Abstract
The molecular analysis of insect hormone biosynthesis has long been hampered by the minute size of the endocrine glands producing them. Expressed sequence tags from the corpora allata of the cockroach Diploptera punctata yielded a new cytochrome P450, CYP15A1. Its full-length cDNA encoded a 493-aa protein that has only 34% amino acid identity with CYP4C7, a terpenoid ω-hydroxylase previously cloned from this tissue. Heterologous expression of the cDNA in Escherichia coli produced >300 nmol of CYP15A1 per liter of culture. After purification, its catalytic activity was reconstituted by using phospholipids and house fly P450 reductase. CYP15A1 metabolizes methyl (2E,6E)-3,7,11-trimethyl-2,6-dodecatrienoate (methyl farnesoate) to methyl (2E,6E)-(10R)-10,11-epoxy-3,7,11-trimethyl-2,6-dodecadienoate [juvenile hormone III, JH III] with a turnover of 3–5 nmol/min/nmol P450. The enzyme produces JH III with a ratio of ≈98:2 in favor of the natural (10R)-epoxide enantiomer. This result is in contrast to other insect P450s, such as CYP6A1, that epoxidize methyl farnesoate with lower regio- and stereoselectivity. RT-PCR experiments show that the CYP15A1 gene is expressed selectively in the corpora allata of D. punctata, at the time of maximal JH production by the glands. We thus report the cloning and functional expression of a gene involved in an insect-specific step of juvenile hormone biosynthesis. Heterologously expressed CYP15A1 from D. punctata or its ortholog from economically important species may be useful in the design and screening of selective insect control agents.
The juvenile hormones (JHs) control insect development, metamorphosis, reproduction, and many other physiological functions (1). These sesquiterpenoids are biosynthesized de novo in specialized endocrine glands, the corpora allata (CA) (2). The early steps of JH biosynthesis follow the mevalonate pathway from acetyl-CoA to farnesyl pyrophosphate. The CA of Lepidoptera also generate and use propionyl-CoA in a parallel homomevalonate pathway to yield rare examples of natural homosesquiterpenoids (2). The steps that follow the elaboration of the sesquiterpenoid skeleton seem to be specific to the JH pathway and are still poorly understood. Studies with cell-free extracts or with isolated CA in vitro indicate that the biosynthesis proceeds from farnesyl pyrophosphate by conversion to farnesol, oxidation to farnesoic acid, esterification, and epoxidation (2). The minute size of the CA has been a major obstacle to classical biochemical approaches such as enzyme purification or even the preparation of subcellular membrane fractions (2, 3). Molecular cloning of the enzymes involved in the early steps of JH biosynthesis has taken advantage of the conservation of the mevalonate pathway between vertebrates and insects (4), but this option is not available for the later, JH-specific steps.
Esterification of farnesoic acid by an S-adenosylmethionine-dependent transferase is thought to occur before epoxidation to JH III in the CA of locusts and cockroaches, but this may not be a general rule. In Lepidoptera, epoxidation may precede esterification, and the latter reaction is under strict developmental control (5). The O-methyl transferase activity disappears from the CA of Lepidoptera during metamorphosis but can be found later in some peripheral tissues, most notably in the accessory sex glands of adult male Cecropia moths (6). This peripheral O-methyl transferase is highly specific, recognizing the correct configuration of the epoxide at the distal end of the molecule, and favoring methylation of JH I acid over JH III acid (6). The farnesoic acid O-methyl transferase has recently been cloned from the CA of the silkworm Bombyx mori by mRNA differential display (7).
Less is known about the epoxidase, reported to be a microsomal cytochrome P450 enzyme in cockroach and locust CA (8, 9). Although cytochrome P450 enzymes are ubiquitous and share common catalytic mechanisms, they are the products of a large family of genes (10) that can be highly divergent in sequence. This divergence has allowed many P450 enzymes to assume specific physiological roles, for instance in steroid hormone biosynthesis in vertebrates, insects (molting hormones), and plants (brassinosteroids) (10). Effective and selective inhibitors of the epoxidase involved in JH biosynthesis would likely represent “biorational” insect control agents (11). Indeed, by disrupting the normal JH titer, such inhibitors would cause precocious metamorphosis in larval stages and would block reproduction in the adult stage of most insect species. Progress in the quest for such inhibitors requires the molecular identification of the epoxidase.
We report here the cloning from the cockroach, Diploptera punctata, the heterologous expression and the characterization of a P450 (CYP15A1) that catalyzes the last step in JH biosynthesis, a highly stereoselective epoxidation reaction.
Materials and Methods
Insects and CA ESTs. D. punctata were reared as described (12). One hundred pairs of CA were dissected from adult mated females aged 3–5 days. The glands were briefly rinsed in saline and collected in lysis solution of the Poly(A)Pure mRNA isolation kit (Ambion, Austin, TX), which was used to prepare mRNA. cDNA was synthesized with the SuperScript plasmid system (Life Technologies, Grand Island, NY), and fractions 7 to 13 of the sizing column were directionally cloned with the appropriate linkers into the NotI/SalI site of the pSPORT I vector. The ligation product was transformed into MAX efficiency DH5α competent cells (BRL). Individual colonies were stored as glycerol stocks in 96-well plates, which were used as templates for plasmid preparation and purification (Qiagen miniprep kit, Qiagen, Valencia, CA). Insert sizes were monitored by electrophoresis on 1% agarose gels after digestion with BamHI, and ESTs sequenced on an Applied Biosystems 377 instrument.
Construction of the Vector for Expression in Escherichia coli. The coding region of a P450 cDNA obtained by screening the EST library (CYP15A1, see below) was cloned into the expression vector pCWori+ (13) after modification by PCR mutagenesis. The pCWori+ vector contains an NdeI site adjacent to the ATG starting codon. Using the forward primer 5′-GGA ATT CCA TAT GGC TAT TGC TCT GAT TGT CAT CAT CAT C-3′, the 5′ end of the CYP15A1 coding region was modified by PCR mutagenesis to introduce an NdeI site upstream of the ATG codon and to replace the second codon Val (GTC) by Ala (GCT) (13). Further PCR mutagenesis with the primer 5′-AGT GCT AAG TTG ATT CCA AGG AAA CAT CAT CAT CAT CAT CAT TAA AAG CTT GGG-3′ was used to add six histidine codons to the 3′ end of the coding region to allow enzyme purification by nickel-chelate chromatography and to introduce an HindIII site downstream of the stop codon. The purified PCR product was subcloned into the pCR2.1 vector (Invitrogen) and sequenced. The NdeI-HindIII fragment was cut out of the pCR2.1 vector and subcloned into the expression vector pCWori+.
Expression in E. coli and Purification of His-Tagged CYP15A1. DH5α E. coli cells were transformed with the pCWori+ vector containing the CYP15A1 insert in the presence of ampicillin. Cells were grown at 37°C for 2 hat 250 rpm in modified TB containing thiamin (1 mM), potassium phosphate buffer, pH 7.7 (100 mM), δ-aminolevulinic acid (1 mM), and a rare salt solution. Isopropyl β-d-thiogalactoside (IPTG, 1 mM) was then added, and the cells were grown at 28°C and 170 rpm. After 48 h, cells were harvested by centrifugation (2,800 × g, 5 min, 4°C). The resulting pellet was resuspended in the lysis buffer, 0.1 M Tris·HCl, pH 7.4, containing 10 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS, Sigma), 20% glycerol, and 1 mM EDTA and used to determine expression levels by CO-difference spectroscopy (14).
For the isolation of the membrane fraction, cells expressing CYP15A1 were collected by centrifugation, and the cell pellets were suspended in TSE buffer (50 mM Tris·acetate, pH 7.6, containing 250 mM sucrose and 0.25 mM EDTA). A solution of freshly prepared lysozyme was added (0.25 mg/ml, final concentration), and the mixture was then gently shaken at 4°C for 30–45 min. After centrifugation (2,800 × g for 12 min at 4°C), the resulting spheroplasts were suspended in 0.1 M sodium phosphate buffer, pH 7.4, containing 20% glycerol, 1 mM PMSF, 0.1 g/ml leupeptin, and 0.04 units/ml aprotinin (all from Sigma), and disrupted by sonication at 4°C. After a 10-min centrifugation at 4,000 × g, the supernatant was centrifuged at 100,000 × g for 1 h. The resulting membrane pellet was suspended in 0.1 M sodium phosphate buffer (pH 7.4) containing 20% glycerol and used for either protein purification or enzymatic characterization.
For protein purification, the membrane fractions were suspended in 100 mM sodium phosphate buffer (pH 7.5) containing 20% glycerol and 0.5 M NaCl and solubilized by the dropwise addition of 10% sodium cholate (wt/vol) and 10% CHAPS (wt/vol) to obtain a final concentration of 0.75% and 0.5%, respectively. This detergent combination was optimized for the amount of P450 solubilized, as measured by CO-difference spectroscopy. After a 12- to 16-h incubation at 4°C, insoluble proteins were removed by centrifugation (1 h, 100,000 × g), and the NaCl, sodium cholate, and CHAPS concentrations of the high-speed supernatant were adjusted to 0.25 M, 0.375%, and 0.25%, respectively. After addition of 5 mM imidazole, the resulting suspension was immediately loaded onto a nickel-nitrilotriacetic acid column (Qiagen) equilibrated with 0.1 M sodium phosphate buffer (pH 7.5) containing 300 mM NaCl, 5 mM imidazole, 20% glycerol, and 0.1% sodium cholate (buffer A). The column was washed with buffer A containing 25 mM imidazole. CYP15A1 was then eluted with 0.1 M Tris·HCl (pH 7.5) containing 300 mM NaCl, 20% glycerol, 200 mM imidazole, and 1% sodium cholate. Fractions of 1 ml were collected and measured for P450 contents. Fractions containing CYP15A1 were dialyzed against two changes of 0.1 M Tris·HCl (pH 7.4) containing 20% glycerol and 0.1% sodium cholate and concentrated in a centrifugal concentrator (Macrosep, Pall Filtron).
Enzymatic Activities and Product Characterization. CYP15A1 activity was assayed in a 0.1-M Tris·HCl buffer (pH 7.7) at 30°C under constant mixing in a final volume of 500 μl. Typical reaction mixture contained 100 μM substrate, NADPH-regenerating system (100 μM NADPH, 2.0 mM glucose-6-phosphate, and 4.0 units/ml glucose-6-phosphate dehydrogenase), 0.2 μM recombinant CYP15A1, and 2 μM house fly P450 reductase.
Before assay, CYP15A1 was incubated 15 min on ice with house fly P450 reductase (15) in the presence of 1 mg/ml l-α-dilauroyl-sn-glycero-3-phosphocholine in 0.1 M Tris·HCl (pH 7.7). The reaction was started by addition of 50 μl of the enzyme mixture, and, after 15–30 min, the reaction was stopped by addition of 2 ml of isooctane and extracted.
E. coli membranes containing expressed recombinant CYP15A1 were mixed with 2 μM house fly P450 reductase to a final volume of 50 μl and a P450 concentration of 0.2 μM. After 15 min on ice, the reaction was started by addition of the membrane mixture to 450 μl of reaction mixture containing 100 μM substrate and the NADPH-regenerating system in 0.1 M Tris·HCl buffer (pH 7.7). Conditions of incubation are identical to the above. The organic phase was concentrated, and the products of the reactions were analyzed by GC by using a Hewlett Packard 5890 series II gas chromatograph with a flame ionization detector and a Ultra1 column (12 m × 0.2 mm × 0.33 μm, Hewlett–Packard).
The identity of the mono-epoxide was verified by GC/MS, on a Varian Saturn GC/MS 2000 and the same column. The standard temperature program was 1 min hold at 100°C followed by an increase of 10°C/min to 250°C. Under our GC conditions, the retention time of the 10,11-epoxide of methyl farnesoate was 6.3 min.
Separation of JH III Enantiomers. We first purified the JH III isomers by reversed-phase HPLC on a 5-μm SupelCosil LC18-DB column (4.6 cm × 250 cm, Supelco) using a solvent mixture composed of 20% CH3CN, 80% H2O, which was isocratic for 5 min, followed by a linear gradient to 80% CH3CN, 20% H2O over 40 min (flow rate 1 ml·min–1; retention time ≈ 35 min for JH III). For separation of the optical isomers, products were injected onto a 50 × 4.6-mm chiralpak AS guard column (Chiral Technologies, Exton, PA) with hexane as a mobile phase and a flow rate of 0.4 ml·min–1. Products were monitored at 224 nm.
Time Course and Tissue Location of the CYP15A1 Transcripts. CA, brains, and fat body were collected from 3- to 7-day-old mated females. For the time course, we used CA from day 0 to day 10 and also days 15 and 20. Total RNA extractions were done on these tissues, by using the RNeasy Mini kit (Qiagen). RT-PCR was done for CYP15A1 and the controls, D. punctata CYP4C7 (AF071072), CYP9E1 (AY509245), and actin. Before RT-PCR, total RNA was treated with RNase-free DNase according to the supplier's instructions (Ambion). The RETROscript kit (Ambion) was used for first-strand cDNA synthesis with the oligo(dT)18 primer and mouse murine leukemia virus (M-MLV) reverse transcriptase. PCRs were performed with the following cycles: hot start 3 min at 95°C, addition of Taq polymerase (FisherBiotech) at 80°C, then 1 min at 95°C followed by 35 cycles of 30 s denaturation (95°C), 30 s annealing (58°C for CYP15A1, 49°C for CYP4C7, 45°C for CYP9E1, 52°C for actin), 1 min extension (72°C), and a final single extension cycle of 4 min at 72°C. Primers used were as follows: CYP15A1 forward primer, TAG GAT CCA TGG TCA TCG CTC TTA TTG TCA TCA TCA TCT TC; CYP15A1 reverse primer, AAT CTA GAT CAT TTC CTT GGA ATC AA; CYP4C7 forward primer, TGG AGC CTA TTT ATC AAC AT; CYP4C7 reverse primer, CTC GGT GTA GTA GGT ATG TG; CYP9E1 forward primer, TGT TTA CGT TGG TGT CTG AAT GTG; CYP9E1 reverse primer, GAT CAA TTT CTT TCT GGA GTC GTG; actin forward primer, GTG ACG AAG AAG TTG CTG; actin reverse primer, CGG TAA GAA GCA CAG GAT. Amplified RT-PCRs were then analyzed on 1% agarose gels.
Results
A Cytochrome P450 Sequence from CA ESTs. A cDNA library was prepared from the CA of mated adult females, at the time of increasing JH synthetic activity (days 3–5) to obtain molecular tools for the analysis of CA physiology. The 5′ end of 1,056 clones was sequenced to establish an EST library. Analysis of the sequences by blast yielded three overlapping sequences with best matches to known cytochromes P450. Sequencing revealed that one EST represented a full-length P450 ORF coding for a protein of 493 aa (GenBank accession no. AY509244). The sequence was significantly different from CYP4C7 that was previously obtained from the CA of this species (16) and was named CYP15A1 by D. R. Nelson (University of Tennessee, Memphis). It has all of the features of a typical microsomal P450 (10).
Functional Expression of CYP15A1 in E. coli. The CYP15A1 cDNA optimized for E. coli expression and purification was subcloned in pCWori+. E. coli cells were transformed, and CYP15A1 was detected in the cell lysate after an induction period of 48 h at 28°C by a characteristic reduced CO/reduced difference spectrum with a peak at 448 nm. The total P450 content was 300 ± 100 nmol P450 per liter of culture (mean ± SD of three preparations).
Recombinant CYP15A1 was present in both soluble (25%) and membrane-bound (75%) fractions. The protein was solubilized from the membrane fraction and purified on a nickel-nitrilotriacetic acid column. Analysis on SDS/PAGE showed a 55-kDa band corresponding to CYP15A1 (Fig. 1).
Fig. 1.
SDS/PAGE analysis of recombinant CYP15A1 produced in E. coli. (A) Membrane fraction. (B) CYP15A1 after purification on a nickel-nitrilotriacetic acid column. The migration distance of molecular weight (MW) markers (Bio-Rad) is indicated (MW × 10–3).
Spectral Characterization of CYP15A1. The absolute spectra of oxidized, reduced, and CO-bound reduced CYP15A1 protein were typical of P450 proteins. The absolute spectrum of oxidized recombinant CYP15A1 showed Soret, α, and β bands at 418, 571, and 537 nm, respectively. The presence of a shoulder at ≈392 nm indicated that the recombinant protein was isolated in a mixed spin state. Reduction with sodium dithionite resulted in the attenuation and displacement of Soret band maxima to 415 nm, and the replacement of the α and β bands for a broad absorption band centered at ≈546 nm. The addition of CO to a solution of reduced CYP15A1 led to the rapid development of a strong absorption band with a maximum at 448 nm and shoulders at 422 and 549 nm, indicative of the formation of the CO-bound complex (14) of reduced CYP15A1.
The difference spectrum of membrane-bound CYP15A1 elicited by the addition of methyl (2E,6E)-farnesoate was characterized by a peak at 389 nm and a through at 423 nm, features typical of a P450 type I interaction (17). Titration of the enzyme gave a spectral dissociation constant (Ks)of6 ± 1.5 μM (mean ± SD of three preparations), indicating that methyl (2E,6E)-farnesoate has a high affinity for CYP15A1. Precocene II did not give a type I spectrum with CYP15A1 even at high concentrations (>200 μM).
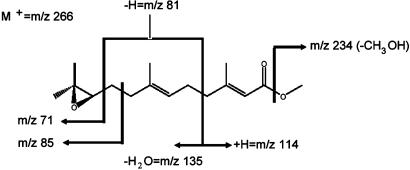
Metabolism of Methyl (2E,6E)-Farnesoate by CYP15A1. Methyl (2E,6E)-farnesoate was metabolized by CYP15A1 to a single product that had the same retention time as JH III on reversed-phase HPLC. The reaction did not require the presence of cytochrome b5. The HPLC-purified product was analyzed further by GC/MS. The mass spectrum of the product showed the characteristic ion fragments (Fig. 2) at m/z 71, 81, 85, 114, 135, and 234 of JH III. These results clearly demonstrated that CYP15A1 catalyzes the epoxidation of methyl farnesoate to JH III.
Fig. 2.
Conversion of methyl (2E,6E) farnesoate to JH III by CYP15A1. Molecular ion and fragmentation patterns observed by mass spectrometry are indicated.
When the purified CYP15A1 enzyme was incubated in the presence of different concentrations of methyl (2E,6E)-farnesoate, we measured a Km of 380 ± 70 μM and a Vmax of 16 ± 3 min–1. However, when E. coli membranes containing expressed recombinant CYP15A1 were tested, the value for the Km was ≈20-fold lower (26 ± 1.5 μM), and the Vmax was slower (2 ± 0.2 min–1; means ± SD of three determinations with six concentrations). These differences are most likely due to the differences in P450 reductase interaction with the P450 in the two different reconstitution assays and/or to the lipophilic nature of the substrate. The critical micelle concentration of methyl farnesoate is ≈10 μM (18), favoring rapid partition into the membrane lipid bilayer, and this lipophilicity has hampered kinetic determinations before (9). Although we have not pursued an optimization of the reconstitution conditions in this study, the values obtained for the enzyme's velocity and affinity for its substrate are consistent with the physiological function of CYP15A1.
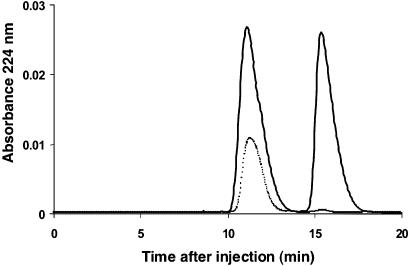
Stereospecificity of the 10,11-Epoxidation Catalyzed by CYP15A1. A comparison of the reaction product of CYP15A1 with optically pure JH III (from CA incubations in vitro and from the plant Cyperus iria; ref. 19) and with racemic, synthetic methyl (2E,6E)-10,11-epoxy-farnesoate allowed us to determine the absolute configuration of the 10,11-epoxide. The (10R)-epoxide produced a single peak by chiral HPLC (Fig. 3), coincident with the faster eluting peak of the racemic material (retention time ≈ 11.2 min). The epoxide of the reaction product from CYP15A1 contained a ratio of ≈98:2 in large favor of the (10R)-epoxide, with only traces of the (10S) enantiomer produced. CYP15A1 therefore converts methyl (2E,6E)-farnesoate to the natural JH III (10R) with a very high degree of specificity.
Fig. 3.
Chiral HPLC elution profile of the CYP15A1 metabolite (dotted line) and of the racemic synthetic methyl (2E,6E)-10,11 epoxy-farnesoate (solid line). The 10R enantiomer is the faster eluting compound.
Substrate Specificity of CYP15A1. Different geometrical isomers of methyl farnesoate (Table 1) were incubated in the presence of purified CYP15A1 to study the specificity of the enzyme. Epoxidation was observed only when the C2,3 double bond of methyl farnesoate was of the trans configuration. However, the best substrate for CYP15A1 was the natural precursor of JH III, methyl (2E,6E)-farnesoate.
Table 1. Conversion of methyl farnesoate isomers to epoxides by CYP15A1.
| Substrate | Epoxide formation* |
|---|---|
| Methyl (2E,6E)-farnesoate | 2.5 ± 0.5 |
| Methyl (2Z,6E)-farnesoate | ND† |
| Methyl (2E,6Z)-farnesoate | 0.3 ± 0.1 |
| Methyl (2Z,6Z)-farnesoate | ND† |
Epoxidation is expressed in nmol of product per min/nmol of recombinant CYP15A1 (mean ± SD, n = 3). The epoxidation of the different isomers of methyl farnesoate was assayed as described in Materials and Methods.
ND, not detectable.
Other terpenoids, including farnesol, farnesal, farnesoic acid, JH III, 3,7,11-trimethyl-dodecanol, geranyl geraniol, farnesyl methyl ether, and geraniol and the fatty acids, lauric acid, and arachidonic acid were tested as substrates. None of these compounds were metabolized by CYP15A1 in our reconstituted system. The synthetic analog of JH III, methoprene, and precocene II were not metabolized by the enzyme, and preincubation of CYP15A1 with precocene II did not lead to any subsequent inhibition of methyl farnesoate epoxidation.
Inhibition by 1,5-Disubstituted Imidazoles. The ability of three substituted imidazole inhibitors of JH biosynthesis to inhibit CYP15A1 activity was examined. These imidazole compounds were chosen to cover a broad range of potency in the in vitro assay for JH biosynthesis (Table 2). The two most potent compounds are also known to cause massive accumulation of methyl farnesoate in the glands whereas the least potent does not (12, 20). We found that these compounds were also effective inhibitors of purified CYP15A1 in the conversion of methyl (2E,6E)-farnesoate to JH III. Moreover, the rank order for inhibition in both assays was identical for the three compounds, and there was a good correspondence of the potencies in the two assays.
Table 2. Imidazole inhibition of JH III production by CA in vitro and by recombinant CYP15A1.
| Imidazole | IC50 CA in vitro, μM* | IC50 CYP15A1, μM† | |
|---|---|---|---|
| TH27 |

|
0.036 | 0.025 ± 0.014 |
| KK96 | 0.77 | 0.2 ± 0.09 | |
| KK71 | 48 | 60 ± 5 |
Concentration for 50% inhibition of JH III biosynthesis by CA in vitro. The IC50 values were taken from ref. 12 (KK96 and TH27) and ref. 20 (KK71).
Concentration for 50% inhibition of methyl (2E,6E)-farnesoate epoxidation by recombinant CYP15A1 in a reconstituted system (mean ± SD, n = 3). Epoxidation of methyl farnesoate by E. coli membranes containing recombinant CYP15A1 was measured as described in Materials and Methods.
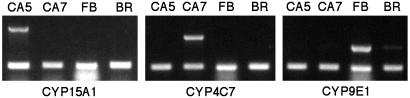
Expression of CYP15A1 in the Cockroach: Tissue and Developmental Specificity. We used RT-PCR to analyze the tissue specificity of CYP15A1 expression. CYP15A1 transcripts were detected only in the CA of day 5-mated females, and not in the brain or fat body (Fig. 4). CYP4C7 transcript was detected in the CA of day 7-mated females as expected (16). In contrast, another cytochrome P450 transcript, CYP9E1, was massively expressed in the fat body, with traces in the brain, but not in the CA, whereas actin was detected in all tissues examined. We then examined the changes in CYP15A1 transcript levels during adult development. Highest CYP15A1 transcript levels were observed on days 3–6, at the time of maximal biosynthetic activity of the glands, whereas the message was barely detectable on days 10, 15, and 20, after oviposition and during pregnancy, when JH synthesis is low.
Fig. 4.
Comparative expression in the CA (day 5, CA5; day 7, CA7), fat body (FB), and brain (BR) of CYP15A1 (1,490 bp), CYP4C7 (781 bp), and CYP9E1 (645 bp) by RT-PCR. The lower migrating band (301 bp) is an amplicon of actin serving as control.
Discussion
CYP15A1 Is the Methyl Farnesoate Epoxidase. This study reports the cloning and functional expression of a previously uncharacterized cytochrome P450, CYP15A1. Based on the following criteria, we propose that this enzyme is the epoxidase of the JH biosynthetic pathway.
First, CYP15A1 is a microsomal P450. The sequence of CYP15A1 is clearly homologous to microsomal cytochromes P450, with a 19-aa hydrophobic N-terminal anchor preceding a single charged residue and a typical “PPGP” hinge (10). Its catalytic activity is supported by NADPH and house fly NADPH cytochrome P450 reductase. The microsomal P450 nature of the enzyme had been demonstrated in Blaberus giganteus and Locusta migratoria (8, 9).
Second, CYP15A1 catalyzes the stereoselective epoxidation of methyl farnesoate. Chiral HPLC separation of the 10R and 10S enantiomers of JH III conclusively showed that CYP15A1 has a very high degree of selectivity in the epoxidation of methyl farnesoate, producing the natural 10R enantiomer. The absolute configuration is known for the JH III produced by the CA of Manduca sexta and Tenebrio molitor (21–23), and for the JH III produced enzymatically with L. migratoria and B. giganteus CA microsomes (9, 18).
Third, CYP15A1 is selectively expressed in the CA. Other enzymes are certainly capable of epoxidation of methyl farnesoate, e.g., house fly CYP6A1 (24), which is expressed in the gut and fat body (25), and D. punctata CYP9E1, which is highly expressed in the fat body (J. F. Andersen, G.C.U., J.F.K., and R.F., unpublished results). Male accessory glands and ovaries of mosquitoes apparently also express an epoxidase (26, 27). CYP15A1 is expressed in the CA, and its transcript was not detected by RT-PCR in either brain or fat body. Although precise quantitation is not possible by RT-PCR, the results suggest that expression is higher at times of active JH III synthesis.
Fourth, the enzyme is highly specific. In addition to the enantioselectivity of epoxidation, we have also shown a very high degree of substrate selectivity. Of the geometrical isomers of methyl farnesoate, only 10% of the activity was retained with the 2E,6Z isomer. We found no reactivity with the 2Z,6E isomer, reported to be actively epoxidized by cockroach CA (18, 28). We could not detect epoxidation of farnesoic acid, which strongly suggests that esterification takes place before epoxidation in the biosynthetic pathway of JH III in cockroaches, as also inferred by the accumulation of methyl farnesoate in glands treated with P450 inhibitors (12, 20). The specificity of the epoxidase contrasts markedly from the specificity of the O-methyl transferase of locusts, which is very loose and readily accommodates the epoxy acid as substrate (29). It also contrasts significantly from the specificity of CYP4C7, an ω-hydroxylase that prefers sesquiterpenoid substrates but is not highly specific for the distal end of the molecule (16).
In our hands, precocene II was not metabolized by the recombinant CYP15A1 and did not inhibit its activity toward methyl farnesoate. Precocenes are anti-juvenile hormones (30) described as a pro-allatocidins (31, 32) acting directly on the CA of sensitive insects to cause anti-JH effects by causing necrosis of the glands. Their mode of action involves metabolic activation within the glands by a cytochrome P450. One potential candidate was methyl farnesoate epoxidase, but our study surprisingly does not support this hypothesis. It seemed logical 20 years ago to assume that the enzyme responsible for the bioactivation of precocenes was methyl farnesoate epoxidase because inhibition and destruction of the CA could be observed in vitro, the effects of precocenes were sensitive to P450 inhibitors, and the allatal epoxidation of precocene I was shown to be stereospecific (32–35). However, inhibition of JH production by isolated glands occurs only at very high precocene concentrations (33, 35), and precocene sensitivity is independent of the intrinsic activity of the glands (33, 36). Also, dihydroprecocene II (which cannot be bioactivated by epoxidation) is only marginally less potent than precocene II in inhibiting JH III production by Blattella germanica CA in vitro (37). Taken together, these results suggest either that we have been unable to reproduce with the recombinant enzyme all of the properties of the enzyme, or that the epoxidation of precocenes is carried out by a P450 enzyme other than CYP15A1.
Fifth, inhibitors of JH III synthesis inhibit CYP15A1 activity. We have explored substituted imidazoles as inhibitors of JH III synthesis by isolated CA of D. punctata (12, 20), and the definition of structure-activity relationships allowed the design of potent photoaffinity labels of the epoxidase in situ (28). Here, we show that the rank order of inhibition by compounds of this type is respected over a 1,000-fold range of potency. The pharmacological profile of the functionally expressed recombinant enzyme is therefore the same as that of the native epoxidase from the CA because the rank order reflects the relative degree of specificity of the inhibitors. Interestingly, the behavior of the purified recombinant enzyme on SDS/PAGE is identical (55 kDa) to that of the native enzyme labeled by the bifunctional imidazole compound BP-IceT (28).
Evolution of the CYP15 Gene. Two insect genomes have been fully sequenced, and we searched them for sequences orthologous to CYP15A1. It is difficult to assign an orthology relationship from a single cockroach sequence to one of the many P450 sequences of Drosophila melanogaster and of Anopheles gambiae because such relationships are best established by comparing all P450 sequences from each organism to each other. When the CYP15A1 peptide sequence was aligned with all A. gambiae-deduced P450 sequences (ref. 38; http://P450.antibes.inra.fr), we found a sequence with 47% identity, significantly higher than the level of identity with any other mosquito P450 sequence. This mosquito sequence did not have a clear ortholog in the fruit fly genome. Instead, the most closely related sequences are CYP303A1 (which has one A. gambiae ortholog), CYP305A (which has three paralogs in A. gambiae), and a CYP305B1 cDNA sequence from ovaries of B. mori. The relationship of the fruit fly, moth, and mosquito sequences to the cockroach P450 sequences and CYP15A1 are shown in Fig. 5.
Fig. 5.
CYP15A1 and related sequences. Sequences were aligned by clustal x, and a tree was generated by phylip protpars. Species abbreviations are as follows: Dpu, D. punctata; Aga, A. gambiae; Bmo, B. mori; Dme, D. melanogaster; Has, Homo sapiens; Mdo, Musca domestica; and Bdi, Blaberus discoidalis.
JH III is the only juvenile hormone produced by cockroaches and mosquitoes. In contrast, higher diptera such as fruit flies and blow flies produce both JH III and its 6,7-epoxide, or JHB3 (39, 40). This “second” epoxidation therefore seems to be a derived trait rather than an ancestral one, and the lack of a clear ortholog in Drosophila might be explained on the basis of this different chemistry of the JH. Perhaps the CYP15 of higher flies evolved to allow the epoxidation at both the 6,7 and 10,11 double bonds, and this evolution resulted in such significant changes that the sequence is no longer recognizable as a CYP15. Another possibility is that CYP15 underwent a gene duplication event with paralogs specializing in the 10,11- and 6,7-epoxidation (product regioselectivity without substrate specificity) or paralogs specialized in mono- or bis-epoxidation of methyl farnesoate (substrate specificity without product regioselectivity). Functional expression of the candidate sequences may shed more light on this question. Even though the situation in Drosophila may be unusual, epoxidation precedes methylation in Lepidoptera whose CA produce JH acids at some developmental stages (5). Therefore, it may not be trivial to “discover” the epoxidase by simple orthology relationships in widely divergent species.
Specificity of Insect Cytochromes P450. Relatively few insect P450 enzymes have been analyzed in sufficient detail to allow a clear distinction between those P450 involved in detoxification and those thought to participate in physiological pathways. Such a distinction is not borne out by our current understanding of P450 phylogeny, and it may not exist at all. Instead, some P450s may be characterized by a narrow specificity whereas others may have a broad catalytic competence. We can now compare four different P450s, each expressed in E. coli and each tested with methyl farnesoate as substrate. This comparison (Table 3) reveals a gradation from a quite unspecific type of P450 (CYP6A1) to a most specific one (CYP15A1). P450 enzymes can be compared from the point of view of the reaction catalyzed, of their substrate specificity, and of their product specificity. For each category, the four P450s differ significantly. All are epoxidases on this substrate, except CYP4C7, which is a hydroxylase. CYP15A1 and CYP4C7 have the highest degree of product stereoselectivity, but CYP15A1 has a much tighter substrate specificity. CYP9E1, abundantly expressed in the fat body (just as CYP9E2 in B. germanica; ref. 41), is not stereoselective in the epoxidation and metabolizes the (2Z) isomer of methyl farnesoate better than the (2E) isomer. Least specific is CYP6A1, which has little substrate or product selectivity and can metabolize a variety of pesticides in addition to terpenoids. The topology of its active site with an unusually available heme group (24) supports this broad substrate specificity.
Table 3. Specificity of insect P450 enzymes towards methyl farnesoate (MF).
| P450 | Substrate | Reaction | Product formed | Ref. |
|---|---|---|---|---|
| CYP15A1 | (2E, 6E)-MF | Epoxidation | (10R)-epoxide (10R):(10S) (98:2) | This work |
| CYP4C7 | Sesquiterpenoids | ω-Hydroxylation | (12E)-alcohol | 16 |
| CYP9E1 | (2Z) > (2E)-MF (1.5:1) | Epoxidation | 10, 11-epoxide (10R):(10S) (1:1) | * |
| CYP6A1 | MF or JH All 4 isomers | Epoxidation | 6, 7 or 10, 11-epoxide (10S):(10R) (3:1) | 24 |
J. F. Andersen, G.C.U., J.F.K., and R.F., unpublished results
In conclusion, our work shows that CYP15A1 is a highly specific epoxidase. It is now available as a molecular tool for dissecting the physiological regulation of JH biosynthesis. The value of these molecular tools has already been demonstrated by another allatal P450, CYP4C7 (42). It should also prove useful as an in vitro target for the screening of JH synthesis inhibitors, and, ultimately, its structure will reveal the determinants of stereoselective epoxidation.
Acknowledgments
We thank P. Evans for the isomers of methyl farnesoate, M. B. Murataliev for a sample of purified P450 reductase, and E. Kuwano for samples of the imidazole compounds. This work was supported by National Institutes of Health Grants DK34549 and GM39094 (to R.F.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: JH, juvenile hormone; CA, corpora allata.
Data deposition: The cDNA sequences reported in this paper have been deposited in the GenBank database (accession nos. AY509244 and AY509245).
References
- 1.Truman, J. W. & Riddiford, L. M. (2002) Ann. Rev. Entomol. 47, 467–500. [DOI] [PubMed] [Google Scholar]
- 2.Schooley, D. A. & Baker, F. C. (1985) in Comprehensive Insect Physiology Biochemistry and Pharmacology, eds. Kerkut, G. A. & Gilbert, L. I. (Pergamon, Oxford), Vol. 7, pp. 363–389. [Google Scholar]
- 3.Feyereisen, R. & Farnsworth, D. E. (1987) Mol. Cell. Endocrinol. 53, 227–238. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Gonzalez, J., Buesa, C., Piulachs, M. D., Belles, X. & Hegardt, F. G. (1993) Eur. J. Biochem. 213, 233–241. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskaran, G., Sparagana, S. P., Barrera, P. & Dahm, K. H. (1986) Arch. Insect Biochem. Physiol. 3, 321–338. [DOI] [PubMed] [Google Scholar]
- 6.Peter, M. G., Shirk, P., Dahm, K. H. & Röller, H. (1981) Z. Naturforsch. 36c, 579–585. [Google Scholar]
- 7.Shinoda, T. & Itoyama, K. (2003) Proc. Natl. Acad. Sci. USA 100, 11986–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammock, B. D. (1975) Life Sci. 17, 323–328. [DOI] [PubMed] [Google Scholar]
- 9.Feyereisen, R., Pratt, G. E. & Hamnett, A. F. (1981) Eur. J. Biochem. 118, 231–238. [DOI] [PubMed] [Google Scholar]
- 10.Werck-Reichhart, D. & Feyereisen, R. (2000) Genome Biol. 1, 3003.1–3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sparks, T. (1990) in Safer Insecticides, eds. Hodgson, E. & Kuhr, R. J. (Dekker, New York), pp. 103–154.
- 12.Unnithan, G. C., Andersen, J. F., Hisano, T., Kuwano, E. & Feyereisen, R. (1995) Pestic. Sci. 43, 13–19. [Google Scholar]
- 13.Barnes, H. J., Arlotto, M. P. & Waterman, M. R. (1991) Proc. Natl. Acad. Sci. USA 88, 5597–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omura, T. & Sato, R. (1964) J. Biol. Chem. 239, 2370–2378. [PubMed] [Google Scholar]
- 15.Murataliev, M. B., Ariño, A., Guzov, V. M. & Feyereisen, R. (1999) Insect Biochem. Mol. Biol. 29, 233–242. [DOI] [PubMed] [Google Scholar]
- 16.Sutherland, T. D., Unnithan, G. C., Andersen, J. F., Evans, P. H., Murataliev, M. B., Szabo, L. Z., Mash, E. A., Bowers, W. S. & Feyereisen, R. (1998) Proc. Natl. Acad. Sci. USA 95, 12884–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jefcoate, C. F. (1978) Methods Enzymol. 52, 258–279. [DOI] [PubMed] [Google Scholar]
- 18.Hammock, B. D. & Mumby, S. M. (1978) Pestic. Biochem. Physiol. 9, 39–47. [Google Scholar]
- 19.Toong, Y. C., Schooley, D. A. & Baker, F. C. (1988) Nature 333, 170–171. [Google Scholar]
- 20.Pratt, G. E., Kuwano, E., Farnsworth, D. E. & Feyereisen, R. (1990) Pestic. Biochem. Physiol. 38, 223–230. [Google Scholar]
- 21.Judy, K. J., Schooley, D. A., Dunham, L. L., Hall, M. S., Bergot, B. J. & Siddall, J. B. (1973) Proc. Natl. Acad. Sci. USA 70, 1509–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judy, K. J., Schooley, D. A., Hall, M. S., Bergot, B. J. & Siddall, J. B. (1973) Life Sci. 13, 1511–1516. [DOI] [PubMed] [Google Scholar]
- 23.Judy, K. J., Schooley, D. A., Troetschler, R. G., Jennings, R. C., Bergot, B. J. & Hall, M. S. (1975) Life Sci. 16, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 24.Andersen, J. F., Walding, J. K., Evans, P. H., Bowers, W. S. & Feyereisen, R. (1997) Chem. Res. Toxicol. 10, 156–164. [DOI] [PubMed] [Google Scholar]
- 25.Cariño, F., Koener, J. F., Plapp, F. W., Jr. & Feyereisen, R. (1992) Am. Chem. Soc. Symp. Ser. 505, 31–40. [Google Scholar]
- 26.Borovsky, D., Carlson, D. A., Hancock, R. G., Rembold, H. & van Handel, E. (1994) Insect Biochem. Mol. Biol. 24, 437–444. [DOI] [PubMed] [Google Scholar]
- 27.Borovsky, D., Carlson, D. A., Ujvary, I. & Prestwich, G. D. (1994) Arch. Insect Biochem. Physiol. 27, 11–25. [Google Scholar]
- 28.Andersen, J. F., Ceruso, M., Unnithan, G. C., Kuwano, E., Prestwich, G. D. & Feyereisen, R. (1995) Insect Biochem. Mol. Biol. 25, 713–719. [DOI] [PubMed] [Google Scholar]
- 29.Pratt, G. E., Stott, K. M., Brooks, G. T., Jennings, R. C., Hamnett, A. F. & Alexander, B. A. J. (1981) in Juvenile Hormone Biochemistry: Action, Agonism and Antagonism, eds. Pratt, G. E. & Brooks, G. T. (Elsevier/North-Holland Biomedical Press, Amsterdam), pp. 107–121.
- 30.Bowers, W. S., Ohta, T., Cleere, J. S. & Marsella, P. A. (1976) Science 193, 542–547. [DOI] [PubMed] [Google Scholar]
- 31.Ellis-Pratt, G. (1983) in Natural Products for Innovative Pest Management, eds. Whitehead, D. L. & Bowers, W. S. (Pergamon, Oxford), pp. 323–355.
- 32.Pratt, G. E., Jennings, R. C., Hamnett, A. F. & Brooks, G. T. (1980) Nature 284, 320–323. [Google Scholar]
- 33.Feyereisen, R., Johnson, G., Koener, J. F., Stay, B. & Tobe, S. S. (1981) J. Insect Physiol. 27, 855–868. [Google Scholar]
- 34.Hamnett, A. F. & Pratt, G. E. (1983) Life Sci. 32, 2747–2753. [DOI] [PubMed] [Google Scholar]
- 35.Pratt, G. E. & Bowers, W. S. (1977) Nature 265, 548–550. [DOI] [PubMed] [Google Scholar]
- 36.Pratt, G. E. & Pener, M. P. (1983) J. Insect Physiol. 29, 33–39. [Google Scholar]
- 37.Belles, X., Camps, F., Casas, J., Messeguer, A. & Piulachs, M. D. (1988) J. Insect Physiol. 34, 457–461. [Google Scholar]
- 38.Ranson, H., Claudianos, C., Ortelli, F., Abgrall, C., Hemingway, J., Sharakhova, M. V., Unger, M. F., Collins, F. H. & Feyereisen, R. (2002) Science 298, 179–181. [DOI] [PubMed] [Google Scholar]
- 39.Lefevere, K. S., Lacey, M. J., Smith, P. H. & Roberts, B. (1993) Insect Biochem. Mol. Biol. 23, 713–720. [DOI] [PubMed] [Google Scholar]
- 40.Richard, D. S., Applebaum, S. W., Sliter, T. J., Baker, F. C., Schooley, D. A., Reuter, C. C., Henrich, V. C. & Gilbert, L. I. (1989) Proc. Natl. Acad. Sci. USA 86, 1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen, Z., Horak, C. E. & Scott, J. G. (2001) Gene 272, 257–266. [DOI] [PubMed] [Google Scholar]
- 42.Sutherland, T. D., Unnithan, G. C. & Feyereisen, R. (2000) J. Insect Physiol. 46, 1219–1227. [DOI] [PubMed] [Google Scholar]