate buffer in the presence of 50 µM GSH was incubated for 30 min and then subjected to ultrafiltration (10,000 g, 4°C, and 30 min) using a centrifugal filter device (Amicon Ultra-4 10K, Millipore, Billerica, MA, USA), which allowed the GSH, but not the GSH bound to the SDO, to pass through the filter. The filtrate was analyzed for GSH using a total glutathione assay kit (Jiancheng, Nangjing, China). The GSH binding ability was calculated by determining the loss of GSH in the filtrate.
Western blot analysis
Mitochondria were isolated from the midgut of U. unicincuts according to the methods described by Schroff and Schöttler [26] with slight modifications. The midgut tissue and mitochondrial total protein were extracted using the tissue protein extraction kit (Cwbio, Beijing, China). SDS-PAGE and western blotting were carried out as described [21]. A polyclonal antibody of U. unicinctus SDO was prepared by injecting purified recombinant SDO into New Zealand white rabbits at a titer of 1∶ 25,600.
Statistical analysis
All data are presented as mean±SE. Significant differences among means were tested by one-way analysis of variance (ANOVA) followed by Duncan's multiple comparison procedure using the SPSS statistical package version 18.0 (IBM SPSS, Chicago, IL, US) at a significance level of p<0.05.
Results
Sequence characteristics of U. unicinctus SDO
A cDNA fragment of 260 bp was obtained by RT-PCR using degenerate primers, and two fragments of 886 bp and 1240 bp were cloned by 5′ and 3′ RACE, respectively, and then assembled to a 1976-bp full-length SDO cDNA (GenBank accession number: HQ730921.1).
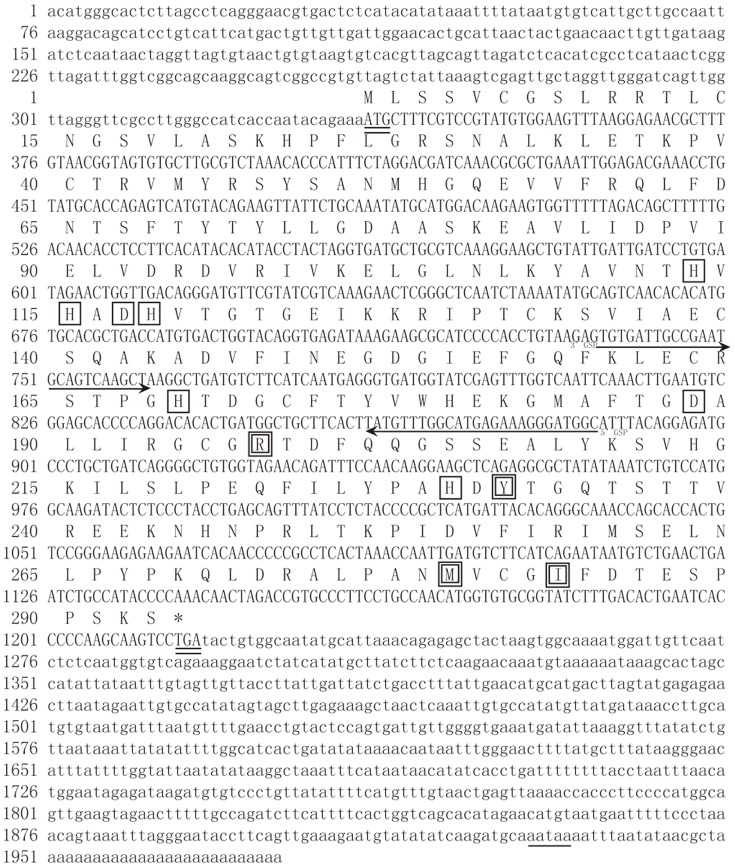
The full length cDNA sequence of SDO as well as the resultant amino acid sequence is shown in Figure 3. The ORF has 882 bp, encoding a 293 amino acids polypeptide with a theoretical pI of 8.03 and molecular mass of 32.61 kDa. The conserved metal I (H113, H115, H169 and D188), metal II (D117, H118, H169 and H229) and potential GSH (R197, Y231, M279 and I283) binding sites are indicated in the amino acid sequence. The western blot analysis shows that the SDO protein is present in the mitochondria (Figure 4).
Figure 3. Nucleotide sequence and predicted amino acid sequence of SDO in U. unicinctus.
Start (ATG) and stop (TGA) codons, double lines; RACE gene specific primers (GSP), indicated by an arrow; the AATAAA polyadenylation signal is underlined; Conserved metal binding sites are boxed in black; Conserved GSH binding sites are double boxed.
Figure 4. Mitochondrial location of SDO in U. unicinctus.
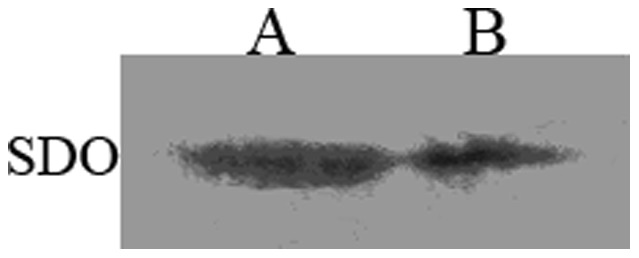
A. Midgut total protein; B. Midgut mitochondrial protein.
According to the three-dimensional model of the U. unicinctus SDO protein (Figure 5), the functional domain is characterized by a typical β-lactamase fold where the metal binding sites locate, a four-layered β-sandwich fold with two mixed β-sheets flanked by α-helices. It is well known that five histidines and two aspartate residues are important for metal binding in the metallo-β-lactamase superfamily [27], [28] and these residues are also conserved in U. unicinctus SDO (Figure 5A). The amino acids presumably involved in the metal I binding site are the conserved H113, H169, and D188 (Figure 5B); no metal ion is predicted to be bound to the conserved metal II binding site [31].
Figure 5. The predicted three-dimensional models of U. unicinctus SDO based on SDO protein in A. thaliana (PDB ID: 2GCU).
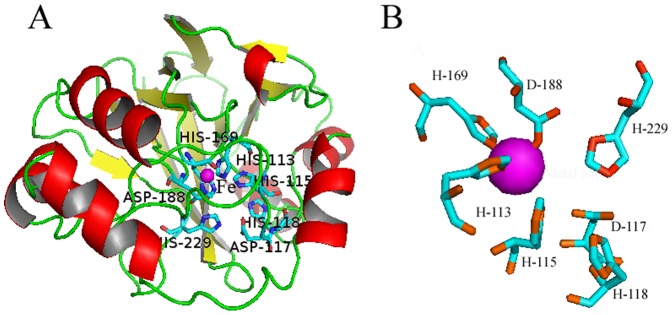
A. The β-strands, α-helices, and loop regions are shown as yellow, red, and green ribbons, respectively. The typical β-lactamase fold and metal binding sites are labeled. B. The iron (magenta sphere) binding amino acids, H113, H169 and D188, in metal binding site I form the 2His:1Asp facial triad, the remaining residues shown are found in the metal binding site II around the iron.
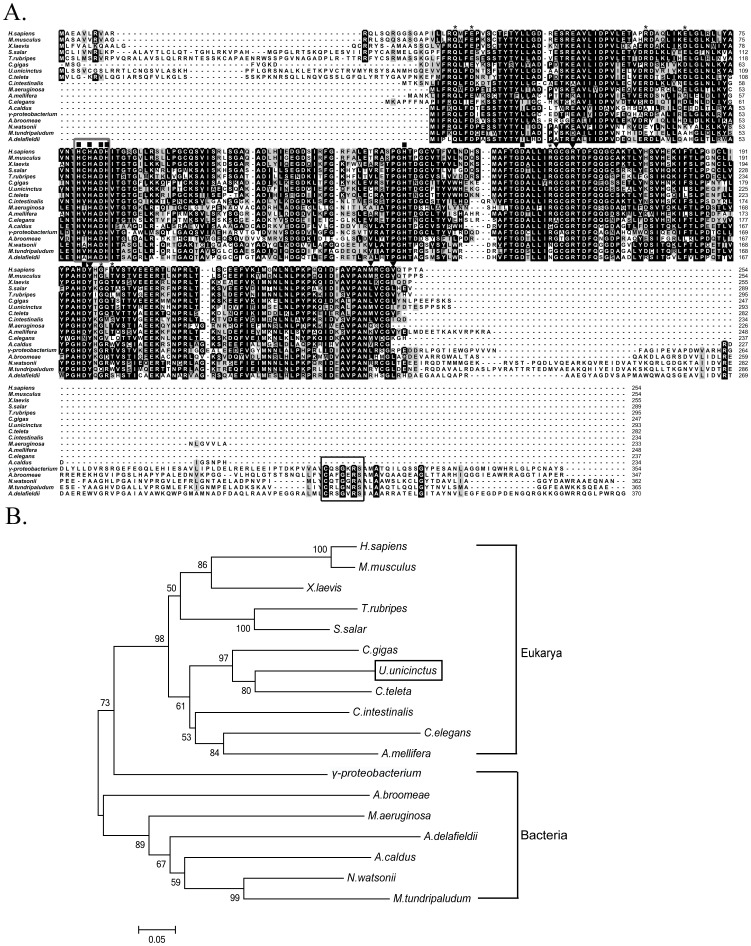
The results from homology analysis using Blastp show that U. unicinctus SDO shares some identity to SDOs from the oyster Crassostrea gigas (69%), the toad Xenopus laevis (63%), the sea squirt Ciona testinalis (62%), the Atlantic salmon Salmo salar (59%), and the mouse Mus musculus (58%). A ClustalX2 alignment was used to determine the overall gene conservation (Figure 6A) and indicates that SDOs among different organisms are highly conserved, especially in terms of their metal ion and GSH binding sites and also in the residues involved in the dimer formation. The phylogenetic tree constructed using the MEGA5 program based on the protein alignment (Figure 6B) shows that U. unicinctus SDO is most closely related to the polychaete Capitella teleta and the relationships displayed in the phylogenetic tree are generally in accordance with classical taxonomy.
Figure 6. Multiple sequence alignment (A) and phylogenetic relationships (B) among the SDO sequences from different species.
Identical and similar residues are highlighted in black and gray, respectively. Conserved relevant metal binding sites, GSH binding sites and dimer formation residues are indicated by ▪, ▾ and *, respectively. The β-lactamase signature motif and rhodanese active-site loop are marked with a black box and grey box, respectively. GenBank accession numbers: Afipia broomeae (ZP_11429388.1), Acidithiobacillus caldus (YP_004749948.1), Acidovorax delafieldii (ZP_04761469.1), Apis mellifera (XP_393510.1), Caenorhabditis elegans (NP_501684.1), Capitella teleta (JGI Genome), Ciona intestinalis (XP_002128021.1), Crassostrea gigas (EKC28487.1), Homo sapiens (NP_055112.2), Methylobacter tundripaludum (ZP_08782165.1), Microcystis aeruginosa (ZP_18834377.1), Mus musculus (NP_075643.1), Nitrosococcus watsonii (YP_003760989.1), γ-proteobacterium HTCC2148 (ZP_05095460), Salmo salar (ACI68458.1), Takifugu rubripes (XP_003977175.1), Urechis unicinctus (AEV92813.1), Xenopus laevis (NP_001079404.1).
Identification of U. unicinctus SDO functional domains
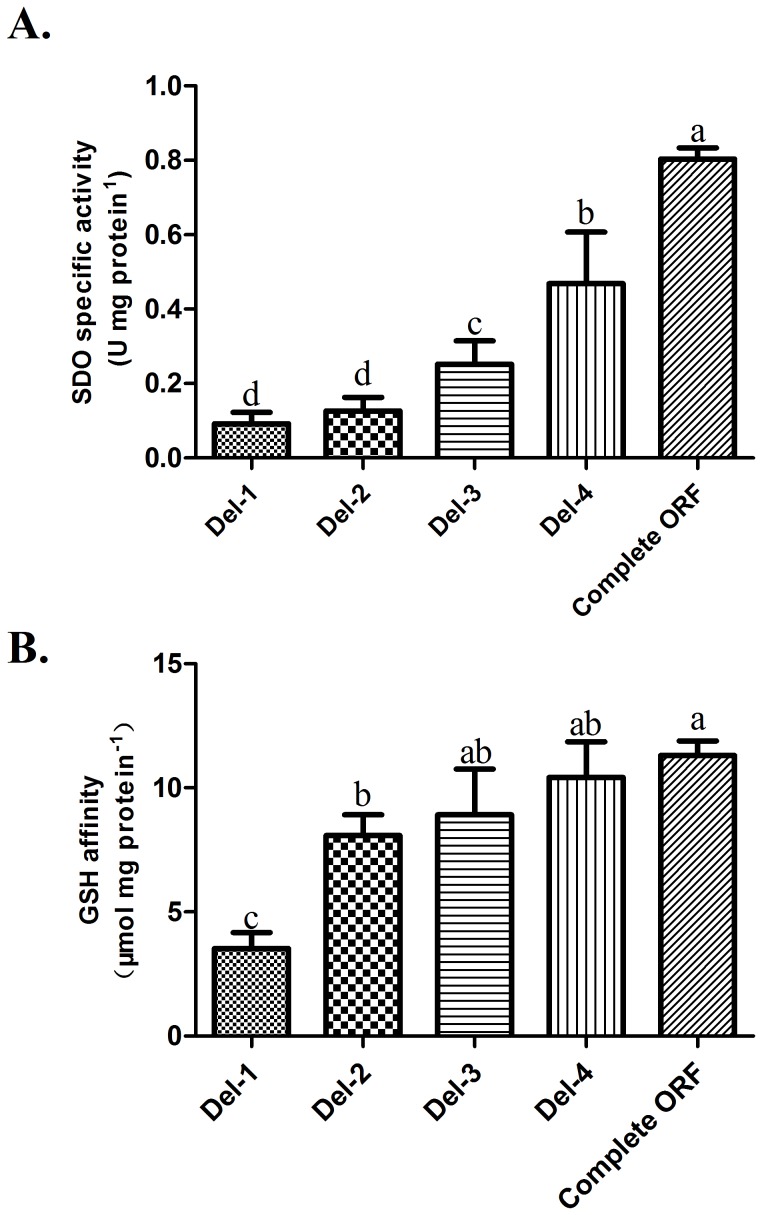
The five sequences of U. unicinctus SDO were expressed successfully in E. coli using a pET28a expression system. The recombinant proteins containing His-tags at both the C-terminal and N-terminal, were purified to 95% purity and their sizes are consistent with what was predicted (Figure 7). The SDO activities of the recombinant proteins were determined by methylene blue reduction (Figure 8A). The specific activity of the complete SDO protein is approximately 0.80 U mg protein−1. However, the specific activities of the truncated SDO proteins are lower than wild type enzyme. In Del-4, the SDO specific activity is significantly decreased (p<0.05) by 41.7% as compared with the wild type protein. No significant difference (p>0.05) was observed between the specific activity of Del-1 (0.09 U mg protein−1) and Del-2 (0.12 U mg protein−1), however, they were both significantly lower (p<0.05) than those measured for the rest of the enzymes.
Figure 7. Purified recombinant SDO proteins detected by SDS-PAGE.
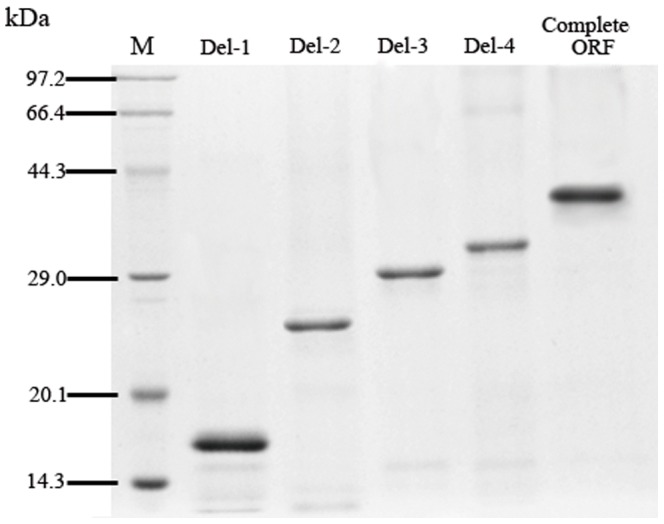
Figure 8. Characteristics of the five sub-segment-expressed recombinant SDO proteins.
A. SDO specific activities (mean±SE, n = 3) of five recombinant SDO proteins. B. GSH affinities (mean±SE, n = 3) of the recombinant SDO proteins. Groups containing the same letters on the bar indicate no significant difference while different letters on the bar indicate a significant difference (p<0.05).
To further understand the characteristics of truncated forms of SDO the kinetic parameters to monitor the rate of oxygen consumption during the conversion of GSS− to sulfite were determined using a Clark oxygen electrode (Table 2). The Vmax is 1.74±0.08 µmol min−1 mg protein−1 and the KM is 82.5±9.9 µM for the recombinant wild type enzyme. The Del-4 protein shows a 29.3% decrease in Vmax while the KM for GSSH is unaffected. For the Del-3 protein, the Vmax decreased to 56% and for both the Del-1 and Del-2 proteins, the Vmax are about 20% that of the obtained for the wild type form. The values of KM are not significantly different (p>0.05) among Del-2, Del-3, Del-4 and wild type enzyme but suffer a steeply increase up to 218.9±33.5 in the Del-1 which represents a two-fold lower affinity to the substrate than the wild type enzyme.
Table 2. Comparison of the kinetic properties of five recombinant SDO proteins.
| Vmax (μmol min−1 mg protein−1) | KM (μM) | kcat (s−1) | kcat/KM (μM−1 s−1) | |
| Del-1 | 0.33±0.014d | 218.9±33.5A | 0.09 | 42.68 |
| Del-2 | 0.42±0.024d | 137.0±17.2B | 0.16 | 116.13 |
| Del-3 | 0.98±0.065c | 104.6±16.7B | 0.45 | 433.72 |
| Del-4 | 1.23±0.063b | 83.0±10.9B | 0.68 | 822.00 |
| Complete ORF | 1.74±0.081a | 82.5±9.9B | 1.09 | 1327.22 |
The values with different superscripts in the same column are significantly different (p<0.05).
The GSH-affinity of the recombinant complete protein is 11.3 µmol mg protein−1 (Figure 8B). The GSH-affinity for Del-1 is the lowest among the truncated enzymes with 3.5 µmol mg protein−1 while for Del-2, Del-3 and Del-4 are significantly higher (p<0.05), GSH-affinities reaching 8.1, 8.9 and 10.4 µmol mg protein−1 were measured, respectively.
U. unicinctus SDO double mutant characterization
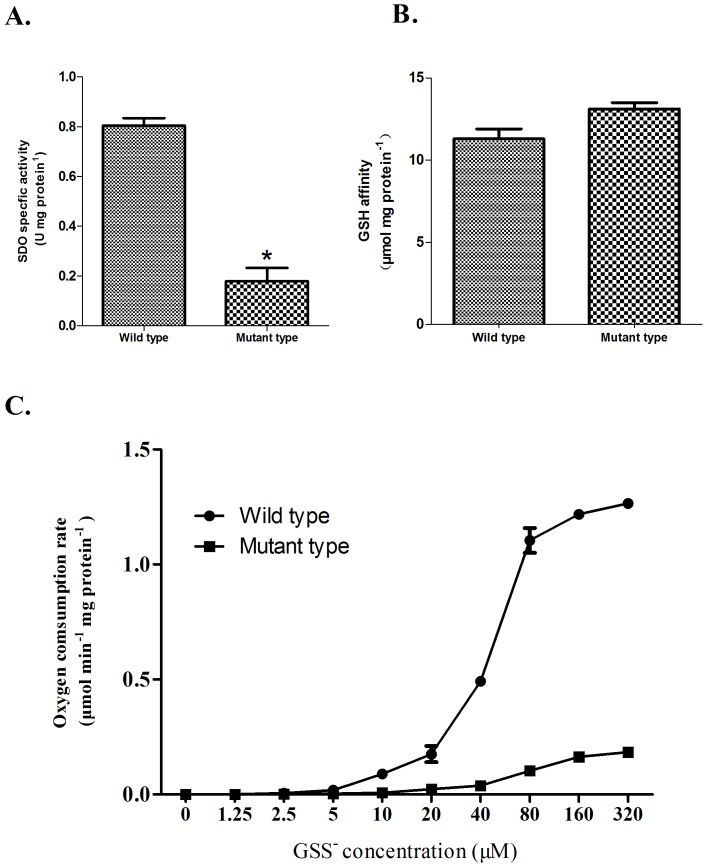
The SDO-specific activity almost double in Del-4 compared with Del-3 when the second metal binding site became intact (Figure 8A). Based on this, D117 and H118 located in the second metal binding site (belonging to the metallo-β-lactamase superfamily signature motif HXHXDH (X for an arbitrary amino acid)) were chosen to be replaced by Glu and Ala, respectively, by double mutagenesis. The analysis of the double mutant D117E/H118A indicates that its SDO specific activity (0.17 U mg protein−1) is significantly decreased compared with wild type (Figure 9A). The reaction rates of wild type and double mutant at different substrate concentrations are shown in Figure 9C. The rates of oxygen consumption are much lower for the mutant than for wild type; a KM for GSS− of 174.4±16.8 µM and a Vmax of 0.30±0.014 µmol min−1 mg protein−1 were determined for the double mutant enzyme. In contrast, the GSH affinity (13.1 µmol mg protein−1) is similar to wild SDO (p>0.05) (Figure 9B), indicating that the loss of SDO activity reflects the replacement of the Asp residues by Glu and His residues by Ala in the double mutant enzyme.
Figure 9. Characteristics of the U. unicinctus SDO mutant.
A. SDO specific activities in wild type and mutant. * indicates a significant difference from the wild type (p<0.05). B. GSH affinities in wild type and mutant. C. Oxygen consumption rate versus GSS− concentration in wild type and mutant.
Discussion
SDO, a novel member of the metallo-β-lactamase superfamily
The members of the metallo-β-lactamase superfamily can catalyze a diversity of reactions, and are divided into 17 groups based on their biological functions [27], [28]. Glyoxalase II (GLX2) belongs to group 2 and can hydrolyze s-D-lactoylglutathione (SLG) into D-lactate and GSH [27], [29]. In this study, the U. unicinctus SDO protein was shown to be highly conserved among known ETHE1 (SDO) proteins, containing the signature motif HXHXDH of the metallo-β-lactamase superfamily and sharing 61% sequence identity to the Ixodes scapularis glyoxalase (XP_002399673.1). Because of its high similarity with GLX2, SDO is believed to be a member of group 2; for example, SDO (ETHE1) from A. thaliana is thought to be the putative glyoxalase II isozyme GLX2-3 [30], [31]. However, it is known that SDO cannot hydrolyze any glutathione thioesters [32], [33], [34] because it lacks several highly conserved residues (N179, Y145, F182 and D253) that participate in the hydrogen bonding of SLG in GLX2. Therefore, SDO (ETHE1) protein is suggested to belong to a new class in the metallo-β-lactamase superfamily playing an important role in GSS− oxidation [13]. In our study, U. unicinctus SDO also lacked the GLX2 conserved amino acids (mentioned above) and instead uses oxygen to oxidize the GSS− to sulfite (Table 2). Most members of the metallo-β-lactamase superfamily only display hydrolase activity, such as β-lactamases [35] and glyoxalases II [36], with the exception of the group 3 member, ROO (flavoproteins and rubredoxin oxygen: oxidoreductase) that contains two domains: a metallo-β-lactamase and a flavodoxin-like which act together to provide the oxidoreductase activity [37]. In this study, the U. unicinctus SDO shows oxidoreductase activity, although it only contains a metallo-β-lactamase domain. This result further supports that SDO is a novel member of metallo-β-lactamase superfamily.
As shown by the western blot, the SDO of U. unicinctus is located in the mitochondria (Figure 4), which was in accordance with previous reports of human and A. thaliana ETHE1s [14], [34]. Interestingly, it seems that separation of the SDO gene and rhodanese gene occurred during the mitochondrial occurrence in eukaryotes during the evolution of the species. Indeed in some bacteria, such as Methylobacter tundripaludum, Nitrosococcus watsonii and γ-proteobacterium, a SDO-like amino acid sequence is linked with a rhodanese-like domain (Figure 6A). However, in eukaryotes, the SDO gene and rhodanese gene are assigned as two separate genes in the NCBI database. In addition, we found that the SDO-like amino acid sequence in γ-proteobacterium was clustered with that of eukaryotes (Figure 6B). Considering that early eukaryote mitochondria are thought to be derived from intracellular bacterial symbionts of proteobacterial origin [38] it is therefore suggested that the SDO gene and rhodanese gene were separated when the endosymbiont genes were integrated into the eukaryote genomes.
SDO catalytic activity
Hildebrandt and Grieshaber [10] reported maximal rates for SDO purified from rat liver and lugworm body-wall tissue of 0.87±0.04 and 0.85±0.24 U mg protein−1, respectively, similar to 0.80 U mg protein−1 measured for U. unicinctus SDO conversion of GSS− to sulfite. However, a significantly lower KM (82.5±9.9 µM) was determined in this latter enzyme as compared to human (340±30 µM [15]) or thiobacilli (120–240 µM [39]) SODs, indicating that SDO from U. unicinctus binds the substrate tighter than those from other animals.
The members of the metallo-β-lactamase superfamily usually possess two potential metal binding sites [28], which in U. unicinctus SDO were predicted to be formed by the residues H113, H115, H169 and D188 for metal I binding site and D117, H118, H169 and H229 for metal II binding site. We show that the two metal binding sites are important for enzyme activity: the activity decreased steeply from the complete form to the increasingly truncated forms of the protein (Figure 8A). For example, the activity increased 2.8-fold and 5.1-fold in Del-3 (completed metal I binding site) and Del-4 (completed both metal binding sites) respectively as compared to Del-1 (no metal binding site). No significant differences (p>0.05) were detected for the KM among the Del-2, Del-3, Del-4 or wild type enzyme (Table 2). Bugg [40] suggested that in the dioxygenase superfamily, the metal iron center is important for substrate binding and activation of the catalytic reaction. Therefore, this may explain the almost invariable affinity for the substrate GSS− in the truncated proteins. In this study, the KM decreased more obviously from Del-3 to Del-2 than from Del-4 to Del-3. Because only one metal ion is found in SDO, these results suggest that the metal ion is most likely located in the metal I binding site, a suggestion in agreement with a recent report where only one Fe2+ was found in the metal I binding site in A. thaliana [34]. McCoy et al. [31] reported that the metal II binding site usually directly coordinate to iron in other metallo-β-lactamase group members with only one metal iron. In our study, the metal II binding site integrity may also affect the SDO catalytic activity as a 388 µM−1 s−1 increase in catalytic efficiency (kcat/KM) was observed indicating the metal II binding site is important for the enzyme activity. Moreover, the double mutant D117E/H118A, in the metal II binding region, shows approximately one-fifth of the wild type specific activity. The KM of the double mutant is 174.4±16.8 µM, two fold higher than that of the wild type, indicating also a reduced affinity for binding the substrate. In many metallo-β-lactamases, substitution of the homologous Asp and His can impair the enzyme activity, as Asp coordinates the metal ion for correct substrate binding [41]. In addition, dissociation of H− from OH2 binding of the metal ion is suppressed in the metallo-β-lactamase (IMP-1) mutant D120E [42]. However, dissociation of H−is the most important step for the binding of the substrate GSS− [15]. Taken together, these are thought to be the major reasons for the increase in the KM and the decrease in Vmax in the double mutant D117E/H118A.
The catalytic efficiency (kcat/KM) is 822.00 µM−1 s−1 and 1327.22 µM−1 s−1 for Del-4 and wild type enzyme, respectively. This difference can be attributed to differences in the number of GSH binding sites [15]. The predicted amino acid for GSH binding in U. unicinctus SDO are Arg197, Tyr231, Met279 and Ile283 according to its sequence homology with human GLX2. In human GLX2, the backbone amino group of Lys143 and the side chain of Tyr175 were shown to bind the cysteine portion of GSH via hydrogen bonds while the side chains of Arg249 and Lys252 establish hydrogen bonds with the glycine portion of GSH but overall [32]. In this study, Del-1 and Del-2 contained no GSH binding site; Del-3 contained Arg197, and Del-4 contained Arg197 and Tyr231. The increase in GSH affinity for Del-4 compared with Del-3 indicated that Tyr231 in U. unicinctus SDO (Figure 8B) is homologous with Tyr175 in human GLX2 the key residue for GSH binding. However, the decreased GSH binding ability of other amino acids such as Arg197, Met279 and Ile283 as compared with that of Tyr231 (Del-3), may be caused by mutations in the homologous human amino acids. In addition the reason behind the significant increase in GSH binding affinity for Del-1 to Del-2 may result from the fact that the metal also combines with GSH because of its similarity in structure to GSS−. The binding to GSH could compete with the binding of GSS−, but may be helpful to promote the oxidation of GSS−. In humans, the Km for GSS− of SDO is slightly lower in the presence of GSH with a higher SDO specific activity [15]. Although the Del-1 construct did not contain the predicted GSH binding site, its GSH binding affinity is still as low as 3.5 µmol mg protein−1. We suppose that an unknown GSH binding region exists in the Del-1 construct, which non-enzymatically enhances GSS− oxidation with an overall increase in SDO specific activity. However, further research into the nature of the GSH binding sites is required.
Funding Statement
This work was supported by the Natural Science Foundation of China (31072191). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carrico RJ, Blumberg WE, Peisach J (1978) The reversible binding of oxygen to sulfhemoglobin. J Biol Chem 253: 7212–7215. [PubMed] [Google Scholar]
- 2. Beauchamp RO, Bus JS, Popp JA, Boreiko CJ, Andjelkovich DA (1984) A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol 13: 25–97. [DOI] [PubMed] [Google Scholar]
- 3. Khan AA, Schuler MM, Prior MG, Yong S, Coppock RW, et al. (1990) Effects of hydrogen sulfide exposure on lung mitochondrial respiratory chain enzymes in rats. Toxicol Appl Pharm 103: 482–490. [DOI] [PubMed] [Google Scholar]
- 4. Julian D, April KL, Patel S, Stein JR, Wohlgemuth SE (2005) Mitochondrial depolarization following hydrogen sulfide exposure in erythrocytes from a sulfide-tolerant marine invertebrate. J Exp Biol 208: 4109–4122. [DOI] [PubMed] [Google Scholar]
- 5. Yang G, Sun X, Wang R (2004) Hydrogen sulfide-induced apoptosis of human aorta smooth muscle cells via the activation of mitogen-activated protein kinases and caspase-3. The FASEB Journal 18: 1782–1784. [DOI] [PubMed] [Google Scholar]
- 6. Joyner-Matos J, Predmore BL, Stein JR, Leeuwenburgh C, Julian D (2010) Hydrogen sulfide induces oxidative damage to RNA and DNA in a sulfide-tolerant marine invertebrate. Physiol Biochem Zool 83: 356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jørgensen BB, Fenchel T (1974) The sulfur cycle of a marine sediment model system. Mar Biol 24: 189–201. [Google Scholar]
- 8. Arp AJ, Hansen BM, Julian D (1992) Burrow environment and coelomic fluid characteristics of the echiurian worm Urechis caupo from populations at three sites in northern California. Mar Biol 113: 613–623. [Google Scholar]
- 9. Grieshaber MK, Völkel S (1998) Animal adaptations for tolerance and exploitation of poisonous sulfide. Annu Rev Physiol 60: 33–53. [DOI] [PubMed] [Google Scholar]
- 10. Hildebrandt TM, Grieshaber MK (2008) Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS Journal 275: 3352–3361. [DOI] [PubMed] [Google Scholar]
- 11. Jackson MR, Melideo SL, Jorns M S (2012) Human sulfide: quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite [J]. Biochemistry 51: 6804–6815. [DOI] [PubMed] [Google Scholar]
- 12. Tiranti V, D'Adamo P, Briem E, Ferrari G, Mineri R, et al. (2004) Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am J Hum Genet 74: 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tiranti V, Viscomi C, Hildebrandt T, Di Meo I, Mineri R, et al. (2009) Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med 15: 200–205. [DOI] [PubMed] [Google Scholar]
- 14. Holdorf MM, Owen HA, Lieber SR, Yuan L, Adams N, et al. (2012) Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol 160: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kabil O, Banerjee R (2012) Characterization of patient mutations in human persulfur dioxygenase (ETHE1) involved in H2S catabolism. J Biol Chem 287: 44561–44567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li N, Song SL, Tang YZ (1995) The Life History of the Urechis Unicinctus (Von. Draschi). Shandong Fisheries 12: 24–27. [Google Scholar]
- 17. Li N, Song SL, Tang YZ (1998) Urechis Unicinctus (Von. Draschi). Bulletin of Biology 33: 12–14. [Google Scholar]
- 18.Ma ZJ, Bao ZM, Kang KH, Yu L, Zhang ZF (2005) The changes of three components in coelomic fluid of Urechis unicinctus exposed to different concentrations of sulfide. Chin J Oceanol Limnol 23:104 –109.
- 19. Ma ZJ, Bao ZM, Wang SF, Zhang ZF (2010) Sulfide-based ATP production in Urechis unicinctus . Chin J Oceanol Limnol 28: 521–526. [Google Scholar]
- 20. Wang SF, Zhang ZF, Cui H, Kang KH, Ma ZJ (2010) The effect of toxic sulfide exposure on oxygen consumption and oxidation products in Urechis unicinctus (Echiura: Urechidae). J. Ocean Univ. China 9: 157–161. [Google Scholar]
- 21. Ma YB, Zhang ZF, Shao MY, Kang KH, Tan Z, et al. (2011) Sulfide: quinone oxidoreductase from echiuran worm Urechis unicinctus . Mar Biotechnol 13: 93–107. [DOI] [PubMed] [Google Scholar]
- 22. Ma YB, Zhang ZF, Shao MY, Kang KH, Shi XL, et al. (2012) Response of sulfide:quinone oxidoreductase to sulfide exposure in the echiuran worm Urechis unicinctus . Mar Biotechnol (NY) 14: 245–251. [DOI] [PubMed] [Google Scholar]
- 23. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 24. Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The Swiss-model repository and associated resources. Nucleic Acids Re 37: D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peitsch MC (1995) Protein modeling by E-mail. Bio/Technology 13: 658–660. [Google Scholar]
- 26.Schroff G, Schöttler U (1977) Anaerobic reduction of fumarate in the body wall musculature of Arenicola marina (Polychaeta). J Comp Physiol B 116: 325 – 336.
- 27. Daiyasu H, Osaka K, Ishino Y, Toh H (2001) Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett 503: 1–6. [DOI] [PubMed] [Google Scholar]
- 28. Bebrone C (2007) Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74: 1686–1701. [DOI] [PubMed] [Google Scholar]
- 29. Thornalley PJ (1993) The glyoxalase system in health and disease. Mol Aspects Med 14: 287–371. [DOI] [PubMed] [Google Scholar]
- 30. Maiti MK, Krishnasamy S, Owen HA, Makaroff CA (1997) Molecular characterization of glyoxalase II from Arabidopsis thaliana . Plant Mol Biol 35: 471–481. [DOI] [PubMed] [Google Scholar]
- 31. McCoy JG, Bingman CA, Bitto E, Holdorf MM, Makaroff CA, et al. (2006) Structure of an ETHE1-like protein from Arabidopsis thaliana . Acta Crystallogr D Biol Crystallogr 62: 964–970. [DOI] [PubMed] [Google Scholar]
- 32. Cameron AD, Ridderström M, Olin B, Mannervik B (1999) Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure 7: 1067–1078. [DOI] [PubMed] [Google Scholar]
- 33. Marasinghe GPK, Sander IM, Bennett B, Periyannan G, Yang KW, et al. (2005) Structural Studies on a Mitochondrial Glyoxalase II. J Biol Chem 280: 40668–40675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holdorf MM, Bennett B, Crowder MW, Makaroff CA (2008) Spectroscopic studies on Arabidopsis ETHE1, a glyoxalase II-like protein. J Inorg Biochem 102: 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dal Peraro M, Vila A, Carloni P (2002) Structural determinants and hydrogen-bond network of the mononuclear zinc(II)-beta-lactamase active site. J Biol Inorg Chem 7: 704–712. [DOI] [PubMed] [Google Scholar]
- 36. Vander Jagt D (1993) Glyoxalase II: molecular characteristics, kinetics and mechanism. Biochem Soc Trans 21: 522–527. [DOI] [PubMed] [Google Scholar]
- 37. Frazão C, Silva G, Gomes CM, Matias P, Coelho R, et al. (2000) Structure of a dioxygen reduction enzyme from Desulfovibrio gigas . Nat Struct Biol 7: 1041–1045. [DOI] [PubMed] [Google Scholar]
- 38. Szklarczyk R, Huynen MA (2010) Mosaic origin of the mitochondrial proteome. Proteomics 10: 4012–4024. [DOI] [PubMed] [Google Scholar]
- 39. Rohwerder T, Sand W (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149: 1699–1709. [DOI] [PubMed] [Google Scholar]
- 40. Bugg TDH (2003) Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron 59: 7075–7101. [Google Scholar]
- 41. Llarrull LI, Fabiane SM, Kowalski JM, Bennett B, Sutton BJ, et al. (2007) Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity. J Biol Chem 282: 18276–18285. [DOI] [PubMed] [Google Scholar]
- 42. Yamaguchi Y, Kuroki T, Yasuzawa H, Higashi T, Jin W, et al. (2005) Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography. J Biol Chem 280: 20824–20832. [DOI] [PubMed] [Google Scholar]