Since Anfinsen (1) first showed that an amino acid sequence defines a structure, physical biochemists have been trying to decode the structural message in the sequence. To accomplish this goal, proteins have been unfolded and refolded under myriad conditions, with increasingly sophisticated folding probes, and at ever finer temporal resolution. However, this work has been largely confined to water-soluble proteins. For membrane proteins, many more hurdles exist and, to a large extent, we are still trying to devise methods to study folding. A protein chemist entering the membrane protein field would not even find a consensus method for measuring something so fundamental as thermodynamic stability. Folding experiments with membrane proteins are difficult because it can be hard to find ways to unfold them reversibly. Ideally, we would like a method to study the thermodynamics of membrane protein folding, in the context of a true bilayer. Just such a method has now been developed by Hong and Tamm (2) and is reported in this issue of PNAS.
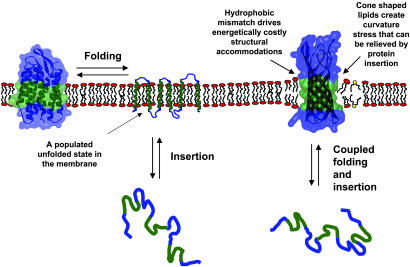
Two distinct classes of membrane proteins have been observed to date: β-barrels and α-helical bundles. Both architectures accommodate the powerful drive to satisfy hydrogen bonds in the apolar core of the bilayer (3). For α-helices, the backbone hydrogen bonds are satisfied locally, so an α-helix can be independently stable within the bilayer (4). Thus, it is reasonable to suppose that helical membrane proteins can lose tertiary structure without concomitant loss of secondary structure (4). We can envision a picture (clearly overly simplistic) of a partially unfolded membrane protein consisting of tethered helices floating around in the bilayer (see Fig. 1). The picture for β-barrel protein folding may be fundamentally different. In a β-sheet, backbone hydrogen bonds are long-range interactions, and for a linear sheet, both ends will have exposed hydrogen bond donors and acceptors. A simple way to satisfy the hydrogen-bonding potential in a sheet is to wrap it up into a barrel, thereby joining and closing off the exposed ends. Thus, it is hard to envision a highly unfolded β-barrel protein within the context of a bilayer. Instead, unfolding and extrusion from the bilayer are likely to be coupled (Fig. 1), leaving the unfolded state in the aqueous phase, or associated with the interfacial region of the bilayer (5). Indeed, kinetic studies suggest that insertion and sheet formation occur simultaneously (6). Because α-helical and β-barrel membrane proteins unfold differently, we need distinct techniques for studying them.
Fig. 1.
Membrane protein unfolding. For helical proteins, shown on the left, tertiary structure can be lost within the membrane environment, leaving stable transmembrane helices. Thus, the insertion of helices and folding (helix packing) can be separated. For β-barrel proteins, secondary structure should not be independently stable in the membrane. Thus, folding and insertion are likely to be coupled. The free energy of this folding and insertion equilibrium has now been measured by Hong and Tamm (2). They were able to study the effects of bilayer properties on this equilibrium. A mismatch between the hydrophobic width of the protein (green region) and the bilayer can induce distortions in either the protein or the bilayer. Cone-shaped lipids (yellow head group) lead to increased packing pressure in the center of the bilayer and decreased packing pressure at the head groups, which can be alleviated by the insertion of an hourglass-shaped protein.
Efforts to measure thermodynamic stability have largely focused on helical membrane proteins. Some of the best and most easily interpretable data have come from measurements of transmembrane helix dissociation constants by using equilibrium sedimentation (7–13). A limitation of this technique is that the measurements must be performed in detergent solution, although there is some evidence that the results in micelles should at least correlate well with the results in a bilayer (9). A promising new technique for obtaining transmembrane helix dissociation constants within a bilayer has been developed by measuring thiol–disulfide interchange (14). These methods are not applicable to large, multipass membrane proteins, however. Chemical denaturant approaches have been developed for larger membrane proteins. Starting with the folded protein in a nondenaturing detergent, the equilibrium is driven in favor of the unfolded state by adding increasing concentrations of urea (15), guanidine (16), or SDS (17–20). The disadvantage of all these methods is that the protein is not retained in a bilayer environment. For helical proteins, however, the SDS method may provide a reasonable mimic of unfolding in the bilayer, because helical secondary structure is largely maintained in SDS.
For β-barrel proteins, insertion into the interfacial region has been examined by using small-model peptides (21), but studying the thermodynamics of the insertion/folding of a large hydrophobic membrane protein seems like a tall order. Nevertheless, Hong and Tamm (2) now show how these measurements can be performed. They start with the protein in small unilammelar vesicles and then drive the extrusion of the protein with increasing concentrations of urea. Urea does not disrupt the membrane itself, but helps to solubilize the hydrophobic membrane protein in water. The extrusion is fully reversible, and for average bilayer thicknesses, fits a two-state model. Data obtained from the unfolding transition zone can then be extrapolated back to zero urea to obtain an unfolding free energy in the absence of denaturant. The beauty of the method is that it allows thermodynamic measurements of insertion/folding into a natural bilayer environment. Thus, it is possible to vary the bilayer itself so that the effect of bilayer properties on the energetics of insertion/folding can be measured.
Membranes have variable compositions, and different membranes can have different effects on the structure and stability of a protein (22, 23). One characteristic of membranes that Hong and Tamm (2) consider is the thickness of the apolar portion (see Fig. 1). If the width of the hydrocarbon region is not the same as the width of the apolar region of the protein, a mismatch occurs. The drive to shield the exposed hydrophobic surface from water creates a stress on either the protein and/or the bilayer. Another important characteristic of bilayers, considered in their paper, is lateral packing pressure. The presence of cone-shaped or nonbilayer lipids can cause an increased packing force in the center of the bilayer and a reduced force near the lipid headgroups (see Fig. 1). Both hydrophobic mismatch and lateral packing pressure are known to have an impact on protein structure and the rate of protein folding (24–26), but the effect on protein stability is hard to quantify. Hong and Tamm (2) use their new technique to evaluate the contribution of bilayer characteristics to protein folding.
When they used saturated lipids and varied the length of the hydrocarbon tails, Hong and Tamm (2) found that unfolding free energy increased with increased hydrocarbon chain length. In other words, insertion and folding became more favorable with longer lipids. This behavior makes sense because more hydrophobic surface can be shielded from water after insertion, thereby increasing the contribution of the hydrophobic effect to insertion/folding. Interestingly, however, the magnitude of the free-energy increase with increasing lipid length is far below what would be expected from the hydrophobic effect alone (approximately six times lower). Presumably, the missing energy was consumed in protein or lipid adjustments driven by the hydrophobic mismatch. To maximize surface burial, either the protein or the lipids need to compress or stretch. From their free-energy measurements as a function of lipid length, Hong and Tamm (2) provide an estimate of the stress energy; ≈1.7 kcal per mol per Å.
Urea denaturation allows thermodynamic measurement of protein insertion into the lipid bilayer.
The opposite trend was observed when cone-shaped lipids were used. In this case, they found a decrease in stability as the lipid length increased. This effect can be attributed to the decreased curvature stress or lateral packing pressure exerted by the longer lipids (27). OmpA is shaped like an hour glass: thinner in the middle. Thus, insertion of the OmpA protein into a bilayer should reduce the curvature stress imposed by cone-shaped lipids. Longer lipids have reduced curvature stress and are therefore less effective in stabilizing OmpA.
The new work opens another door to a more quantitative description of the energetics protein–protein and protein–lipid interactions in the bilayer. It remains to be seen how generally applicable this method will be, but other membrane proteins can be refolded from a urea denatured state (5, 28), so there is reason to be optimistic. Because of methods like the one introduced by Hong and Tamm (2), we are finally beginning to obtain quantitative information about membrane protein structure (8, 9, 12, 13, 19, 29, 30). Clearly, membrane protein folders have many reasons to see a bright future.
See companion article on page 4065.
References
- 1.Anfinsen, C. B. (1973) Science 181, 223–230. [DOI] [PubMed] [Google Scholar]
- 2.Hong, H. & Tamm, L. K. (2004) Proc. Natl. Acad. Sci. USA 101, 4065–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White, S. H., Ladokhin, A. S., Jayasinghe, S. & Hristova, K. (2001) J. Biol. Chem. 276, 32395–32398. [DOI] [PubMed] [Google Scholar]
- 4.Popot, J. & Engelman, D. (1990) Biochemistry 29, 4031–4037. [DOI] [PubMed] [Google Scholar]
- 5.Tamm, L. K., Arora, A. & Kleinschmidt, J. H. (2001) J. Biol. Chem. 276, 32399–32402. [DOI] [PubMed] [Google Scholar]
- 6.Kleinschmidt, J. H., den Blaauwen, T., Driessen, A. J. & Tamm, L. K. (1999) Biochemistry 38, 5006–5016. [DOI] [PubMed] [Google Scholar]
- 7.Noy, D., Calhoun, J. R. & Lear, J. D. (2003) Anal. Biochem. 320, 185–192. [DOI] [PubMed] [Google Scholar]
- 8.Fleming, K. G., Ackerman, A. L. & Engelman, D. M. (1997) J. Mol. Biol. 272, 266–275. [DOI] [PubMed] [Google Scholar]
- 9.Fleming, K. G. & Engelman, D. M. (2001) Proc. Natl. Acad. Sci. USA 98, 14340–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salom, D., Hill, B. R., Lear, J. D. & DeGrado, W. F. (2000) Biochemistry 39, 14160–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, R., Mitra, N., Gratkowski, H., Vilaire, G., Litvinov, R., Nagasami, C., Weisel, J. W., Lear, J. D., DeGrado, W. F. & Bennett, J. S. (2003) Science 300, 795–798. [DOI] [PubMed] [Google Scholar]
- 12.Lear, J. D., Gratkowski, H., Adamian, L., Liang, J. & DeGrado, W. F. (2003) Biochemistry 42, 6400–6407. [DOI] [PubMed] [Google Scholar]
- 13.Gratkowski, H., Lear, J. D. & DeGrado, W. F. (2001) Proc. Natl. Acad. Sci. USA 98, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cristian, L., Lear, J. D. & DeGrado, W. F. (2003) Proc. Natl. Acad. Sci. USA 100, 14772–14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klug, C., Su, W., Liu, J., Klebba, P. & Feix, J. (1995) Biochemistry 34, 14230–14236. [DOI] [PubMed] [Google Scholar]
- 16.Klug, C. S. & Feix, J. B. (1998) Protein Sci. 7, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau, F. & Bowie, J. U. (1997) Biochemistry 36, 5884–5892. [DOI] [PubMed] [Google Scholar]
- 18.Chen, G. Q. & Gouaux, E. (1999) Biochemistry 38, 15380–15387. [DOI] [PubMed] [Google Scholar]
- 19.Faham, S., Yang, D., Bare, E., Yohannan, S., Whitelegge, J. P. & Bowie, J. U. (2004) J. Mol. Biol. 335, 297–305. [DOI] [PubMed] [Google Scholar]
- 20.Otzen, D. E. (2003) J. Mol. Biol. 330, 641–649. [DOI] [PubMed] [Google Scholar]
- 21.Wimley, W. C., Hristova, K., Ladokhin, A. S., Silvestro, L., Axelsen, P. H. & White, S. H. (1998) J. Mol. Biol. 277, 1091–1110. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain, A. K., Faham, S., Yohannan, S. & Bowie, J. U. (2003) Adv. Protein Chem. 63, 19–46. [DOI] [PubMed] [Google Scholar]
- 23.Lee, A. G. (2003) Biochim. Biophys. Acta 1612, 1–40. [DOI] [PubMed] [Google Scholar]
- 24.Curran, A. R., Templer, R. H. & Booth, P. J. (1999) Biochemistry 38, 9328–9336. [DOI] [PubMed] [Google Scholar]
- 25.Perozo, E., Kloda, A., Cortes, D. M. & Martinac, B. (2002) Nat. Struct. Biol. 9, 696–703. [DOI] [PubMed] [Google Scholar]
- 26.Williamson, I. M., Alvis, S. J., East, J. M. & Lee, A. G. (2003) Cell. Mol. Life Sci. 60, 1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate, M. W. & Gruner, S. M. (1987) Biochemistry 26, 231–236. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, J. K., Lonzer, W. L. & Sanders, C. R. (2001) Biochemistry 40, 8971–8980. [DOI] [PubMed] [Google Scholar]
- 29.DeGrado, W. F., Gratkowski, H. & Lear, J. D. (2003) Protein Sci. 12, 647–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard, K. P., Lear, J. D. & DeGrado, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 8568–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]