Abstract
Human papillomaviruses (HPVs) establish long-term infections in patients. The mechanism for extrachromosomal HPV DNA persistence in cycling cells is unknown. We show that HPV origin-containing plasmids partition as minichromosomes, attributable to an association of the viral origin recognition protein E2 with mitotic spindles. α-, β-, and γ-tubulins were pulled down with a tagged E2. The N-terminal transacting and C-terminal protein dimerization/DNA binding domains independently associated with the spindles. We suggest that this E2 property enables these viruses to establish persistence. Its implication for HPV oncogenesis is presented.
For any extrachromosomal DNA virus to establish a persistent infection in cycling host cells, the viral genome must replicate and partition into both daughter cells during division. The E2 origin (ori)-binding protein of bovine papillomavirus type 1 (BPV-1) associates with mitotic chromosomes (1–3), thus providing a mechanism for viral DNA segregation. Comparable mechanisms have been demonstrated for the Epstein–Barr virus through the Epstein–Barr virus-encoded nuclear antigen 1 protein and the Kaposi's Sarcoma virus (human herpesvirus 8) through the latency-associated nuclear antigen 1 protein (4, 5). In contrast, the mechanism by which human papillomavirus (HPV) DNA partitions during cell division has not been elucidated. In this report, we demonstrate that HPV ori-containing DNA segregates as minichromosomes by association with mitotic spindles and this association is mediated by the HPV origin recognition protein E2.
HPVs are medically important pathogens that establish persistent infections in long-living basal keratinocytes. Infections typically cause benign hyperproliferation of squamous epithelia in the form of cutaneous warts, laryngeal papillomas, and anogenital condylomata. Over time, infections can become subclinical, but may reactivate during episodes of immune suppression. The HPV genome is a double-stranded, circular DNA of ≈7,900 bp and replicates extrachromosomally in the nucleus of infected keratinocytes. Low copy numbers of the mucosotrophic HPV DNA plasmids are maintained in the basal and parabasal cells that divide, whereas the productive phase takes place only in postmitotic, differentiated cell strata and progeny virus shed within the sloughing superficial cells (6). Thus, it is paramount that, in either latent or active infections, HPV DNA must partition into the two daughters of dividing cells for viral persistence. To support viral DNA amplification in postmitotic cells, the viral E6 and E7 proteins inactivate the host tumor suppressor proteins p53 and pRB (retinoblastoma protein), respectively, reestablishing an S-phase environment. Elevated transcription of these oncogenes is normally limited to the differentiated compartment. However, if inappropriately expressed in the basal cells, such as during repeated wounding and healing, the high-risk HPV oncoproteins can promote excessive cell cycling and host chromosome instability. Indeed, a small fraction of the persistent infections by high-risk HPV types can progress to high-grade dysplasias and cancers (6–8).
The mechanisms of papillomavirus DNA replication are conserved. Viral DNA replication requires the viral ori sequences, the virus-encoded DNA helicase E1, the ori-recognition protein E2, and the cellular DNA replication machinery (9–12). The viral ori contains multiple copies of a consensus E2 protein binding site (BS) flanking a single E1BS. For its simplicity, papillomavirus DNA replication has been an excellent model for investigating the mechanisms that regulate the initiation of cellular DNA replication. E1 protein is the equivalent of the cellular minichromosome maintenance complex. It binds to E1BS and functions as the replicative dihexameric helicase (13–17). In addition, it recruits DNA polymerase α/primase and the single-stranded DNA binding protein replication protein A to the ori (18–20). Consequently, E1 is required throughout initiation and elongation (21). The E2 protein, on the other hand, is the equivalent of the eukaryotic ORC protein complex; it is the primary viral ori-recognition protein and helps recruit E1 to the ori (9–12). The BPV-1 E2 protein prevents nucleosome formation on the ori in vitro (22), facilitating initiation of replication. Thus, E2 is necessary for establishing the preinitiation complex. However it is not present during elongation (21). Finally, the BPV-1 E2 associates with mitotic chromosomes as fine dots and plays a key role in BPV-1 DNA segregation during cell division (1–3).
Materials and Methods
Cell Culture. We maintained 293, CV1 and COS7 cells in DMEM supplemented with 10% FBS. Transfection was conducted by electroporation as described (11). We established 293–11 E2 and HeLa-11 E2 cell lines as described (refs. 23 and 24 and S.-Y.W. and C.-M.C., unpublished results). Both cell lines were carried in DMEM plus 10% FBS in the presence of 1 μg/ml tetracycline until induction. The BPV-1 E2 cell line was carried and induced as described in ref. 25.
Plasmid Construction and Mutagenesis. pMT2-H11 E2 (previously pMT2 E2) expressing the native HPV-11 E2 protein as well as pGFP-H11 E2, pGFP-H11 E2N, pGFP-H11 E2H, and pGFP-H11 E2C expressing GFP fused to HPV-11 E2 or the E2 N (amino), H (hinge), or C (carboxyl) domain were described in refs. 11 and 26. Plasmids expressing GFP fusions to HPV-16 E2 (pGFP-H16 E2), HPV-18 E2 (pGFP-H18 E2), and BPV-1 E2 (pGFP-B1 E2) were constructed by PCR amplification of the E2 genes from previously described expression vectors (11, 27, 28), and placed in frame into the BglII and EcoR1 sites of the pEGFP-C1 plasmid (CLONTECH). The plasmid expressing GPF fusions to the HPV-11 E2K239A, HPV-11 E2K293R, or HPV-11 E2K293Q mutation (pGFP-H11 E2K239A, pGFP-H11 E2K239R, or pGFP-H11 E2K239Q) was derived by PCR with mutagenic primers. HPV-11 E2K239A-STR was derived from HPV-11 E2K239A by using a reverse primer to introduce a 9-amino acid STR tag (29). pGal4DBD-GFP expressing the Gal4 DNA binding domain (DBD) fused to GFP was constructed by excising the BglII/PstI fragment of plasmid pBXG1[Gal4 (1–147)] (30) and ligating it into the same sites in pEGFP-N1 (CLONTECH). pH11URR-Gal4BS containing 40 copies of Gal4 BS and HPV-11 URR is described in supporting information, which is published on the PNAS web site.
Pull-Down Assay and Western Blots. COS7 cells mock-transfected or transfected with pGFP-H11 E2K239A-STR were grown for 24 h and lysed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) containing protease inhibitors (Sigma). Pull-down assays were performed by using streptavidin-conjugated Sepharose beads (Invitrogen). The samples were resuspended in SDS loading buffer, separated on a 4–12% polyacrylamide/SDS gel, and western blotted. The nitrocellulose membranes were probed separately with monoclonal antibodies specific for α-, β- or γ-tubulins followed by an anti-mouse IgG antibody conjugated with peroxidase and developed with an enhanced chemiluminescence detection system (ECL; Amersham Pharmacia). The membranes were stripped and reprobed by using an anti-GFP antibody to detect the GFP-E2 protein.
Protein Detection by Indirect Immunofluorescence. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in 1× PBS at 4°C. Slides were then incubated with primary antibody for 2 h at 37°C (β-tubulin Cy3 conjugate at 1:200 dilution and γ-tubulin Cy3 conjugate at 1:50 dilution; both from Sigma). The anti-HPV-11 E2 rabbit polyclonal antibody (11) was used at 1:100 dilution. After incubation, slides were washed and incubated with a 1:200 dilution of anti-rabbit-tetramethylrhodamine isothiocyanate (Sigma) for 1 h at 37°C and washed, 4′,6-diamidino-2-phenylindole (DAPI)-stained, and mounted in Antifade. All images were acquired with a 100× objective lens with an Olympus AX70 fluorescence microscope with Speicher filters (Chroma, Rockingham, VT) and a Zeiss Axiocam digital camera. Processing and assembly were accomplished with photoshop (Adobe Systems, Mountain View, CA).
Microtubule Repolymerization Assays. Cells in chamber slides were placed on ice for 45 min and then transferred to 37°C for the specified length of time before immediate fixation as described in ref. 31.
Results
Localization of a GFP-HPV-11 E2 Fusion Protein to Centrosomes and Mitotic Spindles. We previously reported that a fusion between the GFP and HPV-11 E2 (GFP-H11 E2) retains the replication and transcription regulatory functions of the wild-type E2 protein (26). To investigate the mechanism of HPV DNA partitioning critical for persistent infection, we determined the localization of GFP-H11 E2 in transfected cells. COS7 cells (monkey CV1 kidney cells transformed by SV40 T antigen) were used in most of the experiments because of their high transfection efficiency, large size, and strong adherence to glass slides, properties optimal for studies of subcellular localization. Nonetheless, some of the experiments were performed in human 293 cells (Figs. 2 A and B and 3), HeLa (Fig. 2C) and C33A cervical carcinoma cell lines, CV1 cells, or in primary human fibroblasts (data not shown). Identical observations were made.
Fig. 2.
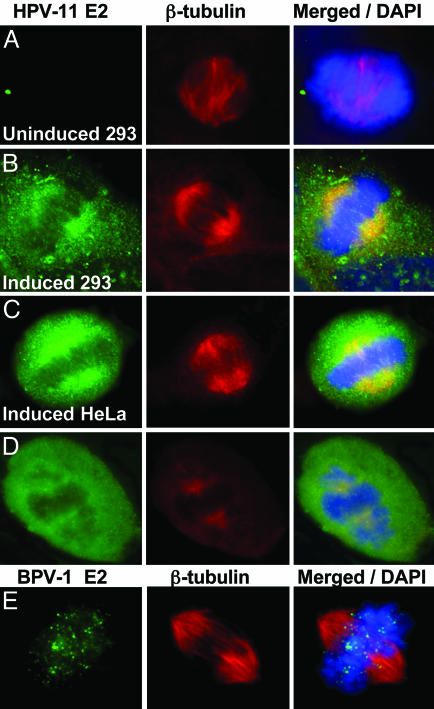
HPV-11 E2, but not BPV-1 E2, associates with mitotic spindles. The cells were probed with antibodies to HPV-11 E2 and β-tubulin (A–D). (A–C) Cell lines harboring epitope-tagged HPV-11 E2 under the control of a tetracycline regulated promoter. Shown are either uninduced 293 cells (A), 293 cells (B), or HeLa cells (C) induced for 24 h. (D) COS7 cells transfected with pMT2-H11 E2 expressing the native HPV-11 E2 protein. (Left) E2 (FITC). (Center) β-Tubulin (Cy-3). (Right) Merged images with DAPI-stained nuclear DNA (blue). (E) Induced CV1 cells harboring BPV-1 E2 under the control of a metallothionein promoter (25). After a 4.5-hour induction with 1 μM CdSO4, BPV-1 E2 (FITC) (Left) and β-tubulin (Cy-3) (Center) were detected with antibodies. (Right) A merged image.
Fig. 3.
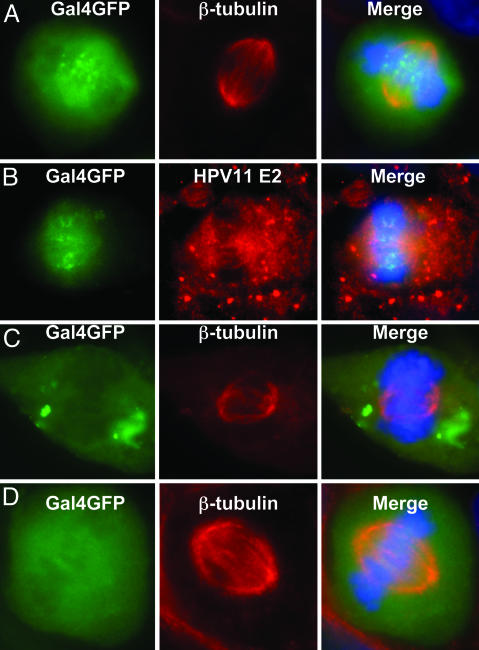
E2 is required for viral ori DNA association with spindles during metaphase. (A–C) The 293–11 E2 cells were transfected with pH11URR-Gal4BS and pGal4DBD-GFP and grown in the absence of tetracycline to induce E2 expression (A and B) or in the presence of tetracycline to inhibit E2 expression (C). (D) The cells were transfected with pH11URR and pGal4DBD-GFP and induced for E2. The cells were examined for Gal4DBD-GFP (labeled Gal4GFP, A–D Left), probed for β-tubulin (A Center, C Center, and D Center; Cy-3) or for E2 (B Center; Cy-3) with antibodies, and stained for chromosomes with DAPI (blue). (Right) Merged images.
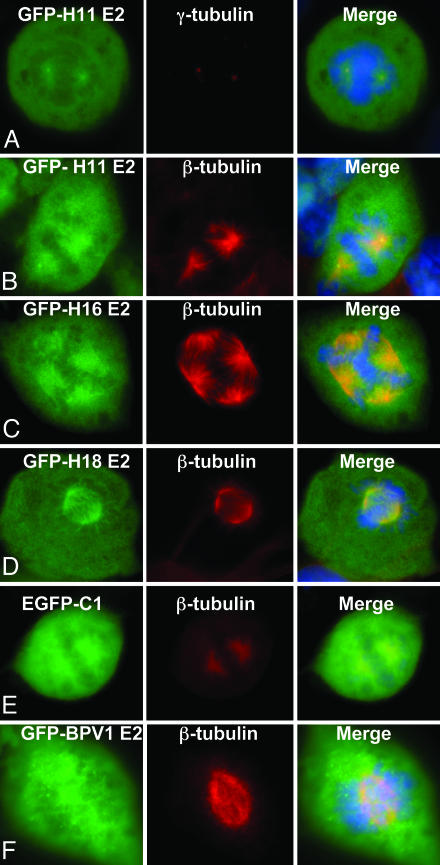
Unlike BPV-1 E2, GFP-H11 E2 was clearly excluded from the metaphase chromosomes (Fig. 1 A and B). The GFP-H11 E2 signals were concentrated in two pairs of closely spaced bright dots in mitotic cells reminiscent of centrosomes, the poles for sister chromosome segregation (Fig. 1 A and B; see also Fig. 4 B and C). The dots indeed reacted with antibody to γ-tubulin (Fig. 1 A; Cy-3, red), a marker for centrosomes (32). Emanating from the centrosome dots were filament-like structures suggestive of mitotic spindles. Centrosomes and mitotic spindles play critical roles in the equal partitioning of chromosomes into the two daughter cells to maintain genomic stability (33). We therefore performed immunofluorescence staining with antibodies to α- and β-tubulins, components of the mitotic spindles. GFP-H11 E2 colocalized with these two tubulins (Cy-3) as well, decorating the mitotic spindles (Fig. 1B and data not shown). The centrosomes positive for GFP-H11 E2 never appeared unduly swollen (Fig. 1 A and B; see also Fig. 4), untypical of large aggresomes that are formed pericentrosomally (34). In addition, after colcemid treatment, the GFP-H11 E2-decorated spindles were lost (data not shown). The control GFP alone was distributed throughout the cells, but the dense metaphase chromosomes reduced the amount of GFP (Fig. 1E). As an additional control, we prepared GFP fused to BPV-1 E2 protein (GFP-B1 E2). GFP-B1 E2 did not localize to centrosomes or mitotic spindles (Fig. 1F, Cy-3). Rather, it appeared as fine dots or was diffuse throughout the cells. Collectively, these results support the notion that HPV-11 E2 protein appears to function through a different mechanism for partitioning the viral genome than BPV-1.
Fig. 1.
HPV E2 proteins associate with the mitotic spindles during mitosis. COS7 cells (A–E) and CV1 cells (F) were transfected with vectors that express GFP fusion proteins with HPV-11 E2 (A and B), HPV-16 E2 (C), HPV-18 E2 (D), BPV-1 E2 (F), or enhanced GFP (E). After transfection (12–16 h), cells were probed with an antibody to γ-tubulin (Cy-3) (A Center)or β-tubulin (Cy-3, red) (B–F Center). Also shown are GFP (Left) and merged images with DAPI-stained chromosomes (Right).
Fig. 4.
HPV-11 GFP-E2K239A colocalizes with MTOC and microtubules at interphase and with centrosomes and mitotic spindles at metaphase. (A–C) COS7 cells were transfected with pGFP-H11 E2K293A. At 16 h, nuclear DNA was DAPI-stained (blue) and the fusion protein was revealed by using GFP. (A) An interphase cell with high fusion protein expression. (B) The bright dot in interphase cells was positive for γ-tubulin (Cy-3), indicative of MTOC. (C) A metaphase cell expressing a very low level of GFP fusion protein. The spindle poles are the only clearly GFP-positive structures. The image is enlarged relative to A and B for clarity. (D–F) A metaphase cell expressing GFP-11 E2K239A (D) was probed for β-tubulin (Cy-3) (E); shown is a merged image (F) with the metaphase chromosomes (DAPI). The centrosomes and spindles were decorated by the GFP fusion, which was clearly excluded from the chromosomes. (G–N) A time course [0 min (G and H), 5 min (I and J), 10 min (K and L), and 15 min (M and N)] of microtubule repolymerization at 37°C after being dissociated at 4°C. β-Tubulin was revealed by indirect immunofluorescence (Cy-3). (H, J, L, and N) Merged images. Aster formation can be seen over time from the MTOC, the bright GFP/tubulin focus. (O) The GFP-H11 E2K239A-STR protein was localized to microtubules and centrosomes in an interphase cell. Shown are GFP (Left), β-tubulin (Cy-3) (Center), and merged images with chromosomal DNA stained with DAPI (blue) (Right). (P) α-, β- and γ-tubulins were pulled down with streptavidin-conjugated Sepharose beads from COS7 cells transfected with an expression vector of GFP-H11 E2K239A-STR (lanes 3) but not from mock-transfected cells (lanes 2). Western blots were probed with individual antibodies to the tubulins (Lower) and then stripped and reprobed for E2 by using a GFP antibody (Upper). Lanes 1 show a small fraction of whole cell lysates from mock-transfected cells.
Localization of Native and Epitope-Tagged HPV-11 E2 to Mitotic Spindles. To verify that our findings are not a result of an overexpression of the GFP fusion protein, we examined 293–11 E2 and HeLa-11 E2 cells that harbor a Tet-off expression cassette of HPV-11 E2 derivatized at the amino terminus with His-6 and FLAG tags. The expression of E2 was monitored by Western blots (data not shown) and indirect immunofluorescence by using an antibody to E2 (FITC, green), and antibody to β-tubulin (Cy-3) for mitotic spindles. In uninduced cells, there was little or no E2 expression above background (Fig. 2A and data not shown). After induction for one or more days, clear colocalization of E2 with β-tubulin was observed (Fig. 2 B and C). Moreover, untagged, native E2 protein in transfected COS cells (Fig. 2D; FITC, green) also localized to the mitotic spindles (Cy-3). In contrast, immunostaining for BPV-1 E2 in an inducible CV1 cell line (Fig. 2E) showed that it appears as fine dots in association with mitotic chromosomes but not with the spindles as described in ref. 25. Collectively, these results show that the localization of HPV-11 E2 to mitotic spindles and centrosomes is not due to misdirection by GFP nor to the formation of aggresomes of misfolded, over-expressed protein transported to the pericentromeric region for degradation (34).
Localization of GFP-HPV-16 and GFP-HPV-18 E2 Fusion Proteins to Mitotic Spindles. We hypothesized that HPV E2/spindle association may not be unique to HPV-11. Thus, we also constructed a GFP fusion protein with HPV-16 (pGFP-H16 E2) or HPV-18 E2 (GFP-H18 E2). Both these proteins were also excluded from the mitotic chromosomes, and they decorated the mitotic spindles (Cy-3) (Fig. 1 C and D). This association was also colcemid-sensitive (data not shown). In the particular example of GFP-H16 E2 shown (Fig. 1C), there are four spindle poles, and E2 was associated with each of the associated spindles. The multiple centrosomes were not an effect of GFP E2, as cells with three or four spindle poles were also observed in untransfected COS7 cells.
Association of a Modified Viral ori Plasmid with Mitotic Spindles in the Presence of E2 as Revealed by a Marker Protein. In situ hybridization suggested HPV ori DNA colocalized with spindles (see supporting information). To verify our hypothesis that E2 protein bridges the viral DNA directly or indirectly to the mitotic apparatus, we constructed a marked HPV-11 ori plasmid (pH11URR-Gal4BS) that contains the HPV-11 URR and 40 copies of the Gal4 DNA BS. The localization of this plasmid can therefore be monitored by the binding of Gal4DBD-GFP, a fusion of the Gal4DBD to GFP. pH11URR-Gal4BS and the pGal4DBD-GFP plasmids were then cotransfected into the 293–11 E2 cells. After transfection, cells were grown in the presence or absence of tetracycline for 24 h or longer. In cells that expressed E2, some of the Gal4DBD-GFP and, by inference, the marked ori plasmids were associated with the spindle fibers (Cy-3) as distinct dots primarily between the two centrosomes (Fig. 3A). Furthermore, antibody to E2 revealed Gal4DBD-GFP dots on E2-positive fibers between the two poles (Fig. 3B). In cells not induced for E2 expression, concentrated Gal4DBD-GFP signals were excluded from the territory occupied by the spindles (Cy-3) and the chromosomal DNA (DAPI) (Fig. 3C). In another control, the cotransfected pH11 URR plasmid did not harbor Gal4BS. No concentrated Gal4DBD-GFP dots were observed to associate with the spindles (Fig. 3D). These experiments demonstrate that E2 protein is responsible for recruiting the HPV ori-containing DNA to the mitotic spindles.
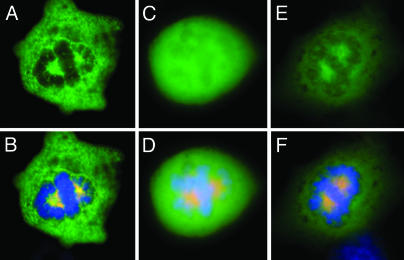
Association of an HPV-11 E2 Nuclear Localization Sequence Mutation with the Microtubule Organizing Center (MTOC) and Microtubules. In asynchronized cultures, the vast majority of the cells are in interphase. In these cells, α- and β-tubulins form microtubules in the cytoplasm, emanating from the perinuclear MTOC, also known as the centrosome. The very low percentage of cells in M phase makes it difficult to demonstrate biochemically a direct or indirect association of E2 with spindles or centrosomes. We tested the hypothesis that an E2 protein without a nuclear localization sequence might retain its ability to associate with mitotic spindles and centromsomes at metaphase but become associated with microtubules or the centrosome during interphase. If true, this ability would greatly facilitate a biochemical approach to prove this association. Residues 238–242 in the hinge domain of HPV-11 E2 were critical for nuclear localization (26). Thus, we constructed an expression vector of GFP-H11 E2K239A, GFP-H11 E2K239R or GFP-H11 E2K239Q. At metaphase, the mutated GFP-11 E2 fusion proteins localize to the centrosomes and mitotic spindles (Fig. 4 C–F and data not shown). At interphase, GFP-H11 E2K239A formed a perinuclear bright small dot positive for γ-tubulin, indicative of the MTOC (Fig. 4 A and B). It also appeared in a network fashion in the cytoplasm, reminiscent of microtubules (Fig. 4A).
Microtubules depolymerize at 4°C but reassemble over time from the MTOC when the cells are returned to 37°C (31). To verify that GFP E2K239A mutation associate with microtubules and MTOC, a microtubule disruption/recovery experiment was conducted. Fig. 4 G–N is a time course experiment in which we followed the distribution of GFP-H11 E2K239A and of β-tubulin during recovery after the cells were placed at 4°C for 45 min. At time 0 after return to 37°C, the centrosome was clearly discernable (Fig. 4H), but there were no detectable organized microtubules based on the distribution of either GFP-H11 E2K239A (GFP, green) or β-tubulin (Cy-3) (Fig. 4G). A time-dependent colocalization of E2 with repolymerizing microtubules was observed. By 10 min, formation of a GFP-H11E2-decorated microtubule aster around the centrosome was evident (Fig. 4 I–L). By 15 min, GFP-H11 E2 colocalized with the extensive microtubule network that had reformed (Fig. 4 M and N). In contrast, control GFP was distributed to both the nucleus and cytoplasm without an apparent association with MTOC or microtubules at any time point (data not shown).
This GFP-H11 E2K239A protein, which was mutated in its nuclear localization sequence, provided us with an opportunity to examine the E2 association with cellular organelles biochemically. We then constructed an expression vector for GFP-H11 E2K239A-STR. STR is a small tag that binds to streptavidin with a high affinity similar to biotin (29). Having established that this protein was localized to microtubules and the MTOC in interphase cells (Fig. 4O), we conducted a one-step pull-down assay in transfected COS7 cells. Twenty-four hours after transfection, streptavidin-Sepharose beads were used to pull down the GFP-H11 E2K239A-STR from cell lysates. After extensive washing, the recovered proteins were Western blotted separately with antibodies to α-, β-, or γ-tubulins. The blots were then stripped and reprobed with monoclonal antibodies to GFP to detect the GFP-H11 E2 protein. Fig. 4P shows that all three tubulins were detected in the pull-down fraction from cell lysates expressing the E2 protein (lanes 3). A small fraction of total lysates from mock-transfected cells were positive for all three tubulins (lanes 1), but streptavidin did not pull down any of the tubulins (lanes 2). These results demonstrated the specificity of the pull-down experiments.
Functional Delineation of HPV-11 E2 Domains. The E2 protein contains three structural and functional domains. The N-terminal half interacts with viral and host proteins, regulating transcription and replication, and the C-terminal portion is responsible for dimerization and ori-binding (35). A hinge domain between N and C is highly divergent among papilloma-virus types. To delineate the domain responsible for spindle association, we examined GFP fused to the N, H, or C domains of HPV-11 E2 protein for their localization during metaphase (Fig. 5). These cells were treated with 17β-estradiol to increase the proportion of mitotic cells (31). Estradiol retards the progression of mitosis by delaying the alignment of condensed chromosomes on the mitotic plate. With the exception of GFP-H11 E2H, the other GFP fusions were clearly seen being strongly excluded from the metaphase chromosomes. Rather, they colocalized with thick spindle cables. Some also appeared as small dots along the fine spindle fibers. Thus, the amino terminal and the carboxyl terminal domains have independent binding sequences but the hinge domain does not. These results also support our conclusion that the E2 association with mitotic spindles is specific and cannot be attributed to GFP or protein overexpression.
Fig. 5.
The HPV-11 E2 N terminus and C terminus independently associate with the spindles. COS cells were separately transfected with pGFP-H11 E2N (A and B), pGFP-H11 E2H (C and D), or pGFP-H11 E2C (E and F). Twenty hours after electroporation, 100 μM 17β-estradiol was introduced for 4 h to enrich for metaphase cells. The cells were probed for β-tubulin (Cy-3), and chromosomes were stained with DAPI (blue). A, C, and E are images of GFP fusion proteins, and B, D, and F are the corresponding merged images.
Discussion
By using a microscopic approach, we have provided evidence that E2 proteins of three common HPV types, HPV-11, HPV-16, and HPV-18, associate with spindle fibers during mitosis and that HPV-11 E2 also associates with centrosomes (Figs. 1 and 2). We also showed that an HPV origin-containing plasmid associated with the mitotic spindles in an E2-dependent manner (Fig. 3). Employing an STR-tagged, GFP-H11 E2 protein mutated in its nuclear localization sequence, we showed that α-, β-, and γ-tubulins, major components of microtubules, mitotic spindles, and MTOC, were pulled down by using streptavidin-Sepharose, substantiating our microscopic observations (Fig. 4). Collectively, these results demonstrate that the E2 protein recruits the HPV ori DNA to the spindles independent of plasmid replication. Recent investigations delineated the responsible region in the C domain of HPV-11 E2 to a span of 14 aa that had significant divergence from the comparable region in BPV-1 E2 (L.D.D., B.A.V.T., T.R.B., and L.T.C., unpublished results). Conversely, the phosphorylation of four serine residues in the BPV-1 E2 hinge domain, some by creatine kinase 2, is critical for its association with mitotic chromosomes (2). These serine residues and the acidic creatine kinase 2 recognition sequence are not present in the hinge domain of HPV E2 proteins. These differences could in part contribute to the differential properties of HPV and BPV-1 E2 proteins.
On the basis of these observations, we propose that this association between mitotic spindles and the E2 protein bound to extrachromosomal viral DNA provides the mechanism by which the viral genomes partition in the long-living basal and parabasal cells to establish persistence when these cells divide. This mode of viral chromosome segregation into daughter cells makes perfect sense, given that it is a tried and proven mechanism used by the cellular chromosomes. Our finding is interesting, because other persistent DNA viruses, such as BPV-1, Epstein–Barr virus, and Kaposi's human sarcoma virus, rely on a mechanism of metaphase chromosome tethering to partition their genomes into daughter cells. A cellular protein has been identified which interacts with Epstein–Barr virus-encoded nuclear antigen type 1, enabling Epstein–Barr virus DNA tethering to metaphase chromosomes (36). The cellular protein responsible for BPV-1 chromosome-tethering has not been found. Presently, we do not know the identity of the cellular proteins that may have mediated the HPV E2/spindle association, and it will be important to identify them in the future.
We propose that this unique property of HPV E2 proteins might also play an important role in HPV-associated carcinogenesis. Cervical cancers and cell lines derived thereof contain integrated HPV DNA and express the viral oncogenes at a high level. The virus-host DNA junction invariably disrupts the expression of the E2 gene and is accompanied by deletion of a portion of the early region, including the viral early polyadenylation site. A stable, downstream host polyadenylation site is invariably hijacked to stabilize the mRNA encoding the upstream viral oncogenes (37, 38). In cases where the viral DNA is integrated as tandem repetitions, only the 3′-junction copy expresses the viral oncogenes, whereas all upstream, internal copies are silenced by DNA methylation (B.A.V.T., J. C. Kappes, N. S. Banerjee, J. Knops, L. Lai, R. D. M. Steenbergen, C. J. L. M. Meijer, P. J. F. Snijders, P. Chatis, T.R.B., P. T. Moen, Jr., and L.T.C., unpublished results). We propose that, shortly after initial viral DNA integration, functional E2 protein may well be expressed. The multiple copies of E2BSs in the integrated viral genomes might then function as a “viro-centromere,” with E2 bridging it to the mitotic spindles during mitosis. If the viro-centromere and the kinetochore perchance link to spindles connected to opposing centrosomes, the possibility of chromosomal breakage may ensue. As described by Barbara McClintock (39), broken ends are highly recombinogenic, and this would lead to successive rounds of breakage-fusion-bridge cycles.
A critical element in this process is the ability of the oncogenic HPV E6 protein, which causes the degradation of p53, “the guardian of the genome,” to suppress the check point activation so that the cell cycle continues despite misaligned spindles or damaged chromosomes, trashing the host genome. However, to avoid mitotic catastrophe, cancer cells that emerge must interrupt the breakage-fusion-bridge cycle. The only way to break this cycle is to inactivate the E2 gene, either through DNA silencing or by DNA deletion. This would explain the genetic and transcriptional portrait of typical HPV-associated cancers or transformed cell lines derived therefrom. Detailed cytogenetic characterization of cervical carcinoma cell lines reveals extensive chromosomal rearrangements as expected of cancer cells. In addition, the transcriptionally active, integrated HPV DNA was cytogenetically mapped to be near the junction of sequences derived from two chromosomes in the cervical carcinoma cell line CaSki (B.A.V.T., J. C. Kappes, N. S. Banerjee, J. Knops, L. Lai, R. D. M. Steenbergen, C. J. L. M. Meijer, P. J. F. Snijders, P. Chatis, T.R.B., P. T. Moen, Jr., and L.T.C., unpublished results). In another cervical carcinoma cell line, HeLa, HPV-18 genome colocalizes with c-myc cytogenetically, and together they translocated to multiple chromosomes, supporting the breakage-fusion model (40). Although these observations do not prove an E2-induced chromosomal breakage, they are consistent with it, especially in view of the observation that, in cancers, HPV DNA is often found integrated near fragile sites (41). However, chromosomal rearrangements were not examined in these cases. In principle, integrated nononcogenic HPV DNA should be equally capable of undergoing a chromosome breakage-fusion cycle in the presence of an E2 protein. However, these HPVs rarely cause cancer because their oncoproteins do not cause p53 to degrade. Thus, cells with misaligned spindles or chromosomal damages are more likely to be arrested or to undergo apoptosis when cell cycle check point mechanisms are activated.
The E2 protein is also a transcription factor that regulates the viral enhancer/promoter responsible for the expression of the viral oncogenes E6 and E7. In the current model for HPV-oncogenesis, the rationale for E2 gene deletion or silencing has been to block a feedback repression of E6–E7 expression from the viral promoter and to stabilize the viral oncogene mRNAs. Our hypothesis would suggest that this concept could now be expanded to include another important rationale: the cessation of breakage-fusion-bridge cycles so that at least some of the cells do not self-destruct and emerge eventually as cancer cells. This hypothesis of E2-induced chromosomal breakage remains to be formally tested.
In summary, HPV genomes appear to behave like true minichromosomes, accounting for the persistent infection in patients. We propose that this E2 property may contribute to oncogenesis by the high-risk HPVs. Finally, we propose that, when properly harnessed, HPV replicons have the potential in the future to be developed into vehicles for introducing and maintaining therapeutic genes without disrupting chromosomal DNA as caused by the integration of retroviral or adeno-associated virus vectors.
Supplementary Material
Acknowledgments
We thank Jen-Sing Liu and Shu-Ru Kuo for the pMT2-H11URR E2 plasmid, Alison McBride for her inducible BPV-1 E2 cell line, Elliot Androphy for monoclonal antibody to BPV-1 E2, and Anindya Dutta, Andy Ball, and Tim Townes for critical reading of the manuscript. This research was supported by U.S. Public Health Service Grants CA83679, CA36200 (to L.T.C. and T.R.B.), and CA81017 (to C.M.C.). B.A.V.T. is partially supported by the University of Alabama at Birmingham Medical Scientist Training Program. L.D.D. is partially supported by University of Alabama at Birmingham Molecular and Viral Oncology Predoctoral Training Grant T32-CA09467.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HPV, human papillomavirus; BPV, bovine papillomavirus; ori, origin; BS, binding site; DBD, DNA binding domain; DAPI, 4′,6-diamidino-2-phenylindole; MTOC, microtubule organizing center.
References
- 1.Skiadopoulos, M. H. & McBride, A. A. (1998) J. Virol. 72, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehman, C. W. & Botchan, M. R. (1998) Proc. Natl. Acad. Sci. USA 95, 4338–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilves, I., Kivi, S. & Ustav, M. (1999) J. Virol. 73, 4404–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanda, T. & Wahl, G. M. (2000) J. Cell Biochem. Suppl. 35, 107–114. [DOI] [PubMed] [Google Scholar]
- 5.Komatsu, T., Ballestas, M. E., Barbera, A. J. & Kaye, K. M. (2002) Front. Biosci. 7, d726–d730. [DOI] [PubMed] [Google Scholar]
- 6.Chow, L. T. & Broker, T. R. (1997) in In Viral Pathogenesis, ed. Nathanson, N. (Lippincott–Raven, Philadelphia, PA), pp. 267–302.
- 7.zur Hausen, H. & de Villiers, E.-M. (1994) Annu. Rev. Microbiol. 48, 427–447. [DOI] [PubMed] [Google Scholar]
- 8.Walboomers, J. M., Jacobs, M. V., Manos, M. M., Bosch, F. X., Kummer, J. A., Shah, K. V., Snijders, P. J., Peto, J., Meijer, C. J. & Muñoz, N. (1999) J. Pathol. 189, 12–19. [DOI] [PubMed] [Google Scholar]
- 9.Yang, L., Li, R., Mohr, I. J., Clark, R. & Botchan, M. R. (1991) Nature 353, 628–632. [DOI] [PubMed] [Google Scholar]
- 10.Ustav, M. & Stenlund, A. (1991) EMBO J. 10, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang, C.-M., Ustav, M., Stenlund, A., Ho, T. F., Broker, T. R. & Chow, L. T. (1992) Proc. Natl. Acad. Sci. USA 89, 5799–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo, S.-R., Liu, J.-S., Broker, T. R. & Chow, L. T. (1994) J. Biol. Chem. 269, 24058–24065. [PubMed] [Google Scholar]
- 13.Seo, Y. S., Müller, F., Lusky, M. & Hurwitz, J. (1993) Proc. Natl. Acad. Sci. USA 90, 702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, L., Mohr, I., Fouts, E., Lim, D. A., Nohaile, M. & Botchan, M. (1993) Proc. Natl. Acad. Sci. USA 90, 5086–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sedman, J. & Stenlund, A. (1998) J. Virol. 72, 6893–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouts, E. T., Yu, X., Egelman, E. H. & Botchan, M. R. (1999) J. Biol. Chem. 274, 4447–4458. [DOI] [PubMed] [Google Scholar]
- 17.Lin, B.-Y., Makhov, A. M., Griffith, J. D., Broker, T. R. & Chow, L. T. (2002) Mol. Cell. Biol. 18, 6591–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, P., Copeland, W., Yang, L., Wang, T., Botchan, M. R. & Mohr, I. J. (1994) Proc. Natl. Acad. Sci. USA 91, 8700–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conger, K. L., Liu, J.-S., Kuo, S.-R., Chow, L. T. & Wang, T. S.-F. (1999) J. Biol. Chem. 274, 2696–2705. [DOI] [PubMed] [Google Scholar]
- 20.Han, Y., Loo, Y. M., Militello, K. T. & Melendy, T. (1999) J. Virol. 73, 4899–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J.-S., Kuo, S.-R., Broker, T. R. & Chow, L. T. (1995) J. Biol. Chem. 270, 27283–27291. [DOI] [PubMed] [Google Scholar]
- 22.Li, R. & Botchan, M. R. (1994) Proc. Natl. Acad. Sci. USA 91, 7051–7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, S.-Y. & Chiang, C.-M. (2001) Curr. Protoc. Mol. Biol. 3, 16.22.1–16.22.17. [Google Scholar]
- 24.Paulus, W., Baur, I., Boyce, F., Breakefield, X. & Reeves, S. (1996) J. Virol. 70, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bastien, N. & McBride, A. A. (2000) Virology 270, 124–134. [DOI] [PubMed] [Google Scholar]
- 26.Zou, N., Lin, B.-Y., Duan, F., Lee, K. Y., Jin, G., Guan, R., Yao, G., Lefkowitz, E. J., Broker, T. R. & Chow, L. T. (2000) J. Virol. 74, 3761–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou, N., Liu, J.-S. Kuo, S.-R., Broker, T. R. & Chow, L. T. (1998). J. Virol. 72, 3426–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, K.-Y., Broker, T. R. & Chow, L. T. (1998) J. Virol. 72, 4911–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerra, A. & Schmidt, T. G. M. (1999) Biomol. Eng. 16, 79–86. [DOI] [PubMed] [Google Scholar]
- 30.Yang, X., Zhang, F. & Kudlow, J. E. (2002) Cell 110, 69–80. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler, W. J., Hsu, T. C., Tousson, A. & Brinkley, B. R. (1987) Cell Motil. Cytoskeleton 7, 235–247. [DOI] [PubMed] [Google Scholar]
- 32.Lange, B. M. H. (2002) Curr. Opin. Cell Biol. 14, 35–43. [DOI] [PubMed] [Google Scholar]
- 33.D'Assoro, A. B., Lingle, W. L. & Salisbury, J. L. (2002) Oncogene 21, 6146–6153. [DOI] [PubMed] [Google Scholar]
- 34.Kopito, R. R. (2000) Trends Cell Biol. 10, 524–530. [DOI] [PubMed] [Google Scholar]
- 35.McBride, A. A., Schlegel, R. & Howley, P. M. (1988) EMBO J. 7, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shire, K., Ceccarelli, D. F. J., Avolio-Hunter, T. M. & Frappier, L. (1999) J. Virol. 73, 2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon, S. & Lambert, P. F. (1995) Proc. Natl. Acad. Sci. USA 92, 1654–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klaes, R., Woerner, S. M., Ridder, R., Wentzensen, N., Dürst, M., Schneider, A., Lotz, B., Melsheimer, P. & von Knebel Doeberitz, M. (1999) Cancer Res. 59, 6132–6136. [PubMed] [Google Scholar]
- 39.McClintock, B. (1951) Cold Spring Harbor Symposia Quant. Biol. 16, 13–47. [DOI] [PubMed] [Google Scholar]
- 40.Macville, M., Schrock, E., Padilla-Nash, H., Keck, C., Ghadimi, B. M., Zimonjic, D., Popescu, N. & Ried, T. (1999) Cancer Res. 59, 141–150. [PubMed] [Google Scholar]
- 41.Thorland, E. C., Myers, S. L., Gostout, B. S. & Smith, D. I. (2003) Oncogene 22, 1225–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.