Abstract
The RlmA class of enzymes (RlmAI and RlmAII) catalyzes N1-methylation of a guanine base (G745 in Gram-negative and G748 in Gram-positive bacteria) of hairpin 35 of 23S rRNA. We have determined the crystal structure of Escherichia coli RlmAI at 2.8-Å resolution, providing 3D structure information for the RlmA class of RNA methyltransferases. The dimeric protein structure exhibits features that provide new insights into its molecular function. Each RlmAI molecule has a Zn-binding domain, responsible for specific recognition and binding of its rRNA substrate, and a methyltransferase domain. The asymmetric RlmAI dimer observed in the crystal structure has a well defined W-shaped RNA-binding cleft. Two S-adenosyl-l-methionine substrate molecules are located at the two valleys of the W-shaped RNA-binding cleft. The unique shape of the RNA-binding cleft, different from that of known RNA-binding proteins, is highly specific and structurally complements the 3D structure of hairpin 35 of bacterial 23S rRNA. Apart from the hairpin 35, parts of hairpins 33 and 34 also interact with the RlmAI dimer.
Keywords: rrmA, methyltransferase, antibiotic resistance, RNA-binding protein
MLS (macrolide, lincosamide, streptogramin B) antibiotics such as erythromycin, tylosin, and spiramycin are used in treating bacterial infections in human and in animals (1). MLS antibiotics bind to the large ribosomal subunit (2) and inhibit translation, possibly by blocking the protein exit channel of the ribosome (3–6). The effectiveness of MLS antibiotics is increasingly limited by the emergence of resistant bacterial strains (1). Certain modifications of bacterial rRNA are known to confer resistance to MLS antibiotics (7, 8). One of the most common forms of bacterial rRNA modification is nucleotide methylation (9); for example, 10 methylations of 16S rRNA and 14 methylations of 23S rRNA nucleotides are reported (10) for Escherichia coli. Although most of these modifications on rRNA occur before the formation of the ribosomal complex (11), they primarily cluster around the catalytic center of the ribosome (12). Methylated nucleotide G748 functions synergistically with a methylated A2058 nucleotide to confer resistance to certain MLS antibiotics (13, 14).
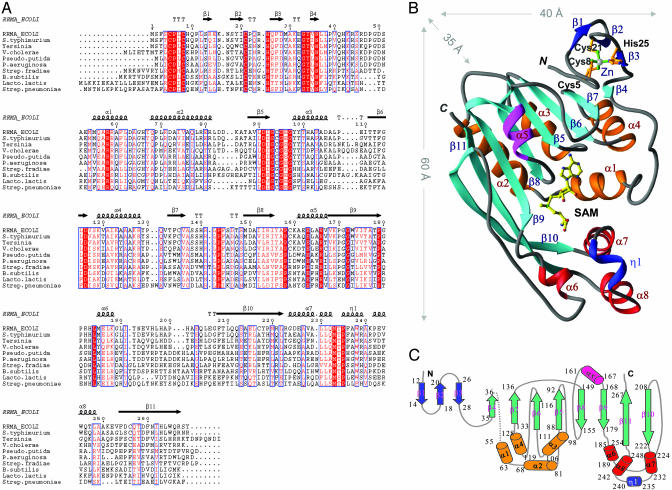
The N1-methylation of nucleotides G745 and G748 is carried out by rRNA large subunit methyltransferases RlmAI and RlmAII (formally known as rrmA and TlrB) enzymes, respectively (14). RlmA enzymes are only present in bacteria (15). However, the methyltransferase (MTase) domains of these enzymes exhibit amino acid sequence similarity with functionally related enzymes from eukaryotic and archea organisms and constitute a large, structurally uncharacterized protein domain family (16). The RlmA class I (RlmAI) enzyme is present in Gram-negative, and the RlmA class II (RlmAII) enzyme is present in Gram-positive bacteria (17, 18). Comparison of the amino acid sequences of RlmAI and RlmAII enzymes indicates that these enzyme classes are homologous (Fig. 1A); ≈29% of residues are conserved (18) across the species. Both the RlmA classes (I and II) contain a conserved MTase domain and use S-adenosyl-l-methionine (SAM) as the methyl group donor (19). Despite functional similarity, RlmA enzymes from Gram-positive bacteria have a characteristic difference from those of Gram-negative bacteria: RlmAI methylates G745 (11, 17), whereas RlmAII methylates G748 (20) at N1 position of the nucleotide bases. Both of these nucleotides, G745 and G748, are located in hairpin 35 of 23S rRNA.
Fig. 1.
(A) Amino acid sequence alignment (40) of selected RlmAI enzymes from Gram-negative (top six sequences) and RlmAII enzymes from Gram-positive (bottom four sequences) bacteria. Conserved amino acids are in red. The secondary structure elements of E. coli RlmAI (RRMA_ECOLI) determined in this work are mapped onto the alignment. (B) A ribbon diagram (41) of the E. coli RlmAI monomer structure. The three-strand small antiparallel β-sheet, a part of the Zn-binding domain, is in blue, and the eight-stranded mixed β-sheet of the MTase domain is in cyan. The helices, except helix α5 (pink), are bundled into two groups (orange and red). The only 310-helix in the structure (η1) is colored blue. The SAM substrate (in yellow) is bound at the center of the MTase domain. (C) A 2D representation of the fold of RlmAI. The color code of the secondary structure elements is the same as in B. The amino acid positions are numbered at the beginning and end of the secondary structure elements.
E. coli RlmAI is one of the best characterized RlmA enzymes (17, 21–23). Modifications to nucleotides of rRNA hairpins 33, 34, and 35 affect methylation by RlmAI (11). A G745-deficient E. coli strain (17) has shown slower growth rate as well as increased resistance to ribosome-binding antibiotic viomycin, which inhibits by blocking translation of peptidyl-tRNA. Here we report the x-ray crystal structure of E. coli RlmAI at 2.8-Å resolution. In addition, we describe modeling of the RlmAI/rRNA complex aimed at understanding the specific recognition of this rRNA fragment, and the mechanism of N1-methylation of G745 and G748.
Materials and Methods
Cloning, Expression, and Purification. E. coli gene rrmA coding for RlmAI (Northeast Structural Genomics Consortium Target ID ER19; www.nesg.org) was cloned into a pET21 (Novagen) derivative, generating plasmid pER19–21. E. coli BL21 (DE3) pMGK, a rare codon-enhanced strain, was transformed with pER19–21. A single isolate was cultured in MJ9 (24) minimal media containing selenomethionine (Se-Met) to produce Se-Met-labeled RlmAI protein (25). Initial growth was carried out at 37°C until the OD600 of the culture reached 1.0. The incubation temperature was then decreased to 17°C, and protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM. After overnight incubation at 17°C, the cells were harvested by centrifugation.
C terminus LEHHHHHH-tagged Se-Met RlmAI was purified by standard methods. Cell pellets were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0/300 mM NaCl/10 mM imidazole/5 mM 2-mercaptoethanol) and disrupted by sonication. The resulting lysate was clarified by centrifugation at 26,000 × g for 45 min at 4°C. The supernatant was loaded onto a nickel-nitrilotriacetic acid column (Qiagen, Valencia, CA) and eluted in lysis buffer containing 250 mM imidazole. Fractions containing the partially purified RlmAI were pooled and loaded onto a gel filtration column (Superdex 75, Amersham Pharmacia), and eluted in Buffer A (10 mM Tris, pH 7.5/5 mM DTT/10 mM NaCl/0.02% sodium azide). The resulting purified RlmAI protein was buffer exchanged and concentrated in 10 mM Tris, pH 7.5/5 mM DTT to 10 mg/ml. Sample purity (>97%) and molecular mass (31.5 kDa) were verified by SDS/PAGE and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry, respectively. The yield of purified protein was ≈100 mg per 1 liter of bacterial culture.
Crystallization. A sample of RlmAI at a concentration of ≈1.0 mg/ml in 10 mM Tris·HCl was used for dynamic light scattering measurements using a Protein Solutions DynaPro light-scattering device. Radius of the sample based on 25 consecutive readings was 344 Å with a polydispersity of ≈43% (a standard value for most crystallizing proteins is <25%). The calculated average molecular mass of the large RlmAI aggregates observed in these measurements (radius ≈334 Å)is ≈1.33 × 104 kDa, whereas the molecular mass of an RlmAI monomer is 31.5 kDa.
Crystallization conditions for the RlmAI protein were surveyed by using hanging drop vapor diffusion techniques and the Hampton Crystal Screen I and II and PEG/ION screen kits. Initial trials with protein concentrations of ≈10 mg/ml did not give any positive indications of crystals, and most of the drops precipitated. Use of a lower concentration of protein (≈6 mg/ml) yielded fiber-like micro crystals using Hampton Crystal Screen II no. 15 (0.5 M ammonium sulfate/1.0 M lithium sulfate/0.1 M sodium citrate, pH 6.5). After numerous optimization attempts, the hanging drop setup with 4 mg/ml protein in 10 mM Tris·HCl, pH 7.5/5 mM SAM/5 mM DTT produced the best crystals when vapor diffused against the above crystallization solution. The crystals grew to optimum size of 0.1 × 0.1 × 0.05 mm3 in ≈4 weeks at 22°C.
Data Collection and Structure Determination. Se-Met E. coli RlmAI crystals were mounted on cryo-loops, cryoprotected by dipping in solution containing 20% ethylene glycol, and flash-cooled in liquid N2. Multiple anomalous diffraction (MAD) data were collected at X12C NSLS, Brookhaven National Laboratory (BNL) from one flash-cooled crystal. The data (Table 1) were processed to 3.2-Å resolution by using denzo/scalepack (26). Another crystal with comparable dimensions was used to collect higher resolution data at the F1 beam line of the Cornell High Energy Synchrotron Source (CHESS), and processed at 2.8-Å resolution. Thirteen Se sites were found by using the Direct Methods implemented in snb 1.0 (27). The phases were calculated at 3.5-Å resolution, by solve 2.03 using the Se sites, and extended to 3.2-Å resolution by using NCS averaging and solvent correction methods implemented in resolve (28). The electron density calculated at 3.2-Å resolution was well defined, and most of the amino acid residues could be modeled manually. Later, 2.8-Å resolution data were used to refine the structure. Cycles of model building (using o; ref. 29) and refinement [initially by using refmac v 5.1.24 (30) implemented in ccp4 v4.2.1, and later by using cns 1.1 (31)] augmented the experimental phases and allowed identification of the remaining amino acid positions. The final model was refined to R = 0.248 and Rfree = 0.296 (Table 1).
Table 1. Crystallographic parameters, x-ray data, and refinement statistics for E. coli RlmAI.
| Se-λ1 | Se-λ2 | Se-λ3 | λhigh resolution | |
|---|---|---|---|---|
| Data collection facility | BNL X12C | BNL X12C | BNL X12C | CHESS F1 |
| Wavelength (λ, Å | 0.97889 | 0.97874 | 0.9500 | 0.9160 |
| Resolution range, Å | 50.0-3.2 | 50.0-3.2 | 50.0-3.2 | 50.0-2.8 |
| Number of reflections (no. of observations) | 28,604 (117,100) | 28,528 (118,011) | 28,354 (102,647) | 21,876 (88,268) |
| Completeness | 95.6 | 95.7 | 94.8 | 93.0 |
| Average I/σ(I) | 4.7 | 3.7 | 4.2 | 11.0 |
| Rmerge on I* | 0.175 | 0.206 | 0.183 | 0.106 |
| σ cutoff | I < -1σ(I) | I < -1σ(I) | I < -1σ(I) | I < -0.5σ(I) |
| Mean figure of merit | 0.40 (40.0-3.5 Å resolution) | |||
| Unit cell constants (space group) | a = 107.10, b = 122.36, and c = 142.68 Å (1222) | a = 107.19, b = 122.28, c = 143.14 Å | ||
| Data set used in structure refinement | ||||
| Resolution range, Å | 20-2.8 Å | |||
| Total number of reflections (Rfree set) | 21,804 (1,138) | |||
| Completeness (Rfree set) | 93% (5%) | |||
| Cutoff criteria | |Fo| ≥ 1.0 σ(Fo) | |||
| Number of atoms refined (nonprotein atoms) | 4,345 (165) | |||
| Rcryst† | 0.248 | |||
| Rfree | 0.296 | |||
| rms deviation | ||||
| Bond length, Å | 0.012 | |||
| Bond angle, ° | 1.7 | |||
Rmerge = ΣhklΣi|I(hkl)i - 〈I(hkl)〉|/ ΣhklΣi〈I(hkl)i〉.
Rcryst = Σhkl|Fo(hkl) - kFc(hkl)|/ Σhkl|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
Results and Discussion
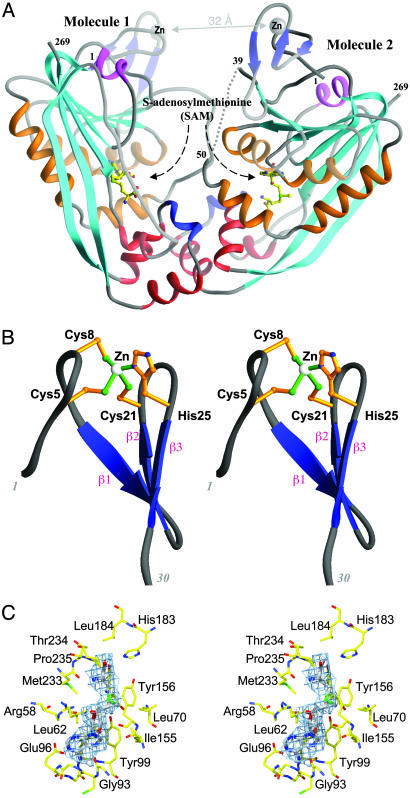
Overall Structure of RlmAI. The crystal structure of E. coli RlmAI is shown in Fig. 1B, and a schematic representation of the arrangement of the secondary structure elements is shown in Fig. 1C. The structure was determined by Se-Met–MAD method and refined to 2.8-Å resolution. RlmAI crystallized as a dimer per asymmetric unit (Fig. 2A), with dimensions 85 × 60 × 35 Å3. The two monomers (each having molecular mass of 31.5 kDa and 269 +8 tag amino acid residues) within the dimer have an unusual asymmetric arrangement in which one monomer relates to the other by ≈160° rotation about a two-fold noncrystallographic symmetry axis. The dimer contains a wide “W-shaped” cleft, a putative binding site for the rRNA substrate. The rms deviation for superposition of Cα-atoms of the two monomers is 1.1 Å.
Fig. 2.
(A) Ribbon representation of an asymmetric dimer, as found in the crystal structure, showing a well defined RNA-binding cleft. The deep W-shaped cleft has two Zn-fingers at the top and two SAM molecules at the bottom. The color code of the ribbon is same as in Fig. 1B. (B) Stereo view of the Zn-finger motif of RlmAI. (C) Stereo view of SAM binding region of an RlmAI molecule. The |Fo| – |Fc| electron density (in cyan) covering the SAM molecule (in gray) was calculated at 2.8-Å resolution based on the phasing by protein atoms only and is displayed at 2.0σ. The surrounding amino acids are in yellow.
The secondary structure of an RlmAI monomer includes eleven β-strands, eight α-helices, and one 310-helix (Fig. 1 B and C). The first three N-terminal β-strands form a small antiparallel β-sheet, a part of a Zn-binding domain (Fig. 2B), and the remaining eight strands form a large twisted mixed β-sheet that contains a characteristic MTase fold. An N-terminal Zn-binding domain (amino acids 1–35) and a C-terminal MTase domain (amino acids 51–269) are connected by a flexible linker of 12–15 aa. This linker is partially ordered in molecule 1 and completely disordered in molecule 2 of the crystallographic dimer. In the MTase domain, the two C-terminal β-strands β10 and β11 are curved and unusually long (≈50 Å in length), each containing 14–15 aa.
The base of the W-shaped RNA-binding cleft is formed by two methyltransferase domains, one per monomer. Two valleys of the W-shaped cleft contain two SAM molecules, one bound to each monomer (Fig. 2A). The helices α6, α7, α8, and η1 (310-helix) as well as parts of helices α1 from each monomer are clustered to form the RlmAI dimer interface. In addition to these interactions between RlmAI monomers, there are extensive interactions between RlmAI dimeric units in the crystal structure. The large β10-strand of molecule 1 interacts with the β10-strand of a crystallographic symmetry related molecule 2 to form an extended 16-strand β-sheet. These two distinct sets of intermolecular interactions for RlmAI molecules, (i) between the monomer units of the dimer and (ii) between these dimers, as seen in the crystal structure, might also exist in solution and could be responsible for formation of the large aggregates (of average radius 344 Å) observed in dynamic light scattering measurements.
Zn-Binding Domain. The N-terminal 35 amino acid residues of RlmAI form a Zn-binding domain that appears to be important in rRNA recognition. Within the Zn-binding domain, conserved amino acids Cys-5, Cys-8, Cys-21, and His-25 coordinate with a single Zn ion. The presence of Zn ion was evident from the crystallographic study and was further confirmed by inductively coupled plasmon resonance spectroscopy. The Zn-binding domain, which is present in all members of both the RlmA enzyme classes (Fig. 1A), has a novel Cys3His Zn-finger fold (Fig. 2B); its amino acid consensus sequence (Cys-Pro-X-Cys-12/13X-Cys-3/4X-His) and 3D structure are different from those of previously characterized Zn-finger structures.
The two Zn ions, positioned at the two top edges of the W-shaped RNA-binding cleft, are ≈32 Å apart; two highly conserved Cys-Pro-Leu-Cys loops (amino acids 5–8, a part of the Zn-finger) are ≈24 Å apart. Based on rRNA docking (as discussed later), the Cys-Pro-Leu-Cys loops and His-25 appear to be involved in recognition and binding of the rRNA substrate, hairpin 35 region of 23S rRNA. The Zn-binding domain of molecule 2 is less ordered, with an average B factor of 82 Å2, compared with an average B factor of 52 Å2 for the corresponding domain in molecule 1. The Zn-binding domains, particularly of molecule 2, and the loops joining them with the MTase domains may adjust their positions upon interacting with the rRNA substrate and could consequently be stabilized by RNA/protein interactions.
SAM Binding. RlmA enzymes use SAM as methyl group donor (19). As mentioned above, SAM molecules are bound to the MTase domains of both the RlmAI monomers (Fig. 2A). Difference electron density maps clearly define the mode of binding of SAM in the RlmAI enzyme structure (Fig. 2C). Relatively higher B factors of the SAM molecules, compared to those of the surrounding protein atoms, indicate partial occupancy (or positional disorder) of these substrate molecules. The amino acid residues that take part in SAM binding, including Arg-58-Leu-62, Tyr-67, Leu-70, Gly-93–Tyr-99, Ile-155–Tyr-156, His-183–Leu-184, and Met-233–Pro-235 (Fig. 2C), are either identical or of similar types in homologous RlmAI and RlmAII enzymes (Fig. 1A). Most of the conserved amino acids interacting with the SAM molecule, except those in α1 helices, are located on structurally flexible regions such as polypeptide loops and the tips of helices pointing toward the SAM-binding regions. The two SAM molecules, although bound in similar regions of the monomers of RlmAI dimer, differ in their precise orientations and specific interactions with protein atoms. Presumably, binding of the RNA substrate is necessary for a SAM molecule to bind to the RlmAI enzyme in a proper orientation for MTase catalysis.
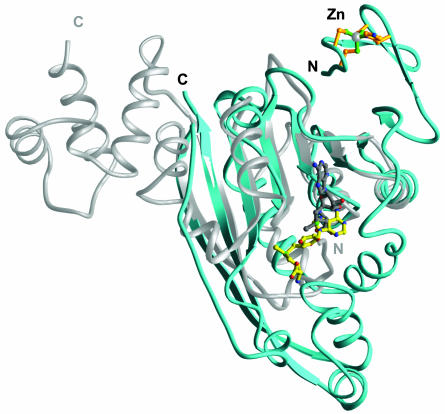
Comparison with Other rRNA Methyltransferase Structures. Crystal structures of bacterial rRNA MTases E. coli RlmB (32), Bacillus subtilis ErmC′ (33, 34), and Streptomyces viridochromogenes AviRa (35) have been described. These enzymes are highly specific to their respective RNA substrates, parts of bacterial rRNA. Although the overall structures of these MTases are different, all three of these enzymes contain MTase domains that have a common Rossmanntype fold. The MTase domain of RlmAI also has this characteristic fold. A Dali structural database search (36) identifies the MTase domain of ErmC′ (33) as one of the top structural analogs (Z = 13.2; 140 Cα atoms superimposed with rms deviation of 1.8 Å) of RlmAI. Despite the structural similarity of the SAM binding/MTase domains of RlmAI and ErmC′ (Fig. 3), the sequence identity in the structurally superimposed regions is only 9%. Because of a low sequence identity with know structures, the fold of the MTase domain of RlmA enzymes could not be recognized before this structure determination.
Fig. 3.
Superposition of the ErmC′ rRNA MTase structure (34) (silver) onto the RlmAI structure (cyan). Despite the fact that both enzymes have superimposable MTase domains, their putative RNA-recognition domains position differently and have different tertiary folds. The SAM molecule bound to ErmC′ is shown in gray, and the SAM molecule-bound RlmA is shown in yellow.
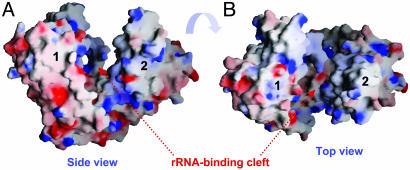
A comparison of the overall structures of RlmAI and ErmC′ provides some valuable insights (Fig. 3). The relative positions and orientations of the bound SAM molecules in RlmAI differ significantly from those of the ErmC′ structure (33). In addition, the putative rRNA-recognizing domains (e.g., the Zn-binding domain of RlmAI) of the two enzymes have different tertiary fold and are positioned differently with respect to superimposed MTase domains (Fig. 3). This structure comparison suggests differences in the mode of rRNA-substrate recognition by the MTase enzymes, despite a plausible common catalytic mechanism. These structural differences provide a basis for these enzymes' specificities to their respective substrates, different parts of bacterial rRNA. Among the above discussed three rRNA MTase structures, the reported structures of ErmC′ (33) and AviRa (35) have no well defined RNA-binding cleft/pocket and the RNA-binding cleft that has been described for dimeric RlmB (32) is very different from that of RlmAI (Fig. 4).
Fig. 4.
Top (A) and side (B) views of the electrostatic potential surface of an E. coli RlmAI dimer plotted by using GRASP (42). The positively charged regions are in blue, and the negatively charged regions are in red. The cleft formed by an RlmAI dimer is largely positively charged and proposed to bind the substrate, hairpin 35 of bacterial 23S rRNA.
Binding of rRNA Substrate. The W-shaped putative rRNA-binding cleft (Fig. 2A) is comprised of conserved amino acid residues from both monomers of an asymmetric RlmAI dimer. Two Zn-fingers are at the top and the two SAM molecules are at the bottom of the cleft. At the bottom of the cleft, helices α1 from each monomer together form a ridge that separates the two SAM-binding pockets. The W-shaped cleft is lined with a positively charged electrostatic surface suitable for interactions with polyanionic nucleic acids (Fig. 4). The unusual asymmetric arrangement of RlmAI molecules in its dimer appears to be functionally relevant in creating the specific shape of the rRNA-binding cleft. The shape of the cleft is unique and different from that of previously reported RNA-binding proteins.
Considering the clearly identifiable rRNA binding cleft of RlmAI, efforts were made to model the structure of its complex with hairpin 35 of 23S rRNA. The relevant parts from structures of the large ribosomal subunit of E. coli [by cryo-electron microscopy at 7.5 Å (37); PDB ID 1C2W], of Haloarcula marismortui [by x-ray at 2.4 Å (3); PDB ID 1FFK], eubacterium Deinococcus radiodurans [by x-ray at 3.1 Å (5); PDB ID 1JZX], Thermus thermophilus [by x-ray at 5.5 Å (38); PDB ID 1GIX], and the structure of hairpin 35 from Streptococcus pneumonia rRNA [by NMR (39); PDB ID 1MT4] were docked into the cleft of the RlmAI dimer (Fig. 4). Manual docking of the portion of the E. coli rRNA structure containing hairpins 33, 34, and 35 (nucleotides 692–770) (37), into the RNA-binding cleft of RlmAI provides a unique complementary match (Fig. 5A). In this modeled complex structure, hairpin 35 is completely buried in the cleft. The RNA-bulge (knot) linking the three hairpins (33, 34, and 35) sits over the Zn-finger regions of the cleft, suggesting that the two Zn-fingers are responsible (i) for recognition of the rRNA substrate structure and (ii) for placing the hairpin 35 in the W-shaped cleft. This model of the rRNA/RlmAI complex (Fig. 5A) is consistent with previously reported biochemical studies (11) by showing that, in addition to the hairpin 35, nucleotides from the adjacent hairpins 33 and 34 interact with RlmAI; most of the interacting nucleotides are from hairpin 35 and the RNA-bulge, whereas the top part of hairpin 34 is not interacting with the RlmAI dimer. Interestingly, in this model of the protein/rRNA complex (Fig. 5A), the base of nucleotide G745 (the target for methylation in Gram-negative bacteria) is positioned in close proximity to the SAM-binding pocket of molecule 1. The excellent unique fit of this rRNA fragment in the dimeric structure of RlmAI suggests that the observed structural asymmetry of the dimer is indeed required for unique recognition and binding of the rRNA substrate.
Fig. 5.
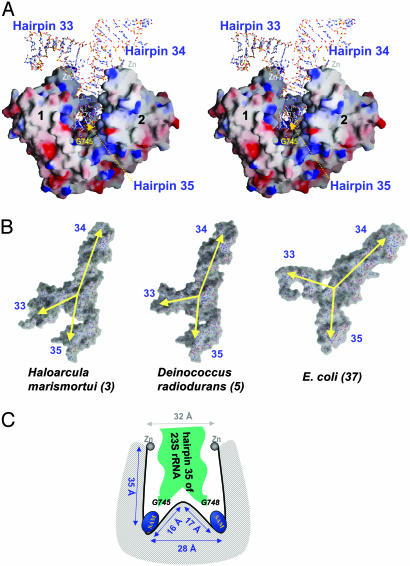
(A) Stereo view of a modeled complex of RlmAI:E. coli 23S rRNA fragment (37) containing hairpins 33, 34, and 35. The 3D structure of this rRNA fragment complements the shape of the RlmAI cleft formed by the MTase and Zn-binding domains. Nucleotide G745, which is methylated by RlmAI, is located near the SAM-binding pocket of molecule 1. (B) Comparison of conformation of the 23S rRNA fragment containing hairpins 33, 34, and 35 in three different structures of ribosomes. The yellow arrows indicate the angles between the domains of the rRNA fragments. (C) A schematic representation of the W-shaped RNA-binding cleft of RlmAI, showing a proposed binding mode of hairpin 35 of 23S bacterial rRNA. The distances indicated are of E. coli RlmAI.
As shown in Fig. 5B, the relative orientations of rRNA hairpins 33, 34, and 35 are somewhat different but related in different ribosome structures. An important difference in the arrangements of these hairpins is the angle between hairpins 33 and 35. This angle appears to be critical for binding of the rRNA fragment to the RlmAI dimer. Docking of the rRNA fragments from highresolution crystal structures of ribosomes [nucleotides 781 to 865 from H. marismortui (3), 704 to 784 from D. radiodurans (5), and 685 to 773 from T. thermophilus (38)] into the RNA-binding cleft of RlmAI showed possible fits of hairpin 35 into the W-shaped RNA-binding cleft; however, hairpin 33 develops steric hindrance with RlmAI when the angle between the hairpins 33 and 35 is small (Fig. 5B). RlmA enzymes do not act on 50S or 70S subunit of ribosome (11), and it is therefore likely that the modeled (Fig. 5A) 23S rRNA fragment (37) more closely reflects its naked conformation that actually binds to RlmA dimer. The two Zn-fingers of the RlmA dimer apparently interact at the hinges between hairpins 33:35 and 34:35 and consequently define the appropriate shape of the rRNA fragment.
Our current structure and modeling study suggests that most of the RNA/protein interactions in this complex are asymmetric; one monomer interacts differently with the RNA substrate than the other. The asymmetric nature of the RNA/protein interactions may be responsible for the unique fit of the substrate to the enzyme. Docking of the rRNA substrate predicts that regions 6–8, 25, 38–52, 117–119, 138–141, 157–162, and 233–235 of molecule 1 and 6–8, 25, 38–52, 115–121, and 136–140 of molecule 2 of the RlmAI dimer are likely to be involved in protein/RNA interactions. The length of the polypeptide linker (amino acids 35–52) between the two domains is 3–4 aa shorter in RlmAII than in RlmAI (Fig. 1A). The amino acid sequences of the linker are also distinct for RlmAI and RlmAII classes of the enzymes. This linker region of RlmA enzymes may play a role in precise positioning of G745 (in RlmAI) or G748 (in RlmAII) appropriately with respect to SAM for methylation.
G745/G748 Methylation. The above analysis suggests that an RlmA dimer is required for binding of its substrate, hairpin 35 of 23S rRNA. However, only one base of the rRNA substrate is methylated, and only one RlmA molecule from the dimer is required to catalyze this N1-methylation. In ribosome structures, rRNA hairpin 35 interacts with the large β-sheet of the ribosomal protein L22 and adopts a complementary inverted “U” shape (Fig. 5B), which is different from the unbound structure of the hairpin determined by NMR (39). Docking of the L22-bound conformation of hairpin 35 from different ribosome structures (discussed in previous section) shows a reasonable match between the hairpin and the ridge of the W-shaped cleft of RlmAI; in these modeled complexes, two nucleotides [U480 and A844 of H. marismortui (3), U760 and A764 of D. radiodurans (5), and A747 and A751 of T. thermophilus (38)], at equivalent E. coli positions 747 and 751, point to two SAM-binding pockets of the RlmAI dimer.
The RlmAI structure, together with sequence comparisons, suggests similar W-shaped conformation of the rRNA binding cleft and mode of binding of the rRNA fragment for both RlmAI and RlmAII. Based on our structure and modeling studies, we speculate that hairpin 35 adopts a shape complementary to the W-shaped rRNA binding cleft, whether bound to RlmAI or to RlmAII, in which nucleotides G745 and G748 point toward the two SAM-binding sites of the enzyme, as shown schematically in Fig. 5C. Alternatively, hairpin 35 may adopt somewhat different conformations when bound to RlmAI or RlmAII, such that either G745 (Fig. 5A) or G748 is pointed toward one SAM-binding pocket. In these two proposed structures of the RlmA/rRNA complex, specific protein/rRNA interactions (e.g., the interactions of the loop connecting the Zn-finger and MTase domains with rRNA hairpin 35) would play decisive roles in proper positioning of the correct nucleotide for N1-methylation catalysis.
Conclusion
The crystal structure of E. coli RlmAI has a well defined and largely positively charged W-shaped RNA-binding cleft formed by asymmetric dimerization (Fig. 4). Structural, functional, and amino acid sequence similarities among RlmAI and RlmAII enzymes (Fig. 1 A) suggest a common fold, as well as similar SAM- and RNA-substrate binding modes, for both classes of RlmA enzymes. It appears that the two Zn-binding domains are responsible for recognition and binding of the hairpin 35 region of 23S rRNA (Fig. 5A). Amino acid sequence comparison of RlmAI and RlmAII and mapping of the conserved regions onto the crystal structure of E. coli RlmAI indicate positioning of some of the key conserved amino acid residues at putative RNA-binding regions, at the SAM-binding pocket, and at the dimer interface. Docking the publicly available atomic coordinates for hairpin 35 of 23S rRNA and surrounding regions (3, 5, 37–39) into the cleft of RlmAI dimer shows complementary RNA/protein structural features. This crystal structure, along with earlier reported biochemical data, provides a basis for detailed investigations aimed at understanding structural features of the specific recognition of rRNA substrates, the role of this type of Zn-finger in RNA recognition, general aspects of RNA/protein interactions, and the mechanism of RNA methylation by RlmA enzymes.
Acknowledgments
This work was carried out as part of the Northeast Structural Genomics Consortium (NESG) study supported by a grant from the Protein Structure Initiative of the National Institutes of Health (P50 GM62413).
Abbreviations: MTase, methyltransferase; RlmAI, RlmA class I; RlmAII, RmlA class II; SAM, S-adenosyl-l-methionine; Se-Met, selenomethionine.
Data deposition: The atomic coordinates and structure factors for the Escherichia coli RlmAI structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1P91).
References
- 1.Roberts, M. C. (2002) Mol. Biotechnol. 20, 261–283. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez, D. (1966) Biochim. Biophys. Acta 114, 277–288. [DOI] [PubMed] [Google Scholar]
- 3.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., Clemons, W. M., Jr., Carter, A. P., Morgan-Warren, R. J., Wimberly, B. T. & Ramakrishnan, V. (2000) Cell 103, 1143–1154. [DOI] [PubMed] [Google Scholar]
- 5.Schlünzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., Yonath, A. & Franceschi, F. (2001) Nature 413, 814–821. [DOI] [PubMed] [Google Scholar]
- 6.Hansen, J. L., Ippolito, J. A., Ban, N., Nissen, P., Moore, P. B. & Steitz, T. A. (2002) Mol. Cell 10, 117–128. [DOI] [PubMed] [Google Scholar]
- 7.Baltz, R. H. & Seno, E. T. (1988) Annu. Rev. Microbiol. 42, 547–574. [DOI] [PubMed] [Google Scholar]
- 8.Vester, B. & Douthwaite, S. (2001) Antimicrob. Agents Chemother. 45, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krzyzosiak, W., Denman, R., Nurse, K., Hellmann, W., Boublik, M., Gehrke, C. W., Agris, P. F. & Ofengand, J. (1987) Biochemistry 26, 2353–2364. [DOI] [PubMed] [Google Scholar]
- 10.Rozenski, J., Crain, P. F. & McCloskey, J. A. (1999) Nucleic Acids Res. 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, L. H., Kirpekar, F. & Douthwaite, S. (2001) J. Mol. Biol. 310, 1001–1110. [DOI] [PubMed] [Google Scholar]
- 12.Brimacombe, R., Mitchell, P., Osswald, M., Stade, K. & Bochkariov, D. (1993) FASEB J. 7, 161–167. [DOI] [PubMed] [Google Scholar]
- 13.Weisblum, B. (1995) Antimicrob. Agents Chemother. 39, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, M. & Douthwaite, S. (2002) Proc. Natl. Acad. Sci. USA 99, 14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, G. E., Stackebrandt, E., Hespell, R. B., Gibson, J., Maniloff, J., Dyer, T. A., Wolfe, R. S., Balch, W. E., Tanner, R. S., Magrum, L. J., et al. (1980) Science 209, 457–463. [DOI] [PubMed] [Google Scholar]
- 16.Liu, J., Hegyi, H., Acton, T. B., Montelione, G. T. & Rost, B. (2003) Proteins Struct. Funct. Genet., in press.
- 17.Gustafsson, C. & Persson, B. C. (1998) J. Bacteriol. 180, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, M. & Douthwaite, S. (2002) Mol. Microbiol. 44, 195–204. [DOI] [PubMed] [Google Scholar]
- 19.Kagan, R. M. & Clarke, S. (1994) Arch. Biochem. Biophys. 310, 417–427. [DOI] [PubMed] [Google Scholar]
- 20.Liu, M., Kirpekar, F., Van Wezel, G. P. & Douthwaite, S. (2000) Mol. Microbiol. 37, 811–820. [DOI] [PubMed] [Google Scholar]
- 21.Bjork, G. R. & Isaksson, L. A. (1970) J. Mol. Biol. 51, 83–100. [DOI] [PubMed] [Google Scholar]
- 22.Isaksson, L. A. (1973) Biochim. Biophys. Acta 312, 134–146. [DOI] [PubMed] [Google Scholar]
- 23.Isaksson, L. A. (1973) Biochim. Biophys. Acta 312, 122–133. [DOI] [PubMed] [Google Scholar]
- 24.Jansson, M., Li, Y.-C., Jendeberg, L., Anderson, S., Montelione, G. T. & Nilsson, B. J. (1996) J. Biomol. NMR 7, 131–141. [DOI] [PubMed] [Google Scholar]
- 25.Doublie, S., Kapp, U., Aberg, A., Brown, K., Strub, K. & Cusack, S. (1996) FEBS Lett. 384, 219–221. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski, Z. & Minor, W. (2001) in The International Union of Crystallography:Crystallography of Biological Macromolecules, eds. Rossmann, M. G. & Arnold, E. (Kluwer Academic, Boston), Vol. F, pp. 226–235. [Google Scholar]
- 27.Howell, P. L., Blessing, R. H., Smith, G. D. & Weeks, C. M. (2000) Acta Crystallogr. D 56, 604–617. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999) Acta Crystallogr. D 55, 247–255. [DOI] [PubMed] [Google Scholar]
- 31.Brunger, A. T., Adams, P. D. & Rice, L. M. (1998) Curr. Opin. Struct. Biol. 8, 606–611. [DOI] [PubMed] [Google Scholar]
- 32.Michel, G., Sauve, V., Larocque, R., Li, Y., Matte, A. & Cygler, M. (2002) Structure (Cambridge, Mass.) 10, 1303–1315. [DOI] [PubMed] [Google Scholar]
- 33.Bussiere, D. E., Muchmore, S. W., Dealwis, C. G., Schluckebier, G., Nienaber, V. L., Edalji, R. P., Walter, K. A., Ladror, U. S., Holzman, T. F. & Abad-Zapatero, C. (1998) Biochemistry 37, 7103–7112. [DOI] [PubMed] [Google Scholar]
- 34.Schluckebier, G., Zhong, P., Stewart, K. D., Kavanaugh, T. J. & Abad-Zapatero, C. (1999) J. Mol. Biol. 289, 277–291. [DOI] [PubMed] [Google Scholar]
- 35.Mosbacher, T. G., Bechthold, A. & Schulz, G. E. (2003) J. Mol. Biol. 329, 147–157. [DOI] [PubMed] [Google Scholar]
- 36.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 37.Mueller, F., Sommer, I., Baranov, P., Matadeen, R., Stoldt, M., Wohnert, J., Gorlach, M., van Heel, M. & Brimacombe, R. (2000) J. Mol. Biol. 298, 35–59. [DOI] [PubMed] [Google Scholar]
- 38.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 39.Lebars, I., Yoshizawa, S., Stenholm, A. R., Guittet, E., Douthwaite, S. & Fourmy, D. (2003) EMBO J. 22, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 41.Carson, M. (1997) Ribbons (Academic, New York).
- 42.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]