Abstract
To explore the function of the giant AHNAK molecule, first described in 1992 [Shtivelman, E., Cohen, F. E. & Bishop, J. M. (1992) Proc. Natl. Acad. Sci. USA 89, 5472–5476], we created AHNAK null mice by homologous recombination. Homozygous knockouts showed no obvious phenotype, but revealed instead a second AHNAK-like molecule, provisionally designated AHNAK2. Like the original AHNAK, AHNAK2 is a 600-kDa protein composed of a large number of highly conserved repeat segments. Structural predictions suggest that the repeat segments of both AHNAKs may have as their basic framework a series of linked, antiparallel β-strands similar to those found in β-propeller proteins. Both AHNAKs appear to localize to Z-band regions of mouse cardiomyocytes and cosediment with membrane vesicles containing the dihydropyridine receptor, which is consistent with earlier reports that the AHNAKs are linked to L-type calcium channels and can be phosphorylated by protein kinase A. The localization of the AHNAKs in close proximity to transverse tubule membranes and Z-band regions of cardiac sarcomeres raise the possibility that they might be involved in regulating excitation/contraction coupling of cardiomyocytes, but other studies indicate that the association of AHNAKs with calcium channel proteins is more widespread. AHNAK2 is predicted to have a PDZ domain within its N-terminal, nonrepeating domain, which may mediate these interactions.
AHNAK is an unusually large polypeptide that was first described by Schtivelman et al. (1) as a differentiation-related protein that localized in interphase nuclei. Others had also identified a large protein in keratinocytes, named desmoyokin (2), which appeared to be membrane-associated, as a consequence of protein kinase C activation. Later studies showed that AHNAK and desmoyokin were the same protein that had a variable subcellular localization, depending on the cell type. Although it had been suggested that AHNAK might exist in cells as an extended polymer composed of multiple short δ-repeat segments (1), electron microscopic studies indicated that the isolated AHNAK/desmoyokin protein was a relatively short, rod-shaped polymer, ≈150 μm long (3).
The first important clue as to AHNAK's physiological function came from the discovery that it binds to and activates phospholipase C-γ (PLC-γ) in the presence of arachadonic acid (4). Recombinantly derived fragments of AHNAK activate PLC and generate diacylglycerol and inositol trisphosphate, the two products of phosphoinositide hydrolysis that are produced when PLCs are activated by ligand-stimulated receptors or G proteins. It was not clear from these in vitro studies where in the cell and under what conditions PLC-γ can be activated by AHNAK, but another recent study (5) showed that stimulation of cardiomyocytes by adrenergic agonists promoted the phosphorylation of a membrane-associated form of AHNAK. This phosphorylated form of AHNAK could be coprecipitated by antibodies directed against two different subunits of the L-type voltage-regulated calcium channel, implying that AHNAK is physically coupled to one or more components of functioning calcium channels. Both sets of observations link AHNAK to metabolic processes that are involved in signal transduction mechanisms, one operating at the cell surface in collaboration with calcium channels, and the second, PLC activation, which is a process that could potentially take place at multiple points throughout the cell.
The arrangement of channel proteins at the cell surface is believed to be controlled by multidomain polypeptides known as scaffolding proteins that link together activated channels at specific points on the membrane surface. Scaffolding proteins also coordinate the activities of multienzyme complexes by physically linking them together, and as in the case with AHNAK, they are often recognized by their capacity to coprecipitate with antibodies against subunits of the enzyme complexes.
To explore the functions of AHNAK in the intact animal, we undertook to knock out the AHNAK gene in mice by homologous recombination. We created mice with the homozygous null genotype (AHNAK–/–) and were surprised to find that we could not detect any obvious phenotype. On further study we realized that the knockout mouse contained at least one additional high molecular weight polypeptide that reacted with our anti-AHNAK monoclonal antibody. A search of Human Genome sequence databases revealed what appears to be a second AHNAK-like protein whose properties are described here. These results suggest that mammalian cells contain a family of AHNAKs that may operate at different subcellular sites. In cardiomyocytes, their localization at both the sarcolemma and at Z-band sites suggests some role in the excitation/contraction coupling mechanism, in other cells they may be involved in processes that depend in part on a calcium-release mechanism. A recent report (6) showing that AHNAK is a specific target for the calcium-binding S100B protein is yet another link between the AHNAKs and calcium homeostasis.
Materials and Methods
Generation of AHNAK Knockout Mice. A BAC library derived from ES-129/SvJ mice (Genome Systems, St. Louis) was screened by PCR to isolate an AHNAK genomic clone using two sets of primers based on the sequence of an EST clone (GenBank accession no. AA530289): the forward primer (FL, 30mer: ttgggggcaccagatgtaacactgaagggac); the reverse primers (BL, 28mer: ccaggcatttggaggtttccttctgggc); the nested-forward primer (FLN, 30mer: tctggctgtgtctggagacatcaaatgtcc); and the nested-reverse primer (BLN, 29mer: ccgaggctcccctttacttttggtccttc).
A 200-kb BAC clone that contained the entire AHNAK genomic sequence was isolated. A targeting vector was designed to replace a 5.3-kb genomic fragment containing a segment of the coding region and putative promoter and 5′ UTR with the loxP-flanked neomycin resistance gene expression cassette. The phosphoglycerokinase promoter driven thymidine kinase gene was appended to the construct to select against nonhomologous recombination. The targeting vector was linearized with NotI and electroporated into W9.5 embryonic stem cells. Clones resistant to G418 and gancyclovir were selected, and homologous recombination was confirmed by Southern blotting. The AHNAK gene was modified in 9 of 192 clones screened. Two clones containing the targeted mutation were injected into C57BL/6 blastocysts, and these were subsequently transferred into pseudopregnant foster mothers. The resulting male chimeric mice were bred to C57BL/6 females to obtain heterozygous AHNAK+/– mice. Germ-line transmission of the mutant allele was verified by Southern blot analysis of tail DNA from F1 offspring with agouti coat color. Interbreeding of the heterozygous mice was performed to generate homozygous AHNAK-deficient mice.
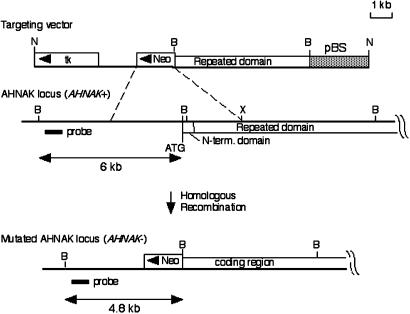
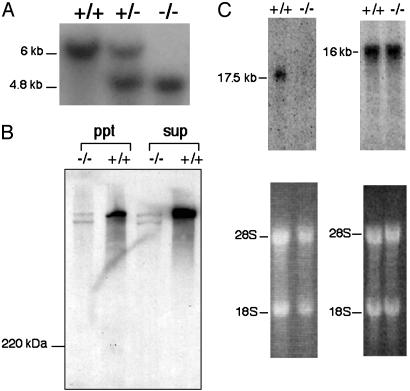
Characterization of AHNAK–/– Mice. For Southern blot analysis, genomic DNA was isolated from mouse tail, was digested with BamHI, and was hybridized with a 500-bp probe located just outside the 5′ short arm of the knockout vector. By using this probe, a 6-kb AHNAK wild-type DNA fragment and a 4.8-kb AHNAK mutant DNA fragment are detected.
For Northern blot analysis of the expression of AHNAK1, total RNA was isolated from heart ventricle by using the RNeasy kit (Qiagen) and a 300-bp AHNAK ApaI cDNA fragment was used as a probe. To assay for AHNAK2 expression, total RNA was isolated from heart ventricle by using TRIzol (GIBCO/BRL), and a probe was prepared by RT-PCR. Four hundred nanograms of total RNA from C57/B6 mouse heart were used for the reverse transcription reaction in a total volume of 20 μl. Reverse transcription was performed for 50 min at 42 C with 10 units of a reverse transcriptase from avian myeloblastosis virus (GIBCO). Two microliters from the reverse transcription reaction mix was used for a subsequent PCR in a total volume of 50 μl by using 50 pmol of each set of primer AN2MF: cagctctgggaggattctg and AN2MR: gggctctggaattttcactttc. Those primers were designed based on the mouse EST clones: BG962217, BF582769, AA619677, AA839034, and AA760494. The 400-bp PCR fragment was labeled with a random priming kit (Stratagene) with [32P]dCTP and was hybridized with the total RNA from AHNAK-null mice or wild-type mice on a hybond-N membrane (Amersham Pharmacia). For Western blot, heart ventricle was homogenized in 20 mM Hepes buffer (pH 7.4) containing 150 mM NaCl, 5 mM MgCl2, a protease inhibitor mixture obtained from Roche supplemented with 1 mM PMSF, 10 μg/ml aprotinin, and 20 μM leupeptine, and was centrifuged at 1,000 × g for 20 min. Aliquots of supernatant were electrophoresed on an SDS/4.5% PAGE gel, were transferred to polyvinylidene fluoride membrane, and were probed with an AHNAK monoclonal antibody.
Fractionation of Heart Homogenate by Sucrose Gradient Ultracentrifugation. Left ventricles from C57/B6 mice (≈400 mg) were homogenized in 1.5 ml of homogenizing buffer (20 mM Hepes, pH 7.4/150 mM NaCl/5 mM MgCl2/2.5 mM EGTA/5% sucrose/25 mM NaF/1 mM Na3VO4/10 μg/ml aprotinin/1 mM phenylmethylsulfonyl fluoride/20 μM leupeptin/17 μg/ml calpain I inhibitor/17 μg/ml calpain II inhibitor/1 μM pepstatin A/0.1 mM benzamidine/2 μg/ml chymostatin) with a Polytron. The nuclear fraction and myofibrillar elements, along with debris and unbroken cells, were removed by centrifugation at 1,000 × g for 15 min. The supernatant was loaded on top of a sucrose gradient (20–40%) and was centrifuged at 50,000 rpm in a TLS-55 rotor (Beckman Coulter) for 12 h. After centrifugation, each fraction was analyzed by Western blot analysis probing with a series of antibodies including anti-AHNAK monoclonal antibody, anti-ryanodine receptor (RyR) monoclonal antibody (Affinity Bioreagents), and anti-dihydropyridine (DHP) receptor monoclonal antibody (Sigma).
Immunostaining of Heart Sections. Left ventricles from wild-type or AHNAK-null mice were embedded in OCT compound (Tissue-Tek) and were frozen in 2-methylbutane with liquid nitrogen. Semithin sections were prepared with a Leica microtome Cryocut 1800 and collected on gelatin-treated glass slides. The sections were fixed with acetone for 10 min and blocked with 5% BSA/PBS for 1 h. The blocked samples were incubated with anti-AHNAK monoclonal antibody, or anti-RyR monoclonal antibody (Affinity Bioreagents), followed by incubation with rhodamine-conjugated anti-mouse IgG, or FITC-conjugated anti-mouse IgG. The images were observed on a fluorescence microscope (Nikon).
Results
AHNAK Homozygous Null Mice Are Viable and Without an Obvious Phenotype. Mouse embryonic stem cells were transfected with the targeting vector (Fig. 1) to eliminate the expression of the AHNAK gene by homologous recombination as described in Materials and Methods. Clones of cells heterozygous for the AHNAK gene were generated and then injected into blastocysts to create knocked-out offspring, which were then mated to produce homozygotes. Such animals were viable, had a normal growth and development pattern, and showed no obvious phenotype. However, Southern analysis of total genomic DNA confirmed that the AHNAK gene had been disrupted, and immunoblot analysis of total cell extracts confirmed that the typical 600-kDa AHNAK band was not present (Fig. 2). The absence of a phenotype suggested that AHNAK was either a nonessential protein or was one of several homologous proteins that shared common functions. Given its large size, remarkably conserved gene sequence, and capacity to interact with proteins involved in calcium homeostasis, we thought it likely that other AHNAK-like proteins were present in the knocked-out mice that compensated for its absence, a view that was confirmed when we searched for cross-reacting proteins in heart and other tissues of AHNAK–/– mice, where we found several high molecular weight bands that reacted with our monoclonal anti-AHNAK antibody. Immunoblots of extracts obtained from hearts of wild-type mice showed a strong immunoreactive band with a mobility that was consistent with a 600-kDa protein that was identical to that found in other cells and tissues but was missing from extracts obtained from AHNAK-null mice. However extracts of AHNAK–/– (Fig. 2A) hearts did contain two immunoreactive bands of much lower intensity, one of which overlapped with the more intense band in the wild-type animals. These same two bands also reacted with polyclonal antibodies prepared against synthetic peptides containing the sequences KISMPDVDLHLKGPK and KEMPKVKMPKFSMPG, which are found in the original AHNAK protein (1).
Fig. 1.
Structure of the targeted vector, the AHNAK locus, and the mutated AHNAK gene after homologous recombination. Relevant restricted enzyme sites are indicated. The corresponding N-terminal domain and repeated domain are indicated on the AHNAK locus. The diagnostic probe used for Southern blot analysis is shown. N, NotI; B, BamHI; X, XbaI.
Fig. 2.
(A) Analysis of AHNAK knockout mice by Southern blot. Genomic DNA from mouse tails was digested with BamHI, and blots were hybridized with the probe shown in Fig. 1. The wild-type allele corresponds to a 6-kb fragment, and the mutant allele is a 4.8-kb fragment. (B) Immunoblot analysis of heart fractions of wild-type and knockout mice. Anti-AHNAK antibodies (both monoclonal and polyclonal) react with a broad ≈700-kDa band that is absent in AHNAK–/– mice. Two faint bands are seen in knockout mice, one of which migrates in the same position as the wild-type AHNAK band. (C) Northern blot analysis. Total RNA (10 μg) isolated from heart ventricles of wild-type (+/+) and null (–/–) mice was hybridized with cDNA fragments derived from AHNAK1 (Left) or AHNAK2 (Right). An ≈17.5-kb signal of AHNAK1 was detected only in wild-type heart, but a 16-kb signal of AHNAK2 was found in both. The original ethidium bromide-stained gels are shown as a reference.
Identification of a Putative AHNAK Homologue. We took advantage of the human genome sequence to search for genes that code for proteins that resembled AHNAK in size and domain composition, and found a segment of genomic DNA, derived from chromosome 14q32 that encoded a 15-kb ORF that resembled AHNAK in a number of ways. Like the original AHNAK gene, which is located on chromosome 11 (7), this DNA fragment was predicted to code for a 600-kDa polypeptide made up of multiple repeat units, some of which contain short peptide segments that were identical with those found in the original AHNAK repeats. These common peptide segments probably account for the ability of our monoclonal anti-AHNAK antibody to recognize this protein, but we have not yet established which peptides are part of the immunoreactive domains. We showed by immunoblotting that our monoclonal antibody reacts with several high molecular weight polypeptides in tissues derived from AHNAK–/– mice, but we have not yet definitely demonstrated that either of these bands actually corresponds to this gene.
This second AHNAK-like protein, which we provisionally refer to as AHNAK2, has the same tripartite organization of AHNAK1, in that it has a short nonrepetitive N-terminal segment that connects to 24 repeats segments and a C segment, not composed of repeating units, that is ≈100 kDa in size. Unlike the reported genomic structure of AHNAK1, AHNAK2 is coded for by at least seven exons, six relatively small, and the seventh, estimated to contain 17,559 bp. Perhaps the most compelling link between the two AHNAKs is the unusual amino acid sequence pattern of their repeating units. The predicted amino acid sequences of each repeat unit are strikingly similar to several well characterized “propeller” proteins as discussed in detail below.
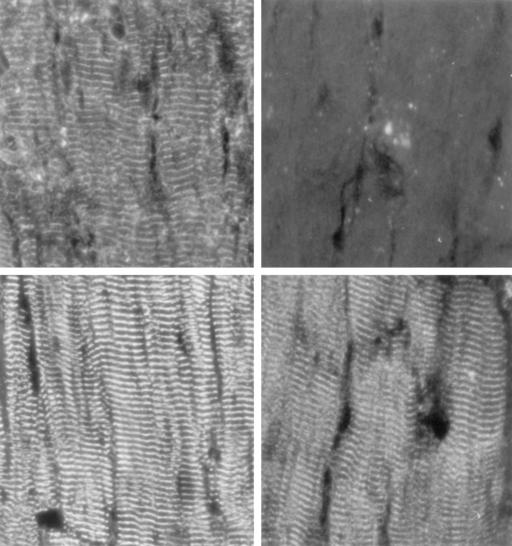
The AHNAKs Localize to Z-Band Regions of Mouse Hearts. Many investigators have reported on the distribution of anti-AHNAK antibodies in a variety of tissues (8, 9). We were particularly interested in their localization in the heart, because AHNAK has been linked functionally to one of the subunits of the voltage-gated calcium channel, and was reported to localize along the inner surface of the sarcolemma of myocardial cells (5). Our monoclonal anti-AHNAK antibody produced the periodic staining pattern of mouse myocardium shown in Figs. 3 and 4. This distribution of anti-AHNAK staining was identical in hearts obtained from either wild-type (AHNAK+/+) or knocked-out (AHNAK–/–) mice. As Fig. 3 clearly shows, the anti-AHNAK staining pattern was essentially identical to the pattern obtained with both anti-α-actinin antibodies and anti-RyR antibodies. α-Actinin is concentrated in Z-band regions, and is considered a useful marker for those sites, but the RyRs are membrane-bound and associated with the sarcoplasmic reticulum (SR) vesicles. The colocalization of the two antibodies suggests that the bulk of the RyRs are bound to SR vesicles that are in immediate contact with the invaginating transverse (T) tubules. Because antibodies to the DHP receptor also localize to these sites (10), it is not possible to identify the structures to which AHNAK is linked by using light microscopic immunofluorescence, because the AHNAKs could be associated with T-tubule membranes, the adjacent SR membranes, the closely associated Z-band material, or all three.
Fig. 3.
Immunocytochemical staining of mouse heart. Identical thin sections of left ventricles were stained with monoclonal anti-AHNAK antibody (Left Upper), polyclonal anti-α-actinin antisera (Left Lower), or monoclonal anti-RyR (Right Lower). All three antibodies stained periodic structures in cardiomyocytes that contain the Z-band material, T-tubule membranes, and SR vesicles enriched with the RyR. (Right Upper) A heart section stained without a primary antibody is shown.
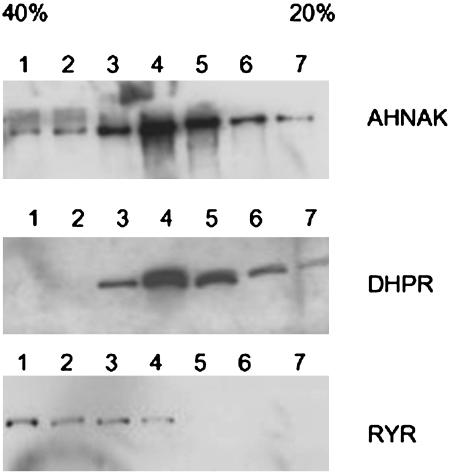
Fig. 4.
Sucrose gradient analysis of mouse heart homogenates. Low-speed supernatants of left ventricles were fractionated on continuous sucrose gradients (20–40%) and were analyzed by SDS/PAGE and immunoblotting with antibodies specific for RyR, DHP receptor, and AHNAK. Most of the AHNAK that was not sedimentable by low-speed centrifugation was found in vesicular fractions that comigrate with the DHP receptor, and presumably represent membrane vesicles derived from the T-tubule system and the sarcolemma. RyR immunoreactivity migrated as more dense membrane fractions derived from the SR. A small fraction of AHNAK cosedimented with the heavy vesicle fraction.
The AHNAKs Cosediment with Membrane Vesicles That Contain Dihydropyridine Receptors. To resolve the ambiguous antibody labeling results described above, subcellular fractions of mouse myocardium were separated by ultracentrifugation on continuous sucrose gradients and individual fractions were analyzed by SDS/PAGE immunoblotting with a battery of antibodies (Fig. 4). The AHNAKs were found to be concentrated in two different subcellular fractions, one a low-speed fraction, composed of nuclei and myofibrillar aggregates, including the bulk of the Z-band material, and the other a less dense vesicular fraction that cosedimented with the DHP receptor, which is consistent with it being bound to a vesicle fraction derived from the T tubules. The bulk of the RyR immunoreactivity sedimented as a more dense membrane fraction, which we assume was derived from the SR. Although small amounts of AHNAK did cosediment with the RyR-containing fractions, we have no way of knowing whether this association is physiologically significant. Based on the fact that a significant faction of the AHNAK-reactive material is present in the low-speed pellet and the remainder in a lighter vesicular fraction containing the DHP receptor, we tentatively conclude that the bulk of the AHNAKs are linked to T-tubule membranes, through the previously described association with the β-subunit of the calcium channel (5), and to some component of the Z-band complex.
A recent study has shown that a subset of myocardial RyRs exist as multiprotein complexes composed of PKA, FK-506-binding protein FKBP 12.6, calcineurin, a kinase-anchoring protein, and an as-yet-unidentified docking protein (11), and its stability is phosphorylation-dependent. Given our immunofluorescence colocalization data, we prepared myocardial extracts in the presence of a variety of phosphatase and protease inhibitors, but were unable to detect any increased association between AHNAK and RyR-containing fractions. If we take into account the technical problems associated with dispersing cardiomyocytes without disrupting functional connections, it is still possible that we have not found the conditions needed to preserve connections between the AHNAKs and the calcium-release apparatus.
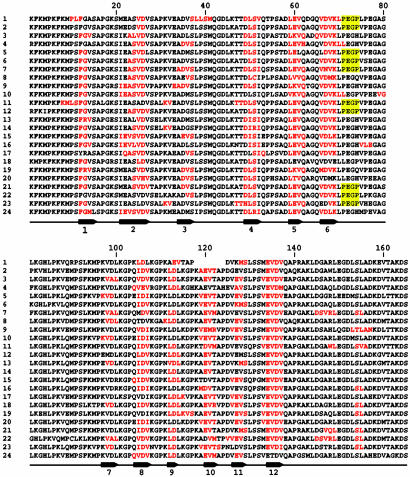
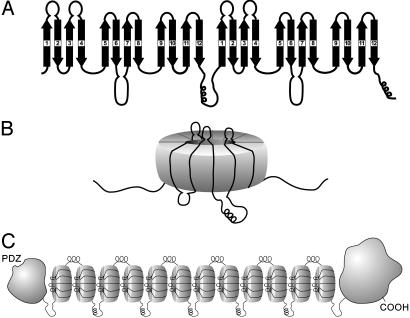
The AHNAKs Are Likely to Be Multimodular Propeller Proteins. In the original description of AHNAK an underlying heptad-repeat motif was noted (1), which was believed to be consistent with an extended β-strand structure for the repeat portion of the molecule. Although the authors predicted that these short β-sheets might form an extended rod, possibly as long as 1 μm, they did also point out that the β-chains could associate together to form seven- or eight-stranded barrels. The repeat structure of AHNAK2 is remarkably conserved and is, on average, 165 aa long. Conformation predictions of the repeat segments, provided by the SOPM secondary structure prediction method (Network Protein Sequence Analysis, Lyon, France) are shown in Fig. 5. The red-colored amino acids are predicted to fold into β-strands, and this prediction is almost uniform throughout all of the repeat segments, which is consistent with the strict conservation of the sequences. Although we are aware that these predictions must be considered provisional, and there may be other β-strands that have not been correctly predicted, the lengths of the β-strands that are predicted are close to those found in other well characterized propeller proteins in which the β-structures have been confirmed by x-ray crystallography (12–14). Because many of the predicted β-strands are linked together by short peptide segments that are characteristic of β-turns, we used these predictions to create the highly schematic model of AHNAK2 that appears in Fig. 6. This model, which assumes 12 β-strands per sequence repeat (Fig. 6A), was created to show how the AHNAK2 repeat sequences could be organized if the underlying basic framework was composed of four antiparallel β-strands packed together in a propeller-like blade (Fig. 6B). All β-propeller proteins that have been reported to date have as their basic motif four short β-strands, each of which appears as a blade of a propeller, but the number of blades varies from four to eight. We show here how the individual β-strands of AHNAK2 might be arranged if AHNAK2 is a seven-bladed propeller, similar to RCCI (12), clathrin (13), and G β-protein (14), but other configurations are also possible.
Fig. 5.
Consecutive sequence alignments of the 24 repeat motifs of AHNAK2. The repetitive sequence region starts at amino acid 460, ends at amino acid 4247, and is comprised almost uniformly of 165 aa. The amino acids in red are predicted to adopt a β-strand conformation (28), and the tetrapeptides highlighted in yellow are potential SH3-binding sites.
Fig. 6.
Because the predicted β-strands, indicated by the black arrows, are strikingly similar in composition, length, and distribution to comparable sequences found in well authenticated propeller proteins, we show how the predicted β-strands might be organized if they were assembled as units of four antiparallel β-strands connected by intervening loops. If we also assume a seven-blade propeller as one possibility, 28 strands could conceivably pack into the disk-shaped structures depicted in B. Based on these predictions, the entire AHNAK molecule is depicted schematically in C with domains of undefined structure at the N and C termini connected by a series of linked, propeller-like structures.
Regardless of the precise number of blades that any individual propeller protein might have, the same basic packing rules seem to apply (15, 16). Individual β-strands interact with each other in a circular array that is stabilized largely by hydrophobic forces, and the assembled peptides create a disk-like structure from which protrude charged and hydrophilic peptide segments, as illustrated in the highly schematic model shown in Fig. 6. Although this model is provisional and speculative, it does provide us with an opportunity to generate testable hypotheses regarding which segments of AHNAK2 are most likely to mediate interactions with other proteins. The predictions shown here suggest that two relatively long peptides, which contain many charged and hydrophilic amino acids, may extend from the “flat” surfaces of the putative discs, possibly even the same side, and are good candidates for further study. The two sets of peptides are structurally distinct from each other, but each has many highly conserved sequences, implying that they might interact with multiple copies of at least two different proteins. One of these peptides contains PxxP sequences that may be putative SH3-binding sites (17). Although the AHNAKs share many features in common with known propeller proteins, they have one distinct difference. If all of the repeat segments have the capacity to fold into multibladed propellers, each AHNAK molecule would contain a large number of tandemly connected propeller units.
The N-terminal domain of AHNAK2 appears to be coded for by multiple short exons, and embedded in it is an ≈90-aa segment, predicted to function as a PDZ domain, which is characteristic of the PSD95 protein and others that are believed to interact with C-terminal peptides of a number of channel proteins, including those involved in calcium transport.
Discussion
We have found that AHNAK null mice contain a high molecular weight protein that is remarkably similar to the original AHNAK molecule, that was first described in 1992 (1). This protein (referred to here as AHNAK2) has the same tripartite structure as AHNAK1, including 24 repeat segments, each ≈165 aa in length. Although the original AHNAK protein was localized to both nuclei and the cytoplasm in a variety of cell types, we found that a monoclonal antibody that reacts with multiple repeat domains of both AHNAKs localizes to Z-band regions of mouse cardiomyocytes when analyzed by light microscopic immunofluorescence. These results are consistent with earlier findings (5) using a polyclonal antibody directed against epitopes that are specific for AHNAK1. Because AHNAK1-null mice show the same antibody staining pattern, which we assume labels AHNAK2 as well, we conclude that both AHNAKs are probably concentrated at these same subcellular sites, a colocalization that may account for the fact that AHNAK1 null mice show no obvious cardiovascular defects. The alternative possibility, that only AHNAK2 is concentrated in cardiomyocytes, must also be considered, because some anti-AHNAK antibodies do not react with cardiac issues to any appreciable extent (18).
Our subcellular fractionation results indicate that a significant fraction of AHNAK is bound to a vesicle population that is largely derived from the sarcolemma and T-tubule membranes, because it cosediments with the DHP receptor when analyzed by sucrose gradient ultracentrifugation. These results confirm an earlier study that showed an association of an AHNAK, presumed to be AHNAK1, with myocardial cell membrane fractions that contain the voltage-gated calcium channel (5). A substantial amount of AHNAK sediments at low speed, and we assume it is associated with Z-band material, which is consistent with the strong immunofluorescence staining of Z-band regions that we found when studying sections of mouse myocardium. At this point, we have no idea how either of the AHNAKs might link up with Z-band material, but a specific association with α-actinin is a possibility, because both α-actinin and the AHNAKs are concentrated at these sites. A much smaller fraction of AHNAK cosedimented with a vesicle fraction enriched with the RyR that was derived from the SR, but we could not rule out nonspecific associations. Because an earlier study (11) suggested that an unknown cytoskeletal protein was part of a multiprotein complex that contained the RyR, PKA, a kinase-anchoring protein, and two different protein phosphatases, further studies are needed to determine whether either of the AHNAKs might be part of this complex. A recent study has shown that a C-terminal fragment of AHNAK1 localizes to the cardiac sarcolemma as well as the T-tubular system and intercalated disk regions, localizations which may involve direct links to the actin-based cytoskeleton (19).
We hypothesize that the repeat segments of both AHNAKs may be organized as β-propeller proteins with a basic structure similar to that found in RCC1, clathrin, and the G β-subunit. We based this interpretation on the number, length, amino acid composition, and distribution of predicted β-strands in each of the repeat sequences of both molecules. If we assume that all of the AHNAK repeat segments fold into anti-parallel β-strands with interconnecting peptide loops, a string of linked propeller-like modules would be created that have the potential to interact with large numbers of ligands or other proteins. Such a structure would be consistent with the electron microscopic images described earlier and make the AHNAKs an ideal class of scaffolding proteins.
If these large polypeptides are indeed composed of multiple, linked propeller-like units (an arrangement yet to be determined experimentally), they would represent proteins that contain multiple propeller-like domains that are linked together, one adjacent to the other. A number of other proteins contain multiple amino acid sequences that are thought to have propeller conformations. One example is another giant protein referred to as p530 (20, 21), which contains three stretches of sequence that are referred to as RCC1-like repeats, but each is thought to be separated by what appear to be nonpropeller intervening segments. X-ray crystallographic studies of RCC1, clathrin, and the G β-protein all depict the propeller modules as being disk-like structures with two “surfaces” and a “periphery” composed of the exposed portions of the antiparallel β-sheets and non-β peptide segments. The two surfaces of RCC1 have distinct binding functions, one surface interacts with DNA and the other with a protein involved with guanine nucleotide exchange activity (12). Studies of the clathrin propeller module also indicate that additional protein–protein interactions are mediated by peptide segments that form the grooves between the individual blades of the propeller. It has been suggested that a specific peptide sequence motif, referred to as the “clathrin box” (22), accounts for the ability of two so-called cargo adaptor proteins, β-arrestin and AP-3, to bind to clathrin.
As of this writing, we know of a number of proteins that interact with AHNAK1, and probably AHNAK2 as well, and each is involved in some way with calcium-related activities. Several recombinantly derived fragments of AHNAK1, that include repeat segments, bind to and activate PLC-γ in the presence of arachadonic acid (4). One consequence of PLC action in vivo is to generate inositol trisphosphate, a potent calcium release agonist in all cells. If similar interactions between an AHNAK and PLC-γ occur in vivo, one or both of the AHNAKs could mediate calcium release reactions at subcellular sites where AHNAK is concentrated. However, recent studies with mutant forms of PLC-γ, that are inactive catalytically (23), suggest that the AHNAKs and PLC-γ may also operate through more direct interactions between PLC-γ and calcium entry mechanisms that operate independently of inositol trisphosphate. Lipase inactive mutants of PLC-γ appear to be fully capable of augmenting agonist-induced calcium entry, possibly through mechanisms that involve the subcellular localization of signaling complexes. Another example of a direct connection between one or both of the AHNAKs and surface membranes is the role that the AHNAKs are believed to play in regulating exocytosis. Through the use of a monoclonal antibody that recognizes AHNAK-like proteins in many, but not all tissues, another type of AHNAK-associated vesicle has been described that appears to undergo rapid calcium-dependent exocytosis (18).
Anti-AHNAK antibodies react with membrane fractions of cardiomyocytes that contain voltage-gated calcium channel proteins, and labeling experiments indicate that one or both AHNAKs interact directly with the β-subunits of the channel complex (5). Because β-subunits are critical for the assembly of complete functional calcium channels (24), AHNAKs may be needed to aggregate enough calcium channels to mobilize an adequate calcium-induced calcium-release response in the RyRs of neighboring SR vesicles.
A recent report (6) describes yet another example of an association between the AHNAKs and a protein involved in calcium-related activities. The S100B protein has been found to bind to AHNAK1 with great specificity that is both calcium- and zinc-dependent (6). Although S100B is abundant in normal brain tissue, it may also be important in the heart, because its synthesis is induced in cardiomyocytes that have been rendered hypoxic (25), an effect that is also true of ischemic human hearts. The S100B protein also has some still unexplained capacity to inhibit the hypertrophic response of hearts to α-adrenergic overstimulation (26).
The pattern of anti-AHNAK antibody staining in heart sections that we describe, and the cosedimentation of AHNAK with DHP receptor-containing vesicles, raise the possibility that the AHNAKs may be important players in the excitation/contraction coupling process that drives myocardial contractility and normal cardiac function. Although mice lacking AHNAK1 show no obvious changes in cardiac function, we think it is likely that AHNAK2, located at T-tubule junction sites, is part of the coupling mechanism.
Recently published studies (27) show that the C-terminal domain of AHNAK1 is phosphorylated by protein kinase B, and they provide convincing evidence that this posttranslational modification influences the ability of AHNAK1 to partition between the nucleus and cytoplasm of epithelial cells (27). We show here that the N-terminal domain of AHNAK2 has a peptide sequence that is characteristic of PDZ domains, and as such, has the potential to bind to short C-terminal peptides of a variety of channel proteins, including the α-1 subunit of the L-type voltage-gated calcium channel.
With a PDZ domain on AHNAK2, a PI3 kinase-related regulatory site on AHNAK1, and both with multiple, linked modules, each capable of interacting with a variety of ligands, it is hard to imagine a family of molecules more suitably designed to function as a multipurpose scaffolding network.
Abbreviations: PLC, phospholipase C; SR, sarcoplasmic reticulum; T, transverse; DHP, dihydropyridine; RyR, ryanodine receptor.
References
- 1.Shtivelman, E., Cohen, F. E. & Bishop, J. M. (1992) Proc. Natl. Acad. Sci. USA 89, 5472–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimoto, T., Gamou, S., Shimizu, N., Kitajima, Y. & Nishikawa, T. (1995) Exp. Cell. Res. 217, 258–266. [DOI] [PubMed] [Google Scholar]
- 3.Hieda, Y., Tsukita, S. & Tsukita, S. (1989) J. Cell Biol. 109, 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekiya, F., Bae, Y. S., Jhon, D. Y., Hwang, S. C. & Rhee, S. G. (1999) J. Biol. Chem. 274, 13900–13907. [DOI] [PubMed] [Google Scholar]
- 5.Haase, H., Podzuweit, T., Lutsch, G., Hohaus, A., Kostka, S., Lindschau, C., Kott, M., Kraft, R. & Morano, I. (1999) FASEB J. 13, 2161–2172. [DOI] [PubMed] [Google Scholar]
- 6.Gentil, B. J., Delphin, C., Mbele, G. O., Deloulme, J. C., Ferro, M., Garin, J. & Baudier, J. (2001) J. Biol. Chem. 276, 23253–23261. [DOI] [PubMed] [Google Scholar]
- 7.Kudoh, J., Wang, Y., Minoshima, S., Hashimoto, T., Amagai, M., Nishikawa, T., Shtivelman, E., Bishop, J. M. & Shimizu, N. (1995) Cytogenet. Cell Genet. 70, 218–220. [DOI] [PubMed] [Google Scholar]
- 8.Shtivelman, E. & Bishop, J. M. (1993) J. Cell Biol. 120, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto, T., Amagai, M., Parry, D. A. D., Dixon, T. W., Tsukita, S., Tsukita, S., Miki, K., Sakai, K., Inokuchi, Y., Kudoh, J., et al. (1993) J. Cell Sci. 105, 275–286. [DOI] [PubMed] [Google Scholar]
- 10.Carl, S. L., Felix, K., Caswell, A. H., Brandt, N. R., Ball, W. J., Jr., Vaghy, P. L., Meissner, G. & Ferguson, D. G. (1995) J. Cell Biol. 129, 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marx, S. O., Reiken, S., Hisamatsu, Y., Jayaraman, T., Burkhoff, D., Rosemblit, N. & Marks, A. R. (2000) Cell 101, 365–376. [DOI] [PubMed] [Google Scholar]
- 12.Renault, L., Nassar, N., Vetter, I., Becker, J., Klebe, C., Roth, M. & Wittinghofer, A. (1998) Nature 392, 97–101. [DOI] [PubMed] [Google Scholar]
- 13.Ter Haar, E., Musacchio, A., Harrison, S. C. & Kirchhausen, T. (1998) Cell 95, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sondek, J., Bohm, A., Lambright, D. G., Hamm, H. E. & Sigler, P. B. (1996) Nature 379, 369–374. [DOI] [PubMed] [Google Scholar]
- 15.Paoli, M. (2001) Prog. Biophys. Mol. Biol. 76, 103–130. [DOI] [PubMed] [Google Scholar]
- 16.Fulop, V. & Jones, D. T. (1999) Curr. Opin. Struct. Biol. 9, 715–721. [DOI] [PubMed] [Google Scholar]
- 17.Mayer, B. J. (2001) J. Cell Sci. 114, 1253–1263. [DOI] [PubMed] [Google Scholar]
- 18.Rosa, J. L., Casaroli-Marano, R. P., Buckler, A. J., Vilaro, S. & Barbacid, M. (1996) EMBO J. 15, 4262–4273. [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa, J. L. & Barbacid, M. (1999) Oncogene 15, 1–6. [DOI] [PubMed] [Google Scholar]
- 20.Ter Haar, E., Harrison, S. C. & Kirchhausen, T. (2000) Proc. Natl. Acad. Sci. USA 97, 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunemann, M., Gerhardstein, B. L., Gao, T. & Hosey, M. M. (1999) J. Biol. Chem. 274, 33851–33854. [DOI] [PubMed] [Google Scholar]
- 22.Tsoporis, J. N., Marks, A., Kahn, H. J., Butany, J. W., Liu, P. P., O'Hanlon, D. & Parker, T. G. (1997) J. Biol. Chem. 272, 31915–31921. [DOI] [PubMed] [Google Scholar]
- 23.Tsoporis, J. N., Marks, A., Kahn, H. J., Butany, J. W., Liu, P. P., O'Hanlon, D. & Parker, T. G. (1998) J. Clin. Invest. 102, 1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunemann, M., Gerhardstein, B. L., Gao, T. & Hosey, M. M. (1999) J. Biol. Chem. 274, 33851–33854. [DOI] [PubMed] [Google Scholar]
- 25.Tsoporis, J. N., Marks, A., Kahn, H. J., Butany, J. W., Liu, P. P., O'Hanlon, D. & Parker, T. G. (1997) J. Biol. Chem. 272, 31915–31921. [DOI] [PubMed] [Google Scholar]
- 26.Tsoporis, J. N., Marks, A., Kahn, H. J., Butany, J. W., Liu, P. P., O'Hanlon, D. & Parker, T. G. (1998) J. Clin. Invest. 102, 1609–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sussman, J., Stokoe, D., Ossina, N. & Shtivelman, E. (2001) J. Cell Biol. 154, 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]