Abstract
The neural F-box 42-kDa protein (NFB42) is a component of the SCFNFB42 E3 ubiquitin ligase that is expressed in all major areas of the brain; it is not detected in nonneuronal tissues. We previously identified NFB42 as a binding partner for the herpes simplex virus 1 (HSV-1) UL9 protein, the viral replication-initiator, and showed that coexpression of NFB42 and UL9 in human embryonic kidney (293T) cells led to a significant decrease in the level of UL9 protein. We have now found that HSV-1 infection promotes the shuttling of NFB42 between the cytosol and the nucleus in both 293T cells and primary hippocampal neurons, permitting NFB42 to bind to the phosphorylated UL9 protein, which is localized in the nucleus. This interaction mediates the export of the UL9 protein from the nucleus to the cytosol, leading to its ubiquitination and degradation via the 26S proteasome. Because the intranuclear localization of the UL9 protein, along with other viral and cellular factors, is an essential step in viral DNA replication, degradation of the UL9 protein in neurons by means of nuclear export through its specific interaction with NFB42 may prevent active replication and promote neuronal latency of HSV-1.
Herpes simplex virus 1 (HSV-1) is a clinically important, neurotropic human pathogen (1–4). After replication in primary infected epithelial cells of the skin or buccal mucosa, HSV-1 gains access to sensory nerve termini and establishes a latent infection in sensory neurons, primarily of the trigeminal and dorsal root ganglia (5, 6). The mechanism that leads to the establishment of latency is not understood.
The UL9 protein, which is the product of the HSV-1 UL9 gene, is strictly required for HSV-1 DNA replication. This protein performs multiple functions as a DNA replication initiator that binds to and unwinds DNA at origins of HSV-1 DNA replication, and also recruits the replication complex proteins required for viral DNA replication (7). The UL9 protein is a limiting component on which HSV-1 DNA replication depends, and may be the site at which HSV-1 DNA replication is regulated. We showed previously (8) that the appropriately phosphorylated form of the HSV-1 UL9 protein interacts with the human neural F-box 42-kDa protein (NFB42 or FBX2), a component of the ubiquitin ligase complex, is polyubiquitinated, and is then degraded via the 26S proteasome pathway (8).
Ubiquitin-mediated proteolysis has been shown to play a crucial role in a variety of cellular processes, including control of the cell cycle, regulation of gene expression, differentiation, signal transduction, apoptosis, DNA repair, DNA replication, and the immune response (9, 10). Degradation of target proteins via the ubiquitin-proteasome pathway requires at least three components: the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the ubiquitin-protein ligase E3. The ubiquitinated proteins are degraded by the 26S proteasome. The E3 proteins play a key role in the ubiquitin-proteasome pathway as specific recognition factors for the proteins to be degraded. One of these, NFB42, is a component of SCFNFB42 E3 ubiquitin ligase (Skp1-Cullin1-NFB42-Roc1) that is linked to the SCF complex by binding to Skp1 through its N-terminal F-box motif. NFB42 is expressed in all major areas of brain but is not detected in nonneural tissues (11). The factors required for ubiquitination and subsequent degradation of target proteins are found throughout the cell, including the cytosol, nucleus, endoplasmic reticulum, and cell-surface membranes (9, 12).
Because NFB42 is found primarily in the cytosol (11), whereas the UL9 protein is located predominantly in the nucleus (13), it was important to determine the mechanism that permits their interaction. We report here that HSV-1 infection promotes the shuttling of NFB42 between the cytosol and the nucleus in both 293T cells and in primary hippocampal neurons, and that NFB42 mediates the specific nuclear export of the UL9 protein.
Materials and Methods
Cell Culture, Transfection, and HSV-1 Infection. Human embryonic kidney 293T cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) heat-inactivated FBS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) at 37°C with 5% CO2 in a humidified incubator. For transfection, cells were plated in Lab-Tek chambered coverglasses (Nalge Nunc) coated with 1 mg/ml poly-d-lysine (Sigma). After 1 day, the cells were transfected with 500 ng of each DNA by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Hippocampal neurons were obtained from 1-day-old postnatal Sprague–Dawley rats and were cultured as described (14). Briefly, neurons were dissociated by trypsinization and were plated on the coverglasses described above. The cell culture media consisted of minimum essential medium (Invitrogen) supplemented with 5% FCS (Invitrogen)/20 mM glucose/10 mM Hepes/0.5 mM sodium pyruvate/2% B-27 supplement (Invitrogen)/1 μl/ml serum extender (Becton Dickinson). Cultures were maintained at 37°C in an atmosphere of 5% CO2 and were transfected 7 days after plating. Each coverslip was treated for 30 min at 37°C with a mixture of 500 ng of each DNA and 2 μl of Lipofectamine 2000 in Opti-MEM (Invitrogen) buffer (50 μl of transfection volume per coverslip). The neurons were subsequently returned to their original media. For HSV-1 infection, cells were infected with HSV-1 strain Δ305 (15) at a multiplicity of infection of 20 at 18 h after transfection.
Plasmid Constructs. The DNA sequences corresponding to the full-length UL9, ICP8, and NFB42 were amplified by PCR from HSV-1 genomic and HeLa cDNA, respectively, using the appropriate pairs of primers, and were subcloned into pECFP-N1 to generate pUL9-CFP and pICP8-CFP, and into pEYFP-N1 to generate pNFB42-YFP (Becton Dickinson). Each protein was expressed as a fusion to the N terminus of the fluorescent protein [cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP)]. The pcDNA3.1-NFB42 and pcDNA3.1-UL9 DNA constructs are described elsewhere (8).
Western Blot Analysis. Samples containing 7 × 106 293T cells were transiently transfected with 3 μg of pcDNA3.1-UL9, with or without 5 μg of pcDNA3.1-NFB42. Where indicated, samples cotransfected with NFB42 and UL9 were infected with HSV-1. To prepare a total cell extract, 293T cells were harvested 6 h after infection by centrifugation, were washed twice with PBS (140 mM NaCl/2.7 mM KCl/0 mM Na2HPO4/1.8 mM KH2PO4, pH 7.4), and were lysed in lysis buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1% Triton X-100) supplemented with the Complete Protease Inhibitor Mixture (Roche). The stepwise separation of cytosolic and nuclear extracts from 293T cells transfected with pcDNA3.1-NFB42 was performed by using NE-PER nuclear and cytoplasmic extraction reagents kit (Pierce). Protein concentrations were determined by the Bradford method (16) with BSA as a standard. Equal amounts of protein from each lysate were mixed with Laemmli sample buffer (60 mM Tris·HCl, pH 6.8/2% SDS/10% glycerol/100 mM DTT/0.005% bromophenol blue) and were subjected to SDS/PAGE. The proteins were transferred electrophoretically onto nitrocellulose membranes (Amersham Pharmacia Biosciences). After blocking and extensive washing of the membranes, the proteins were probed with rabbit polyclonal anti-UL9 antibody, rabbit anti-NFB42 antibody (a generous gift from R. Pittman, University of Pennsylvania School of Medicine, Philadelphia), or monoclonal anti-β-actin (Sigma). The immunoblots were examined by enhanced chemiluminescence (Amersham Pharmacia Biosciences).
Live Cell Microscopy and Fluorescence Quantitation. A spinning disk Nipkow confocal mircroscope (Perkin–Elmer) was used for dual color imaging by using the CFP- and YFP-labeled constructs. Cells were viewed by using an inverted Olympus I ×70 microscope with a 40× oil-immersion Olympus objective (1.35 numerical aperture), and images were acquired with a Hamamatsu charge-coupled device camera. CFP was excited with the 442-nm laser line of a helium-cadmium laser (Kimmon, Tokyo), while YFP was imaged with the 514-nm line of an argon ion laser (Melles Griot, Irvine, CA). Images were exported as 16-bit TIFF files. For quantitation of fluorescence, three or more individual cells for each condition were analyzed. After subtraction of background, regions of interest were selected in the nucleus and in the cytosol of each cell and the average intensity measured by using metamorph analysis software (Universal Imaging, Media, PA).
Results
UL9 Protein and ICP8 Are Localized Predominantly in the Nucleus, and NFB42 Is Localized in the Cytosol. To determine the subcellular localization of the UL9 protein, ICP8, the HSV-1 encoded single-stranded DNA-binding protein, and NFB42, the UL9 and ICP8 genes were fused to the pECFP-N1 vector to construct pUL9-CFP and pICP8-CFP, respectively, and the NFB42 gene was fused to the pEYFP-N1 vector to yield pNFB42-YFP. The subcellular localization of the three proteins after transfection was then analyzed by fluorescence confocal microscopy.
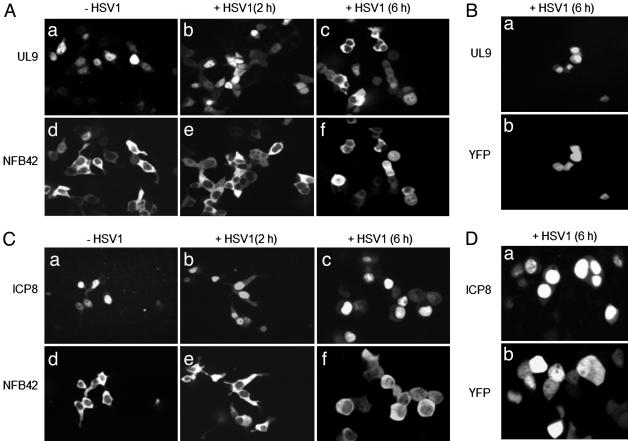
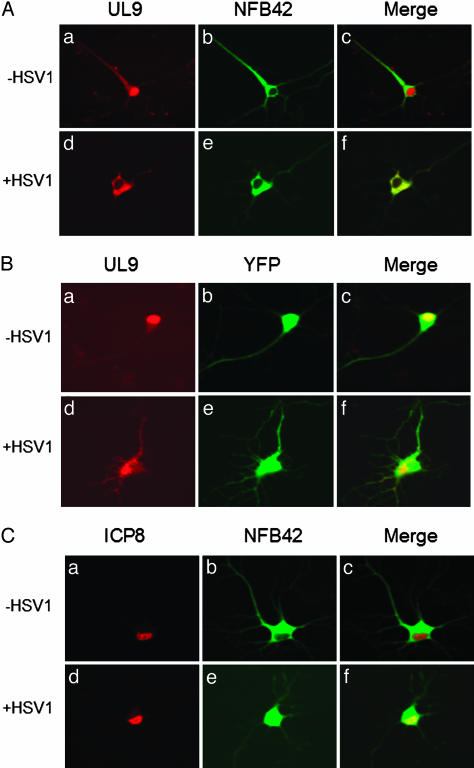
The UL9 protein and ICP8, both expressed from a CMV immediate-early promoter in transiently transfected 293T cells as well as in hippocampal neurons, were localized predominantly in the nucleus (Figs. 1Aa and 2 Aa and Ba for UL9 protein, and Figs. 1Ca and 2Ca for ICP8). In contrast, NFB42 expressed in both 293T cells and hippocampal neurons was localized primarily in the cytosol (Figs. 1 Ad and Cd and 2 Ab and Cb). NFB42 was highly enriched in dendrites (Fig. 2 Ab and Cb), which is consistent with a previous report showing that axons and dendrites are strongly stained with polyclonal anti-NFB42 antibody in pyramidal neurons (11).
Fig. 1.
The UL9 protein and NFB42 shuttle between the nucleus and cytosol in 293T cells after HSV-1 infection. Cells were cotransfected with expression plasmids containing UL9 and NFB42 (A), UL9 and vector (B), ICP8 and NFB42 (C), and ICP8 and vector (D), followed by superinfection of HSV-1, as described in Materials and Methods. Before infection with HSV-1, NFB42 is localized mainly in the cytosol (Ad and Cd), whereas the UL9 protein is predominantly localized in the nucleus (Aa). Nucleocytosolic shuttling of UL9 protein was observed in cells cotransfected with expression plasmids containing UL9 and NFB42, followed by HSV-1 infection (A), but not with expression plasmids containing ICP8 and NFB42 (C). In the absence of exogenous NFB42, viral infection had no effect on translocation of the UL9 protein (B).
Fig. 2.
Shuttling of UL9 protein and NFB42 between the nucleus and cytosol in primary hippocampal neuronal cells after HSV-1 infection. Cells were cotransfected with expression plasmids containing UL9 and NFB42 (A), UL9 and vector (B), and ICP8 and NFB42 (C), followed by infection with HSV-1, as described in Materials and Methods. Similar results were observed for hippocampal neurons and 293T cells, except for the case of cotransfection with the plasmid expressing UL9 protein and pEYFP vector. Under these conditions, UL9 was mainly found in the cytosol (B).
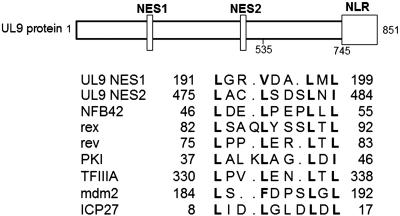
Proteins that contain a nuclear localization signal (NLS) can be transported into the nucleus unless the NLS is masked or blocked within the protein. NFB42 lacks an NLS, which is consistent with its cytosolic localization. In contrast, both the UL9 protein and ICP8 contain NLSs. The NLS for the UL9 protein maps within the C-terminal 107 amino acid residues (Fig. 3 and ref. 13), and NLS for ICP8 maps within the C-terminal region between amino acid residues 1080 and 1135 (17).
Fig. 3.
Putative NESs in UL9 protein and NFB42. The primary structure of UL9 protein is shown. The two putative NESs are positioned within the first 535 amino acids of UL9 protein, which are required for dimerization. The nuclear localization region (NLR) is positioned between residues 745 and 851. The sequence is aligned with the known NESs based on NES consensus L-x (2, 3)-[LIVFM]-x (2, 3)-L-x-[LI] (37). Numbers indicate the position of the amino acids within the proteins. The hydrophobic residues critical for NES function are bold. Dots are inserted to equalize the pairing between conserved hydrophobic residues. The references for the NES elements are as follows: human T cell lymphotrophic virus (HTLV) rex (38); HIV-1 rev (39); cAMP-dependent protein kinase inhibitor (PKI) (40); transcription factor TFIIIA (41); oncoprotein mdm2 (29); and HSV-1 immediate-early protein ICP27 (42).
HSV-1 Infection Promotes the Nuclear Import of NFB42 and the Nuclear Export of UL9 Protein. To examine the effect of HSV-1 infection on the intracellular localization of the UL9 protein and NFB42, 293T cells and hippocampal neurons were superinfected with HSV-1 at 18 h after transfection with pNFB42-YFP and pUL9-CFP. Translocation of NFB42 from the cytosol into the nucleus was observed in both types of cells as early as 6 h after infection (Figs. 1 Af and Cf and 2 Ae and Ce). Because it lacks an NLS, translocation of NFB42 from the cytosol to the nucleus is most likely due to HSV-1 infection, either as a consequence of a conformational change in NFB42, or by its association with another NLS-containing protein capable of nuclear translocation.
Translocation of the UL9 protein from the nucleus to the cytosol was also observed at 6 h after tinfection of both 293T cells and hippocampal neurons that had been cotransfected with pUL9-CFP and pNFB42-YFP (Figs. 1 Ac and 2 Ad). In contrast to the UL9 protein, HSV-1 infection did not result in translocation of ICP8 from nucleus to cytosol after transfection with pNFB42-YFP and pICP8-CFP in either 293T cells or hippocampal neurons (Figs. 1Cc and 2Cd). Thus, the translocation of the UL9 protein from nucleus to the cytosol appears to be specific.
A significant difference in UL9 protein translocation was found between 293T cells and hippocampal neurons transfected with only pUL9-CFP, followed by HSV-1 infection. Translocation of the UL9 protein from the nucleus to the cytosol was observed in hippocampal neurons (Fig. 2Bd), whereas no change in UL9 protein localization occurred in 293T cells in the absence of transfection with pNFB42-YFP (Fig. 1Ba). Because NFB42 is known to be expressed in all major areas of brain but is not detected in nonneural tissues (11), this result is very likely due to endogenous NFB42 present in the hippocampal neurons. This finding strongly implicates NFB42 in the nuclear export of the UL9 protein. Table 1 summarizes the intracellular distribution of the UL9 protein and NFB42 before and after HSV-1 infection.
Table 1. Ratio of cytosolic to nuclear signals.
| 293T cells
|
Primary hippocampal neurons
|
|||
|---|---|---|---|---|
| No infection | HSV-1 infection | No infection | HSV-1 infection | |
| Single transfection | ||||
| UL9 | 0.1 | 0.2 | 0.3 | 5.0 |
| NFB42 | 3.6 | 1.6 | 4.9 | 2.1 |
| Cotransfection | ||||
| UL9 | 0.2 | 3.7 | 0.3 | 5.5 |
| NFB42 | 3.7 | 1.6 | 7.6 | 5.2 |
For quantitation of fluorescence, three or more individual cells for each condition were analyzed. After subtraction of background, regions of interest were selected in the nucleus and in the cytosol of each cell, and the average intensity was measured by using metamorph analysis software.
A Nuclear Export Signal Is Present in Both NFB42 and the UL9 Protein. Similar to nuclear import, nuclear export of proteins depends on the presence of a specific amino acid sequence, the nuclear export signal (NES) within the protein. The NES is a short leucine-rich sequence that has been identified in a variety of proteins, the best studied of which is the HIV Rev protein (18). Nuclear export is mediated by the chromosome maintenance region 1 protein, which binds to the leucine-rich NES directly and mediates the export of NES-containing proteins in a RanGTP-dependent manner (19–21). The UL9 protein contains two sequences that resemble several of the previously characterized NESs; NFB42 contains one such sequence (Fig. 3). The UL9 protein exists in solution either as a homodimer (22) or as multimer (probably a tetramer) in the presence of hTid-1, a human homologue of the Escherichia coli chaperone DnaJ (23). The two putative NESs within the UL9 protein form part of the UL9 protein dimerization domain. Thus, dimerization of the UL9 protein may lead to masking of the NESs, resulting in its nuclear localization. The UL9 protein in the nucleus may therefore be exported into the cytosol through an interaction with the nuclear NFB42.
HSV-1 Infection Accelerates Degradation of the UL9 Protein. Our earlier studies (8) had shown that cotransfection of 293T cells with constructs expressing the UL9 protein and NFB42 led to substantial degradation of the UL9 protein through the ubiquitin ligase-26S proteasome system, even in the absence of HSV-1 infection. This result suggests that transfected 293T cell nuclei do contain some NFB42. We therefore examined the intracellular distribution of transiently expressed NFB42 in 293T cells. Cytosolic and nuclear fractions were prepared from 60-mm culture dishes of 293T cells transfected with pcDNA3.1-NFB42. As shown in Fig. 4, a significant amount of NFB42 (24% of the total) expressed in the 293T cells was present in the nucleus.
Fig. 4.
The intracellular distribution of transiently expressed NFB42. Cytosolic and nuclear fractions were determined for 293T cells transfected with pcDNA3.1-NFB42, as described in Materials and Methods.
To examine the effect of HSV-1 infection on the degradation of the UL9 protein, 293T cells were transiently transfected with constructs expressing the UL9 protein alone, or expressing NFB42 and the UL9 protein, with or without HSV-1 infection. As shown in Fig. 5, and consistent with our earlier findings (8), cotransfection of cells with constructs expressing UL9 and NFB42 decreased the UL9 protein level to 54% of that with UL9 alone (lanes 1 and 2). HSV-1 infection of cells expressing both UL9 and NFB42 decreased the level of UL9 protein to <4% of that in cells transfected with UL9, alone in the absence of infection (lane 3).
Fig. 5.
Effect of HSV-1 infection on the level of UL9 protein in 293T cells cotransfected with UL9 and NFB42. The 293T cells were transiently transfected with pcDNA3.1-UL9 (lane 1) or were cotransfected with pcDNA3.1-UL9 and pcDNA3.1-NFB42, and were followed by HSV-1 infection (lane 3) or mock-infection (lane 2). The cells were lysed and the lysates (50 μg) were analyzed by SDS/PAGE, transferred to a nitrocellulose membrane, and then blotted with polyclonal anti-UL9 antibody as described in Materials and Methods. Equivalence of loading was controlled with anti-β-actin antibody.
Discussion
HSV-1 employs replication compartments within the nucleus to concentrate the viral and cellular factors required for viral replication (17). Specifically, the UL9 protein and ICP8 are colocalized in replication compartments, which are sites of viral synthesis (13). The intranuclear localization of the UL9 protein and ICP8 appear to be an essential step in HSV-1 DNA replication (24, 25). We have shown that HSV-1 infection of both 293T cells and primary hippocampal neurons causes the shuttling of NFB42, a component of SCFNFB42 E3 ubiquitin ligase, between the cytosol and the nucleus, thereby leading to the nuclear export of the UL9 protein to the cytosol, where it can be polyubiquitinated and degraded by 26S proteasome.
Degradation of the p53 tumor suppressor is an example of the nucleocytosolic shuttling of an E3 ubiquitin ligase to promote the degradation of target proteins. The mouse homolog of Mdm2 (HDM2) is known to be an E3 ubiquitin ligase for p53 (26–28). It has been suggested that HDM2 functions as a shuttle protein that binds to p53 in the nucleus and ferries it to the cytosol through the NES of HDM2 (29, 30). The nuclear export of p53 by interaction with HDM2 is a key step in its degradation. However, some HDM2-mediated p53 degradation does occur in the nucleus to permit a tighter and faster control during the down-regulatory phase, when an active p53 program needs to be turned off quickly (31). Nuclear export signals are found in both HDM2 and p53 (29, 32). One p53 NES located within the C-terminal tetramerization domain is masked and inactive when p53 forms a tetramer (32); the other NES located within the N terminus is inhibited by DNA-damage-induced phosphorylation, suggesting that this NES is active only in unstressed cells (33). It is, therefore, believed that the NES of MDM2 is essential for the export of p53 (34, 35). p53 is also degraded by the human papillomavirus E6 protein. HPV E6 recruits the cellular E6AP, and together they act as a potent E3 ubiquitin ligase for p53 (36).
By analogy to p53 degradation, there are two plausible models by which NFB42 mediates the nuclear export of UL9 protein. The first is that the NFB42 imported into the nucleus resulting from HSV-1 infection simply binds to the UL9 protein, thereby facilitating the nuclear export of UL9 protein through the NES of NFB42. The other is that the binding of NFB42 to UL9 protein in which the NES is masked results in a conformational change within the UL9 protein, leading to the unmasking of the NES of the UL9 protein, thereby facilitating nuclear export. It remains to be determined whether UL9 protein degradation occurs exclusively on cytosolic proteasomes.
In summary, we have shown that HSV-1 infection promotes the nuclear import of NFB42, and that the NFB42 then mediates nuclear export specifically of the UL9 protein, leading to its cytosolic degradation. We suggest that the rapid degradation of the UL9 protein in neurons by means of nuclear export through a specific interaction with NFB42 is related to the establishment of the neuronal latency of HSV-1.
Acknowledgments
We thank Dr. Randall Pittman (University of Pennsylvania School of Medicine) for providing the polyclonal anti-NFB42 antibody and Dr. K. J. Huang for helpful discussion. This work was supported by National Institutes of Health Grants AI26538 (to I.R.L.) and MH064801 (to M.L.C.).
Abbreviations: HSV-1, herpes simplex virus type 1; NLS, nuclear localization signal; NES, nuclear export signal; NFB42, neural F-box 42-kDa protein; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; HDM2, the mouse homolog of Mdm2.
References
- 1.Croen, K. D., Ostrove, J. M., Dragovic, L. J., Smialek, J. E. & Straus, S. E. (1987) N. Engl. J. Med. 317, 1427–1432. [DOI] [PubMed] [Google Scholar]
- 2.Nahmias, A. J. & Roizman, B. (1973) N. Engl. J. Med. 289, 781–789. [DOI] [PubMed] [Google Scholar]
- 3.Whitley, R. J. & Gnann, J. W. (2002) Lancet 359, 507–513. [DOI] [PubMed] [Google Scholar]
- 4.Jones, C. (2003) Clin. Microbiol. Rev. 16, 79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, C. (1998) Adv. Virus Res. 51, 81–133. [DOI] [PubMed] [Google Scholar]
- 6.Wagner, E. K. & Bloom, D. C. (1997) Clin. Microbiol. Rev. 10, 419–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, S. S. & Lehman, I. R. (1997) Proc. Natl. Acad. Sci. USA 94, 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eom, C. Y. & Lehman, I. R. (2003) Proc. Natl. Acad. Sci. USA 100, 9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickman, M. H. & Ciechanover, A. (2002) Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- 10.Hershko, A., Ciechanover, A. & Varshavsky, A. (2000) Nat. Med. 6, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 11.Erhardt, J. A., Hynicka, W., DiBenedetto, A., Shen, N., Stone, N., Paulson, H. & Pittman, R. N. (1998) J. Biol. Chem. 273, 35222–35227. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz, A. L., Trausch, J. S., Ciechanover, A., Slot, J. W. & Geuze, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5542–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik, A. K., Shao, L., Shanley, J. D. & Weller, S. K. (1996) Virology 224, 380–389. [DOI] [PubMed] [Google Scholar]
- 14.Shen, K. & Meyer, T. (1999) Science 284, 162–166. [DOI] [PubMed] [Google Scholar]
- 15.Post, L. E., Mackem, S. & Roizman, B. (1981) Cell 24, 555–565. [DOI] [PubMed] [Google Scholar]
- 16.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 17.Taylor, T. J., McNamee, E. E., Day, C. & Knipe, D. M. (2003) Virology 309, 232–247. [DOI] [PubMed] [Google Scholar]
- 18.Pollard, V. W. & Malim, M. H. (1998) Annu. Rev. Microbiol. 52, 491–532. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod, M., Ohno, M., Yoshida, M. & Mattaj, I. W. (1997) Cell 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M. & Nishida, E. (1997) Nature 390, 308–311. [DOI] [PubMed] [Google Scholar]
- 21.Askjaer, P., Jensen, T. H., Nilsson, J., Englmeier, L. & Kjems, J. (1998) J. Biol. Chem. 273, 33414–33422. [DOI] [PubMed] [Google Scholar]
- 22.Bruckner, R. C., Crute, J. J., Dodson, M. S. & Lehman, I. R. (1991) J. Biol. Chem. 266, 2669–2674. [PubMed] [Google Scholar]
- 23.Eom, C. Y. & Lehman, I. R. (2002) Proc. Natl. Acad. Sci. USA 99, 1894–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan, M. P. & Knipe, D. M. (1983) Mol. Cell. Biol. 3, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrington-Lawrence, S. D. & Weller, S. K. (2003) J. Virol. 77, 4237–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. (1997) Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. (1997) Nature 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 28.Honda, R., Tanaka, H. & Yasuda, H. (1997) FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- 29.Roth, J., Dobbelstein, M., Freedman, D. A., Shenk, T. & Levine, A. J. (1998) EMBO J. 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, W. & Levine, A. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3077–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirangi, T. R., Zaika, A. & Moll, U. M. (2002) FASEB J. 16, 420–422. [DOI] [PubMed] [Google Scholar]
- 32.Stommel, J. M., Marchenko, N. D., Jimenez, G. S., Moll, U. M., Hope, T. J. & Wahl, G. M. (1999) EMBO J. 18, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Y. & Xiong, Y. (2001) Science 292, 1910–1915. [DOI] [PubMed] [Google Scholar]
- 34.Boyd, S. D., Tsai, K. Y. & Jacks, T. (2000) Nat. Cell Biol. 2, 563–568. [DOI] [PubMed] [Google Scholar]
- 35.Geyer, R. K., Yu, Z. K. & Maki, C. G. (2000) Nat. Cell Biol. 2, 569–573. [DOI] [PubMed] [Google Scholar]
- 36.Talis, A. L., Huibregtse, J. M. & Howley, P. M. (1998) J. Biol. Chem. 273, 6439–6445. [DOI] [PubMed] [Google Scholar]
- 37.Bogerd, H. P., Fridell, R. A., Benson, R. E., Hua, J. & Cullen, B. R. (1996) Mol. Cell. Biol. 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmeri, D. & Malim, M. H. (1996) J. Virol. 70, 6442–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer, U., Huber, J., Boelens, W. C., Mattaj, I. W. & Luhrmann, R. (1995) Cell 82, 475–483. [DOI] [PubMed] [Google Scholar]
- 40.Wen, W., Meinkoth, J. L., Tsien, R. Y. & Taylor, S. S. (1995) Cell 82, 463–473. [DOI] [PubMed] [Google Scholar]
- 41.Fridell, R. A., Fischer, U., Luhrmann, R., Meyer, B. E., Meinkoth, J. L., Malim, M. H. & Cullen, B. R. (1996) Proc. Natl. Acad. Sci. USA 93, 2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin, R. M. (1998) Genes Dev. 12, 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]