Abstract
Cancer is one of the leading causes of death worldwide. Current therapeutic strategies are predominantly developed in 2D culture systems, which inadequately reflect physiological conditions in vivo. Biological 3D matrices provide cells an environment in which cells can self-organize, allowing the study of tissue organization and cell differentiation. Such scaffolds can be seeded with a mixture of different cell types to study direct 3D cell-cell-interactions. To mimic the 3D complexity of cancer tumors, our group has developed a 3D in vitro tumor test system.
Our 3D tissue test system models the in vivo situation of malignant peripheral nerve sheath tumors (MPNSTs), which we established with our decellularized porcine jejunal segment derived biological vascularized scaffold (BioVaSc). In our model, we reseeded a modified BioVaSc matrix with primary fibroblasts, microvascular endothelial cells (mvECs) and the S462 tumor cell line. For static culture, the vascular structure of the BioVaSc is removed and the remaining scaffold is cut open on one side (Small Intestinal Submucosa SIS-Muc). The resulting matrix is then fixed between two metal rings (cell crowns).
Another option is to culture the cell-seeded SIS-Muc in a flow bioreactor system that exposes the cells to shear stress. Here, the bioreactor is connected to a peristaltic pump in a self-constructed incubator. A computer regulates the arterial oxygen and nutrient supply via parameters such as blood pressure, temperature, and flow rate. This setup allows for a dynamic culture with either pressure-regulated pulsatile or constant flow.
In this study, we could successfully establish both a static and dynamic 3D culture system for MPNSTs. The ability to model cancer tumors in a more natural 3D environment will enable the discovery, testing, and validation of future pharmaceuticals in a human-like model.
Keywords: Cancer Biology, Issue 78, Biomedical Engineering, Bioengineering, Medicine, Anatomy, Physiology, Molecular Biology, Cellular Biology, Tissue Engineering, Tumor Cells, Cultured, Biotechnology, Culture Techniques, Cell Engineering, Cellular Microenvironment, Equipment and Supplies, Decellularization, BioVaSc, primary cell isolation, tumor test system, dynamic culture conditions, bioreactor, 3D in vitro models, cell culture
Introduction
New pharmaceuticals must be validated in regard to their quality, safety, and efficacy before market authorization. To date, animal experiments are the standard method for drug testing and validation. However, due to species-specific differences, animal experiments often do not comprehensively evaluate the effect of the compounds in humans 1. For this reason, it is important to generate human tissue models that can be used for in vitro tests of new drugs and substances.
One of the focuses of our group is the creation of in vitro test models with our biological vascularized scaffold (BioVaSc) 2,3. The BioVaSc can be used as a static or dynamic 3D matrix system. For static culture, the decellularized porcine jejunal segment (Small Intestinal Submucosa SIS-Muc) is placed in a metal insert for cell reseeding. Various cells, such as cancer and endothelial cells can be cultured on the scaffold.
For dynamic culture, the BioVaSc is attached to a bioreactor system that applies flow throughout the vasculature or across the surface of the scaffold. Current bioreactors implement biological, mechanical, or electrical stimuli that act upon the differentiation or proliferation of cells 4. For bioreactors in the field of Tissue Engineering, the basic concept is to simulate the conditions in the human body. Wherein, cells are provided a natural environment in which they can interact with each other and their surrounding extracellular matrix. For the production of in vitro test systems or transplants, the ability to mimic the natural environment of cells with an appropriate carrier structure and bioreactor system is critical 5. Therefore, more complex and technically demanding devices must be developed in order to fulfill these tasks 6.
It is furthermore possible to use our scaffold for the establishment of a vascularized model due to the preserved tubular structures, which include the feeding artery, vein, and the connecting capillary bed. All porcine cells need to be removed by chemical, mechanical and enzymatic decellularization, and the scaffold gamma-sterilized. The restored tubular vascular structures can subsequently be reseeded with human microvascular endothelial cells using a recirculation perfusion bioreactor 7, which mimics the biomechanical and/or biochemical parameters such as pH, temperature, pressure, nutrient supply and waste removal 6. The re-endothelialization of the tubular structures creates a human blood vessel equivalent within the collagenous scaffold 3,7. In the following step, the surface of the former lumen (mucosa) can be seeded with primary human cells to establish co-cultures 3,7,8.
In this study a 3D tumor test system is set up by co-culturing a tumor cell line with primary stromal cells under static and dynamic conditions on the SIS-Muc.
Protocol
1. Decellularization of the BioVaSc
Rinse the vascular system of the porcine jejunal segment via cannulated arterial access and the intestinal lumen with PBS-. Repeat until it is completely clean.
Prepare a 200 mm diameter glass tank with 4 adapters and connect them via silicon tubes to the peristaltic pump (Ismatec). The pressure controlling unit can be monitored via a pressure sensor that is connected to a sterile disposable dome (see Figure 1).
Fill reservoir bottles with decellularization (DZ) solution and check the tubing system for possible air bubbles.
Connect the intestinal lumen with cable ties to the glass connectors for luminal flow. Pump 500 ml DZ solution into the red arterial access (Figure 1B) of the vascular system. Interrupt the pumping process every 15 min shortly to manually press out the entire intestinal lumen.
During the decellularization process, the pressure of the buffer solution should be between 80 - 100 mm Hg, modeled after the natural blood pressure.
Wash the BioVaSc with PBS- until it is free of cell remnants ("completely white").
Fill the BioVaSc completely with DZ solution and incubate it submerge in DZ solution over night at 4 °C on rocking shaker.
Repeat step 1.6.
Place the BioVaSc in DNase solution and incubate over night at 4 °C on rocking shaker.
Remove DNase solution and rinse with washing buffer.
γ-sterilization with 25 kGy
2. The Different Cell Types
2.1 Isolation of primary human dermal microvascular endothelial cells (mvECs) and fibroblasts
Cut skin biopsy (preferably preputium) into strips of 2 - 3 mm width with scalpel and rinse them 3x with PBS- solution.
Cover the tissue with Dispase solution and incubate it for 16 - 18 hr at 4 °C.
Separate epidermis from dermis with 2 tweezers and transfer both separately into petri dishes filled with PBS+.
Rinse dermis strips 1x with Versene.
Add 10 ml Trypsin/EDTA solution to the dermis strips and incubate it in the incubator for 40 min.
Stop the enzyme reaction immediately with 1% FCS.
Transfer skin strips to a petri dish filled with VascuLife and scratch out each strip with the scalpel 8x each side, adding a little pressure.
Transfer the produced cell suspension via a cell strainer to a centrifuge tube and rinse cell strainer 3x with VascuLife.
Centrifuge the tube at 1,200 x g for 5 min and resuspend the cell pellet with VascuLife.
To isolate the fibroblasts, chop dermis strips into little pieces using the scalpel.
Add 10 ml of collagenase solution to dermis pieces.
Incubate it for 45 min in the incubator, then centrifuge it and carefully remove supernatant.
Wash pellet 1x with DMEM + 10% FCS + % PenStrep, centrifuge it and carefully remove supernatant.
Resuspend the pellet in culture medium and transfer it to a T75 culture flask to allow cells to grow out of the tissue.
2.2 Tumor Cell Line S462
The tumor cell line S462 (kindly provided by Dr. Nikola Holtkamp, Charité University Medicine Berlin) was generated from a malignant peripheral nerve sheath tumor of a female patient with the hereditary tumor predisposition syndrome neurofibromatosis type 1 9. S462 are cultured in DMEM supplemented with 10% FCS. Medium has to be changed every 2 - 3 days. Once a week the cells have to be split.
3. Tumor Test System: Static Culture Conditions Compared with Dynamic Culture In Bioreactor Systems
Cut the tubular SIS-Muc open on one side and fix it between two metal rings (cell crowns, 10 mm diameter, self constructed). Cover the SIS-Muc in cell culture medium overnight.
- Seed isolated cells in defined cell numbers (see below) on one or both sides of the SIS (mono- or co-culture set-up).
- Seed 8,000 cells/cm2 of primary mvECs in a total volume of 100 μl onto the basolateral surface of the SIS (former serosa). 3 hr later fill the well with medium to ensure a submersed culture.
- Allow endothelial cells to adhere for 3 days. Flip the static culture system by 180° and transfer it to a 12-well plate.
- Seed a mixture of primary dermal fibroblasts (8,000 cells/cm2) and tumor cells (15,000 cells/cm2) within a total volume of 500 μl on the apical surface of the SIS (the side of the former lumen).
- Allow cells to adhere for 3 hr and fill the well with medium (submersed culture, medium: 50% Vasculife + 50% DMEM supplemented with 10% FCS).
- The tumor test system is cultured under static conditions at 37 °C, 5% CO2 in the incubator for additional 14 days. Change culture medium every 2 - 3 days.
For the dynamic culture, fix the SIS-Muc between two metal rings and seed primary mvECs as described in 3.2.1. After 3 days remove the SIS-Muc from the metal rings and insert the membrane into the flow reactor (see Figures 2C and 2D). Apply the primary dermal fibroblasts and tumor cells with a syringe and cannula onto the matrix in the bioreactor, and allow the cells to adhere for 3 hr before filling the bioreactor system with culture medium.
On the following day dynamic culture conditions with constant medium flow (3.8 ml/min, 37 °C, and 5% CO2) can be initiated. The dynamic culture is maintained for 14 days, culture medium is changed after 7 days.
The dynamic test setup is cultured in a self-constructed incubator that provides media flow through a pump and the necessary temperature and CO2 content.
4. Characterization Methods for Analysis
4.1 Fixing and paraffin-embedding of the seeded collagenous matrix
For (immuno-) histological characterizations remove the culture medium and fix the tissue with 4% paraformaldehyde for 2 hr.
Remove 4% paraformaldehyde and transfer the SIS from the metal insert to a tissue embedding cassette. Water the tissue to remove remaining fixative and dehydrate for paraffin infiltration.
Before embedding in a paraffin block, cut the SIS in 2 - 3 slices and place them in a paraffin-filled metal base mold so that the cut surfaces face downwards. Add the tissue cassette on top of the mold as a backing.
Cut 5 μm slices and float them on a 40 °C water bath for straightening, then mount slides onto suitable glass slides. Uncoated slides are used for H&E stains, polylysine-coated slides for immunohistological staining to improve attachment. Let slices dry thoroughly.
Melt paraffin, remove it with xylene and rehydrate slices for following staining.
4.2 Staining
The rehydrated slices can be stained with Hematoxylin/Eosin as a standardized overview staining.
For immunhistological staining, the fixed and paraffin-infiltrated tissue slices must undergo an antigen retrieval to allow antibodies to recognize and bind to their specific epitopes. Therefore, deparaffinized and rehydrated slides are placed in a steam cooker with heated citrate buffer (pH 6.0) for 20 min.
Transfer slides to washing buffer (0.5 M TBS buffer + 0.5% Tween) and circle slices with a PAP pen to minimize the required volume for staining solutions.
Place the slide in a moisture chamber. To secure a specific horseradish peroxidase-mediated visualization of antigen-antibody-binding, the endogenous peroxidase must be saturated with 3% hydrogen peroxide.
Primary antibody dilutions are applied to the slices, incubated for 1 hr at RT and carefully washed off with washing buffer.
For detection of specific antigen-antibody bindings the Ultra Vision Quanto Detection System HRP DAB (Thermo Scientific) is used according to the recommended protocol.
Nuclei are counterstained with Hematoxylin for 1 min.
Slides are mounted with an aqueous medium, dried, and imaged using an inverse microscope.
Representative Results
As shown in Figure 1B, we decellularized the porcine jejunal segment (about 2 m in length and 20 mm in diameter) with preserved tubular structures of the capillary network. After chemical, enzymatic and mechanical decellularization, we obtained a collagen I/III scaffold, which can be used for 3D cell culture. A Feulgen test was performed to demonstrate the purity (no DNA remnants) of the matrix (data not shown).
Figures 2A and 2B show the static culture of SIS-Muc secured by the cell crowns. We fixed the SIS-Muc in an in-house designed bioreactor (Figure 2C) for dynamic culture. Figure 2D illustrates the simulated dynamic flow through the chamber of the bioreactor. The bioreactor is placed in a self-constructed incubator system and connected with a peristaltic pump. This setup allows a dynamic culture with either pressure-regulated pulsatile flow or constant flow.
Figure 3 gives an overview of the statically cultured S462 tumor cell line in 2D monoculture (Figure 3A) and in 3D coculture (Figures 3B-3D). Figure 3B shows the triple culture of tumor cells S462 and primary fibroblasts on the apical side of SIS-Muc (former inner lumen side) and mvEC on the basolateral side (former serosa side). The identification of different cell types is possible by staining cell-type specific markers, such as von Willebrand factor to label mvEC (Figure 3C). The p53-positive S462 cells can be distinguished from the p53-negative primary fibroblasts (Figure 3D) and the 3D distribution of cells can be analyzed. Figure 4 shows stainings equivalent to Figure 3 of the dynamically cultured triple culture.
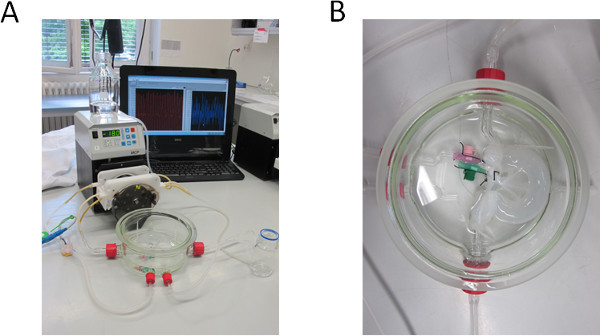 Figure 1. Decellularization set-up. (A) Bioreactor and pump set-up for decellularizing the BioVaSc, monitored by a PC. (B) Decellularized BioVaSc in glass tank. The lumen and the arterial inlet are connected to the adapters. .
Figure 1. Decellularization set-up. (A) Bioreactor and pump set-up for decellularizing the BioVaSc, monitored by a PC. (B) Decellularized BioVaSc in glass tank. The lumen and the arterial inlet are connected to the adapters. .
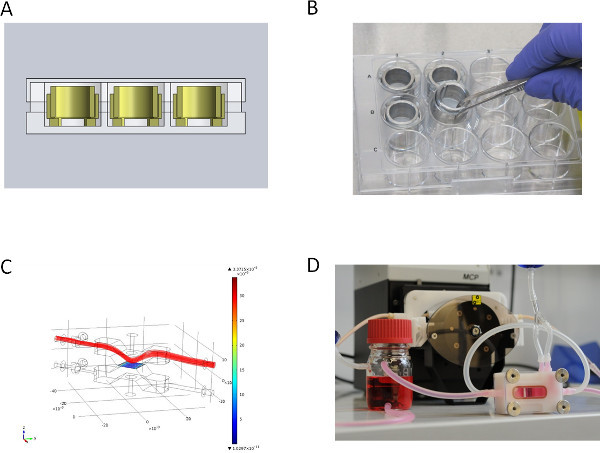 Figure 2. Over view of the different culture set-ups. (A) CAD section view of static culture system in Microtiter well plate, (B) metal inserts for static culture, (C) medium flow simulation (velocity field [m/sec]) for dynamic culture of the upper lid of the flow bioreactor, (D) flow bioreactor for dynamic culture connected to the peristaltic pump. Click here to view larger figure
Figure 2. Over view of the different culture set-ups. (A) CAD section view of static culture system in Microtiter well plate, (B) metal inserts for static culture, (C) medium flow simulation (velocity field [m/sec]) for dynamic culture of the upper lid of the flow bioreactor, (D) flow bioreactor for dynamic culture connected to the peristaltic pump. Click here to view larger figure
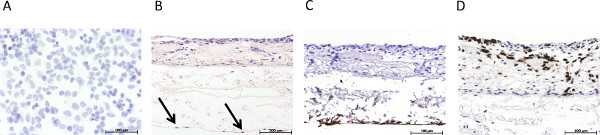 Figure 3. Overview of the immunohistological characterization of the static 3D tumor model. (A) Hematoxylin stain of the statically cultured 2D mono culture of S462 cells on a Permanox slide, (B) H&E stain of the statically cultured 3D triple culture, arrows mark endothelial cells, (C) immunohistological staining for von Willebrand factor, (D) immunohistological staining for p53. Click here to view larger figure
Figure 3. Overview of the immunohistological characterization of the static 3D tumor model. (A) Hematoxylin stain of the statically cultured 2D mono culture of S462 cells on a Permanox slide, (B) H&E stain of the statically cultured 3D triple culture, arrows mark endothelial cells, (C) immunohistological staining for von Willebrand factor, (D) immunohistological staining for p53. Click here to view larger figure
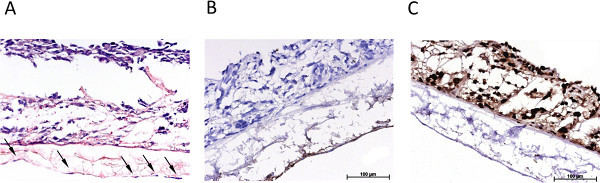 Figure 4. Overview of the immunohistological characterization of the dynamic 3D tumor model. (A) H&E stain of the dynamically cultured 3D triple culture, arrows mark endothelial cells, (B) immunohistological staining for von Willebrand factor, (C) immunohistological staining for p53. Click here to view larger figure
Figure 4. Overview of the immunohistological characterization of the dynamic 3D tumor model. (A) H&E stain of the dynamically cultured 3D triple culture, arrows mark endothelial cells, (B) immunohistological staining for von Willebrand factor, (C) immunohistological staining for p53. Click here to view larger figure
Discussion
When comparing 2D and 3D culture systems in tumor research, 3D systems, despite being the more expensive approach, have proven to mimic the conditions in biological microenvironments better. It could be shown that some tumor cells grow much slower in a 3D culture than in a common 2D culture 12, which is in accordance to the situation in a real tumor. Bissell and coworkers showed in their work that the behavior of carcinogenic breast cells reflects the in vivo situation, including cell morphology and signaling, more accurately when a 3D culture within a matrix offers cell-ECM interactions. Furthermore, they emphasized the importance of the extracellular environment in 3D by demonstrating that changes in the environmental interactions led to the reversion of the malignant cells to a normal phenotype. Additionally and most importantly, these outcomes could also be confirmed in in vivo animal models 10,11.
The direct comparison of in vivo animal experiments and in vitro tissue models reveals advantages and drawbacks in both systems. One advantage of in vitro models is the permission of a much better real-time or fixed imaging by microscopy. A limitation is that they mimic static or short-term conditions, whereas in vivo systems often progress. The current lack of vasculature and normal transport of small molecules, host immune responses, and other cell-cell interactions are further disadvantages of in vitro models 12. Therefore, 3D in vitro systems as presented in this study offer a promising addition to animal experiments. They provide a better comparability to the human organism and therefore minimize experimental misinterpretations. Biomimetic in vivo model systems will hence become more relevant to study how cancer and metastatic spread is dependent on microenvironmental conditions that regulate tumorigenesis 11.
Our study shows that the 3D environment provided by the SIS-Muc leads to a more tumor-like tissue formation of cells, which is not observed in the common 2D cell culture (see Figure 3A). Moreover, the use of primary cells derived from tumor biopsies is a very important step towards personalized medicine, a discipline that aims at identifying the best treatment depending on a patient's individual needs. Incorporating primary patient-specific tumor cells isolated from biopsy material will allow in vitro testing of therapeutic strategies. Such test systems will make it possible to investigate different drugs and combinations thereof in a time- and cost-saving high-throughput screening. Additionally, the integration of tumor-associated stromal cells as shown in this study is important for the personalized approach, since a tumor's microenvironment influences tumor progression 13 and might prove as suitable therapeutic target.
Alternatively to a personalized approach, our tumor model can be modified to serve as a generalized tumor test system by the incorporation of established tumorigenic cell lines. This is a promising adaptation for basic research purposes. For both drug testing approaches the presence of a vascular structure is required to test the distribution and uptake of therapeutic substances. The SIS-Muc matrix allows the basolateral seeding with primary mvEC for barrier uptake studies, the reseeding of the preserved vascular structures of the BioVaSc will further improve the study of drug delivery.
In order to create tissue models, a 3D biodegradable matrix can be used as framework for a co-culture of different cell types 14. The use of such 3D matrices is often limited by the absence of a functional vascularization. This problem can be solved by the use of the BioVaSc, which offers preserved blood vessel structures, which can be reseeded with endothelial cells. Furthermore, the BioVaSc provides extracellular components, which ensure the adhesion of the cells and facilitate tissue differentiation. It also enables the long-time tissue specific function of bioartificial 3D tissues 7,8,15. The prerequisite for the engineering of functional vascular substitutes is the mimicking of human physiological and biomechanical conditions. Therefore, bioreactor systems, which can implement these requirements in vitro, are of extreme interest for creating biological tumor models.
The combination of the BioVaSc, the bioreactor technology and co-culturing of different cell-types is a very promising method to generate vascularized tumor tissues, which will allow the study of mechanisms relevant for cancer progression such as angiogenesis and metastasis. We see such tumor models as a promising approach for complementing animal studies by providing an equivalent to the human tumor physiology.
Disclosures
Authors have nothing to disclose.
Acknowledgments
The authors would like to thank Jan Hansmann (Fraunhofer IGB, Stuttgart) for his technical support to develop bioreactors and the bioreactor incubator.
References
- Schenke-Layland K, Nerem RM. In vitro human tissue models--moving towards personalized regenerative medicine. Adv. Drug Deliv. Rev. 2011;63:195–196. doi: 10.1016/j.addr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Pusch J, Votteler M, et al. The physiological performance of a three-dimensional model that mimics the microenvironment of the small intestine. Biomaterials. 2011;32:7469–7478. doi: 10.1016/j.biomaterials.2011.06.035. [DOI] [PubMed] [Google Scholar]
- Schanz J, Pusch J, Hansmann J, Walles H. Vascularised human tissue models: a new approach for the refinement of biomedical research. J. Biotechnol. 2010;148:56–63. doi: 10.1016/j.jbiotec.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Lanza R, Langer R, Vacanti J. Principles of tissue engineering. 3rd. Burlington, MA: Elsevier Academic Press; 2007. [Google Scholar]
- Barron V, Lyons E, Stenson-Cox C, McHugh PE, Pandit A. Bioreactors for cardiovascular cell and tissue growth: a review. Ann. Biomed. Eng. 2003;31:1017–1030. doi: 10.1114/1.1603260. [DOI] [PubMed] [Google Scholar]
- Martin I, Wendt D, Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004;22:80–86. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Mertsching H, Walles T, Hofmann M, Schanz J, Knapp WH. Engineering of a vascularized scaffold for artificial tissue and organ generation. Biomaterials. 2005;26:6610–6617. doi: 10.1016/j.biomaterials.2005.04.048. [DOI] [PubMed] [Google Scholar]
- Linke K, Schanz J, Hansmann J, Walles T, Brunner H, Mertsching H. Engineered liver-like tissue on a capillarized matrix for applied research. Tissue Eng. 2007;13:2699–2707. doi: 10.1089/ten.2006.0388. [DOI] [PubMed] [Google Scholar]
- Holtkamp N, Atallah I, et al. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia. 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J. Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutmacher DW, Horch RE, et al. Translating tissue engineering technology platforms into cancer research. J. Cell. Mol. Med. 2009;13:1417–1427. doi: 10.1111/j.1582-4934.2009.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Yang S-T, Zhang X, Wen Y. Microbioreactors for high-throughput cytotoxicity assays. Curr. Opin. Drug Discov. Devel. 2008;11:111–127. [PubMed] [Google Scholar]
- Schultheiss D, Gabouev AI, et al. Biological vascularized matrix for bladder tissue engineering: matrix preparation, reseeding technique and short-term implantation in a porcine model. J. Urol. 2005;173:276–280. doi: 10.1097/01.ju.0000145882.80339.18. [DOI] [PubMed] [Google Scholar]


