Abstract
A biological membrane is conceptualized as a system in which membrane proteins are naturally matched to the equilibrium thickness of the lipid bilayer. Cholesterol, in addition to lipid composition, has been suggested to be a major regulator of bilayer thickness in vivo because measurements in vitro have shown that cholesterol can increase the thickness of simple phospholipid/cholesterol bilayers. Using solution x-ray scattering, we have directly measured the average bilayer thickness of exocytic pathway membranes, which contain increasing amounts of cholesterol. The bilayer thickness of membranes of the endoplasmic reticulum, the Golgi, and the basolateral and apical plasma membranes, purified from rat hepatocytes, were determined to be 37.5 ± 0.4 Å, 39.5 ± 0.4 Å, 35.6 ± 0.6 Å, and 42.5 ± 0.3 Å, respectively. After cholesterol depletion using cyclodextrins, Golgi and apical plasma membranes retained their respective bilayer thicknesses whereas the bilayer thickness of the endoplasmic reticulum and the basolateral plasma membrane decreased by 1.0 Å. Because cholesterol was shown to have a marginal effect on the thickness of these membranes, we measured whether membrane proteins could modulate thickness. Protein-depleted membranes demonstrated changes in thickness of up to 5 Å, suggesting that (i) membrane proteins rather than cholesterol modulate the average bilayer thickness of eukaryotic cell membranes, and (ii) proteins and lipids are not naturally hydrophobically matched in some biological membranes. A marked effect of membrane proteins on the thickness of Escherichia coli cytoplasmic membranes, which do not contain cholesterol, was also observed, emphasizing the generality of our findings.
Cell membranes are complex and dynamic systems composed of numerous types of lipids and integral membrane proteins. Hydrophobic forces dominate the interactions between these components. Because it is energetically unfavorable for membrane proteins to expose their hydrophobic regions to water, or to embed hydrophilic portions in the hydrocarbon core of the lipid bilayer, the length of the hydrophobic regions of membrane proteins and lipids has been assumed to be hydrophobically matched (1). Hydrophobic matching has been extensively invoked to explain the dependence of enzyme activity, protein oligomerization, and protein and lipid segregation on bilayer thickness (reviewed in ref. 2).
Hydrophobic matching modulates the activity of several purified integral membrane proteins, such as Na,K-ATPase (3, 4), cytochrome c oxidase (5), Ca-ATPase (6, 7), melibiose permease (8), and diacylglycerol kinase (9), when they are reconstituted into pure phospholipid bilayers. The segregation and partition of proteins and lipids into distinctively functional areas in the membrane might also be influenced by hydrophobic matching (10). Moreover, the sorting of membrane proteins in the exocytic pathway of eukaryotic cells has been explained on the basis of matching the hydrophobic portion of transmembrane domains to the different hydrophobic thicknesses of the membranes along this pathway (11–13).
The bilayer thickness of the membrane is critical in hydrophobic matching. Lipid properties, such as the degree of unsaturation and acyl chain length, have been demonstrated to modulate membrane thickness in both simple model systems and biological membranes (14–17). Additionally, cholesterol has also been shown to affect the thickness of artificial bilayer systems in vitro. Extensive experimental and computational data for pure phospholipid/cholesterol systems have demonstrated that, under certain circumstances, cholesterol increases bilayer thickness (18–20), presumably due to the ordering of the acyl chains of phospholipids. Because cholesterol is ubiquitous in eukaryotic cell membranes, it has been suggested that cholesterol is a principal modulator of bilayer thickness in these cells. In particular, the increase in cholesterol content along the membranes of the endoplasmic reticulum (ER), the Golgi, and the plasma membrane(s), as measured by lipid analysis of cell fractions (21–23) and electron microscopy analysis using the cholesterol-binding antibiotic filipin (24), has been taken to indicate a progressive increase in bilayer thickness along this route.
However, the bilayer thicknesses of eukaryotic cell membranes and the effect of cholesterol have not been directly measured, but merely inferred by extrapolation from model lipid bilayers, systems that may be oversimplified considering the large complexity of cell membranes. We directly test the effect of both cholesterol and resident membrane proteins on the thickness of cell membranes by using solution x-ray scattering (SXS) on membrane fractions purified from rat hepatocytes and Escherichia coli. SXS has been successfully implemented in directly measuring the average bilayer thickness of liposomes and the plasma membrane of Mycoplasma laidlawii (14, 15, 17). Our data provide direct values for the bilayer thicknesses of eukaryotic cell membranes and show, contrary to expectation, that, in biological membranes, the average bilayer thickness is modulated by the transmembrane domains of proteins rather than cholesterol and that lipid and protein components may not be naturally hydrophobically matched in a biological membrane.
Materials and Methods
Chemicals. Chemicals were purchased from Sigma. Lowry protein assay kit was purchased from Bio-Rad. All research grade phospholipids were obtained from Avanti Polar Lipids. Amplex red cholesterol assay kit was purchased from Molecular Probes.
Purification of Membranes. Exocytic pathway membranes were isolated from rat hepatocytes. Rough ER membranes were purified according to Bergstrand and Dallner (21). Purified rough ER membranes were further treated with puromycin to strip ribosomes from the membranes according to Görlich et al. (25). Golgi membranes were purified following the protocol of Hui et al. (26). Purified membranes were assayed for β 1,4-galactosyltransferase (27). Apical and basolateral plasma membrane vesicles were purified as in Meier et al. (23). Apical and basolateral membrane fractions were routinely assayed for 5′-nucleotidase activity (28). Total protein concentration in all samples was determined according to Lowry et al. (29). Total phospholipid content of the membrane fractions was determined by phosphorous analysis according to Rouser et al. (30). All membranes were flash-frozen in liquid N2 and stored at –80°C. Bacterial inner and outer membranes were isolated according to Ishidate et al. (31) from Dh5α cells derived from the E. coli K-12 strain. The isolated membranes were tested for NADH oxidase (an inner membrane marker) according to Osborn and Munson (32).
Electron Microscopy. Membranes were fixed in PBS, 0.2 M sucrose, and 2% glutaraldehyde at room temperature for 30 min, and spun in a swinging bucket rotor (5417R, Eppendorf) at 10,000 rpm for 20 min. The resultant pellet was extensively washed with PBS, postfixed for 30 min with 1% osmium tetroxide, 1.5% cyanoferrate, and 0.1 M cacodylate buffer (pH 7.4), and then dehydrated and embedded in Epon 812. Ultrathin sections (50–70 nm) were cut by using an ultramicrotome 2E, placed on a nickel grid, and stained with 2% uranylacetate and lead citrate.
Sample Preparation for SXS Measurements. Purified plasma membranes stored at –80°C were thawed and pelleted by centrifugation at 15,000 × g for 5 min. The pellet was washed three times with 1 ml of buffer A (25 mM Hepes-KOH/5 mM MgCl2, pH 7.4). Subsequently, the membranes were incubated with 1 ml of buffer A containing 3 mg of proteinase K and 3 mg of trypsin for 2.5 h. To quench the proteolytic reaction, the protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) was added at a final concentration of 20 mM, and the samples were incubated for 30 min followed by three washing steps with buffer A. Golgi membranes were treated similarly, except that the protease incubation time was prolonged to 4.5 h. ER membranes were treated as plasma membranes with the exception that, after protease treatment, membranes were incubated with 1 ml of RNase A (3 mg/ml) in buffer B (25 mM Hepes-KOH/100 mM NaCl, pH 7.4) to degrade ribosomal RNA. Membranes before and after protease treatment were analyzed by SDS/PAGE using precast 4–12% [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane Bistris NuPAGE (Invitrogen) gels. The running buffer contained 50 mM Mes, 50 mM TrisBase, 3.4 mM SDS, and 1 mM EDTA (pH 7.4). Membrane samples were loaded onto the gel after heat denaturation. The protein content of all membranes was determined according to Lowry et al. (29). Protein bands were visualized by using silver staining. SXS intensity measurements were obtained by subtracting buffer scatter from that of the membrane suspension. The supernatant of the last wash of a particular membrane sample was used as the buffer. Buffer was centrifuged at 120 × g for 1 min into a glass capillary held in a specially constructed sample holder. After the diffraction measurement, buffer was centrifuged out of, and a 10-μl membrane sample was transferred into, the same glass capillary.
Preparation of Cholesterol-Depleted Membranes and Liposomes of Extracted Lipids. Protease-treated membranes were depleted of cholesterol by incubation, for 15–60 min depending on the membrane, with 10 mM 2-hydroxypropyl-β-cyclodextrin or methyl-β-cyclodextrin in buffer consisting of 25 mM Hepes-KOH and 5 mM MgCl2 (pH 7.4). Cholesterol content of membranes was measured by total lipid extraction according to Bligh and Dyer (33) and subsequent enzymatic determination of cholesterol using a fluorescence assay (34). Total lipid extracts of untreated membranes were obtained according to Bligh and Dyer (33). The solvent was evaporated under a stream of N2 gas, and the lipid film was dried under vacuum for 12 h. The lipid mixture was then hydrated in buffer (20 mM Hepes/100 mM NaCl, pH 7.4) at 40°C, and the vesicle solution was subjected to extrusion at 40°C by using an extruder (Lipex Biomembranes, Vancouver) coupled to an external water bath according to Hope et al. to produce unilammellar liposomes (35). Total phospholipid content of the liposomes was determined by phosphorous analysis according to Rouser et al. (30).
SXS Instrumentation and Data Processing. An R-axis IV x-ray crystallography work station was modified and adapted for SXS measurements following a setup similar to the one described by Bu et al. (36). All measurements were done under the following conditions: detector to sample distance, 615 mm; temperature, 38°C; exposure time, 360 min or 480 min. Measurements on a buffer-sample pair were performed by using the same capillary. Glass capillaries were purchased from the Charles Supper Company (Natick, MA). Data were collected by using control software from Molecular Structure (The Woodlands, TX). The direct beam position was calculated by using the program display, also from Molecular Structure. Buffer subtraction and subsequent data processing were performed by using programs written by A. Perlo (Yale University). After buffer subtraction, I(S) was calculated from the raw data by summing the intensities lying in a ring at radius S (from the position of the direct beam), and of finite thickness, ΔS. Here, S = 2sin(θ)/λ, where θ is one-half the scattering angle. The corrected intensity, C(S), was calculated by applying a correction factor, which includes the Lorentz factor, a dimensionality factor and the polarization correction. The combined correction factor is thus 4sinθ/{λ[cos2(2θ)][1 + cos2(2θ)]}. The second maximum in the corrected scattering curve was fit with a Gaussian distribution to best measure S = 2/d, where d is the separation of phosphate layers in a bilayer.
Results
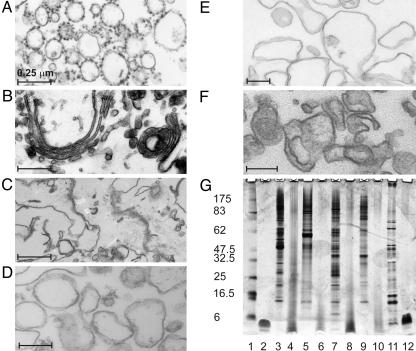
Purification and Characterization of Membranes. Membranes from the rough ER and Golgi apparatus, and the basolateral and apical plasma membranes were purified from rat liver by using established protocols. Electron micrographs of the isolated membrane fractions showed characteristic rough ER, Golgi, and plasma membrane morphology in line with published data (Fig. 1) (21, 23, 26). The rough ER preparation was almost entirely composed of vesicles coated with ribosomes (Fig. 1A). Few, if any Golgi, plasma membrane, or mitochondrial structures were observed, indicating the high purity of the preparation. Golgi membranes were purified 80- to 100-fold over homogenate, as determined by β-1,4-galactosyltransferase activity. Golgi stacks were clearly visible (Fig. 1B), indicating that the structure of this organelle was at least partially preserved during purification. The total plasma membrane fraction contained typical membrane sheets with gap junctions and desmosomes (Fig. 1C). Intracellular organelles were rarely detected. After tight homogenization of this sample, a large proportion of the membranes was vesiculated, and subsequent high-speed centrifugation allowed the separation of basolateral (Fig. 1D) and apical (Fig. 1E) plasma membrane fractions. Apical plasma membranes were enriched ≈50-fold over the initial homogenate and basolateral plasma membranes 25- to 35-fold, as determined by 5′-nucleotidase and alkaline phosphodiesterase activity, respectively. Cytoplasmic membrane fractions from E. coli were purified according to Ishidate et al. (31) and determined to be enriched ≈100-fold over homogenate, as determined by NADH oxidase activity (an inner membrane marker) according to Osborn and Munson (32).
Fig. 1.
Characterization of membrane samples. Electron micrographs of embedded, sectioned, and stained membranes isolated from rat hepatocytes. Shown are rough ER microsomes (A), Golgi membranes (B), mixed plasma membranes (C), basolateral (D) and apical (E) plasma membrane vesicles, and protease-treated ER microsomes (F). Scale bar corresponds to 0.25 μm. (G) Silver-stained 4–12% Bistris SDS/PAGE of membrane fractions. Shown are molecular mass markers (lane 1); 3.5-kDa transmembrane peptide, Gp55 (lane 2); ER microsomes before (lane 3) and after (lane 4) protease treatment; Golgi membranes before (lane 5) and after (lane 6) protease treatment; basolateral plasma membranes before (lane 7) and after (lane 8) protease treatment; apical plasma membranes before (lane 9) and after (lane 10) protease treatment; and E. coli cytoplasmic membranes before (lane 11) and after (lane 12) protease treatment.
The Bilayer Thickness of Exocytic Pathway Membranes. To measure the bilayer thickness of exocytic pathway membranes we used SXS techniques. SXS permits direct determination of the distance (d) between the highly electron-dense phosphate groups across the bilayer of membranes in solution (14, 15, 17). A limitation of this technique is that extramembranous protein domains and cell components containing highly electron-dense groups, such as ribosomes, can affect the quality of the scattering data (37). The rat hepatocyte membranes were therefore treated with proteinase K and trypsin under hypoosmotic conditions to both digest extracellular domains and vesicularize membranes. In the case of the rough ER fraction, puromycin and RNase A treatment was also carried out to detach and degrade ribosomes. On treatment, membranes remained intact as demonstrated by electron microscopy (Fig. 1F). Determination of the protein-to-phospholipid ratio before and after treatment clearly showed a large decrease in protein content (Table 1). SDS/PAGE analysis confirmed that protease treatment effectively degraded proteins such that the majority of the remaining peptides demonstrated gel mobilities similar to that of an appropriate standard, the 3.5-kDa transmembrane domain of a membrane receptor (Fig. 1G). To ensure that the lipid composition of membranes remained unaltered during our study, lipid extracts were routinely analyzed by TLC, and phospholipid spots were quantified (data not shown).
Table 1. Protein-to-phospholipid mass ratios of membranes used in SXS experiments.
| Membranes | Protein/phospholipid, mg/mg |
|---|---|
| ER membranes*† | 2.6 |
| Untreated ER | 2.4 ± 0.2 |
| Protease-treated ER‡ | 0.12 ± 0.01 |
| Cholesterol-depleted ER§ | 0.15 ± 0.02 |
| ER liposomes | ND¶ |
| Golgi membranes* | 1.8 |
| Untreated Golgi | 1.7 ± 0.09 |
| Protease-treated Golgi | 0.08 ± 0.01 |
| Cholesterol-depleted Golgi§ | 0.09 ± 0.01 |
| Golgi liposomes | ND |
| BPM*∥ | 2.2 |
| Untreated BPM | 2.8 ± 0.3 |
| Protease-treated BPM | 0.16 ± 0.2 |
| Cholesterol-depleted BPM§ | 0.17 ± 0.1 |
| BPM liposomes | ND |
| APM*,** | 1.5 |
| Untreated APM | 1.7 ± 0.2 |
| Protease-treated APM | 0.11 ± 0.01 |
| Cholesterol-depleted APM§ | 0.11 ± 0.01 |
| APM liposomes | ND |
| E. coli cytoplasmic membranes* | 2.0 ± 0.2 |
| Untreated E. coli membranes | 1.8 ± 0.2 |
| Protease-treated E. coli membranes | 0.13 ± 0.2 |
| E. coli membrane liposomes | ND |
Values for smooth ER were used because these corresponded most closely to rough ER membranes depleted of ribosomes.
Protein values for the digested membranes are artificially low because colorimetric assays do not quantitatively measure short hydrophobic peptides.
Membranes were protease treated before cholesterol depletion.
Protein content was below the detection limit of the assay. ND, not determined.
BPM, basolateral plasma membrane.
APM, apical plasma membrane.
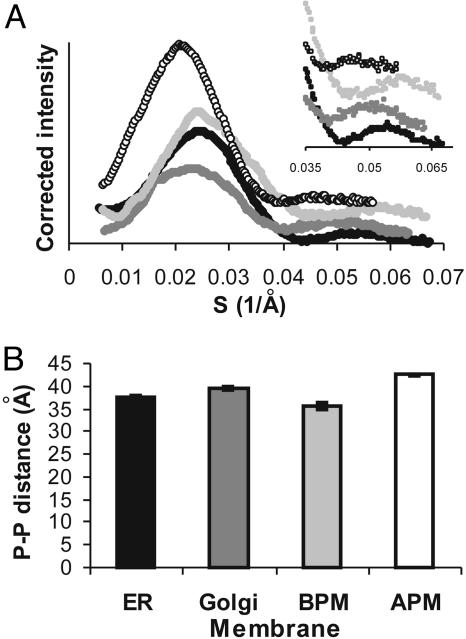
Scattering curves, corrected for background and intensity, were generated in triplicate for all of the treated membranes (Fig. 2A). The curves are characteristic of single bilayer sheets or vesicles, indicating (in addition to EM visualization) that the bilayer structure of the membranes was retained after protease treatment. The average bilayer thickness, d, can be calculated from the position of the second peak (14) and was found to be highly reproducible for each type of membrane. The second peak in each scattering curve was fitted to a Gaussian distribution to define the maximum. Fig. 2B illustrates the change in bilayer thickness [phosphate-to-phosphate (P-P) distance = d] for membranes of the exocytic pathway. Rough ER membranes have a thickness of 37.5 ± 0.4 Å. Golgi membranes (39.5 ± 0.4 Å) are 2.0 ± 0.8 Å thicker than ER membranes. Basolateral plasma membranes (35.6 ± 0.6 Å) are 3.9 ± 1.0 Å thinner than Golgi membranes whereas apical plasma membranes (42.5 ± 0.3 Å) are 6.9 ± 0.9 Å thicker than basolateral plasma membranes. Based on the cholesterol content of these membranes, the bilayer thickness would have been predicted to increase progressively from the ER, to the Golgi, to the basolateral and apical plasma membranes. However, the basolateral membrane is the thinnest of all membranes despite having cholesterol levels comparable to that of the apical plasma membrane, suggesting that cholesterol might not determine bilayer thickness.
Fig. 2.
Bilayer thicknesses of membranes along the exocytic pathway of rat hepatocytes. (A) Corrected scattering curves from SXS measurements of ER membranes (filled circles), Golgi membranes (dark gray circles), basolateral plasma membranes (BPM, light gray circles), and apical plasma membranes (APM, open circles). (Inset) Second maxima. (B) Mean phosphate-to-phosphate (P-P) distances calculated from the second maxima in A, with error bars from repeated measurements.
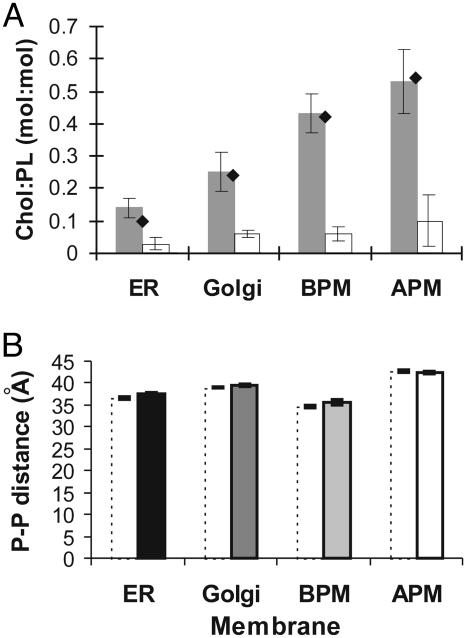
Effect of Cholesterol on the Bilayer Thickness of Exocytic Pathway Membranes. We directly measured the effect of cholesterol on bilayer thickness by depleting cholesterol from the exocytic pathway membranes using cyclodextrins. The cholesterol content of the membranes before and after the extraction procedure was measured by using a fluorometric assay (34) and is shown in Fig. 3A. The cholesterol content of the purified membranes corresponds well with published values (21–23). Between 70% and 90% of cholesterol was removed from the membranes, a figure that is in line with cholesterol depletion experiments on comparable membranes (38–40). Most strikingly, our measurements on cholesterol-depleted membranes show that cholesterol depletion has little or no effect on the bilayer thickness of any of the membranes (Fig. 3B). Golgi and apical plasma membranes retain their respective bilayer thicknesses on cholesterol depletion. In the case of ER membranes, bilayer thickness decreases by only 1.0 ± 0.6 Å and, similarly, basolateral plasma membrane thickness decreases by 1.0 ± 1.0 Å. Given the marginal effect of cholesterol depletion on these membranes, we decided to investigate whether the membrane proteins resident in these exocytic pathway membranes might be able to modulate bilayer thickness.
Fig. 3.
Effect of cholesterol depletion on bilayer thickness. (A) Cholesterol to phospholipid (Chol:PL) molar ratios of membranes before (shaded columns), and after (open columns) incubation with cyclodextrins. For comparison, the cholesterol content of intact membranes as calculated from the literature (21–23) is given (filled diamonds). Standard deviations from repeated measurements are indicated. BPM, basolateral plasma membrane; APM, apical plasma membrane. (B) Mean phosphate-to-phosphate (P-P) distances of cholesterol-depleted membranes (dotted columns) of the ER and Golgi, and basolateral (BPM) and apical (APM) plasma membranes. Distances are calculated from second maxima, with error bars shown. The thickness values before cholesterol depletion are shown for reference (filled columns).
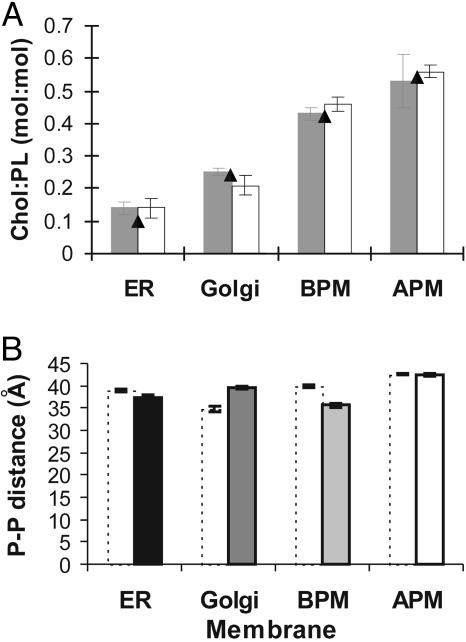
Effect of Membrane Proteins on the Bilayer Thickness of Exocytic Pathway Membranes. To establish the effect of membrane proteins on membrane thickness, we measured the average bilayer thickness of liposomes formed from total lipid extracts of each type of membrane. The protein, cholesterol, and phospholipid content of the reconstituted liposomes was analyzed, indicating that, on lipid extraction, all proteinaceous components were absent and that we were able to reproduce the cholesterol to phospholipid ratio of the original membranes (Table 1 and Fig. 4A). TLC lipid analysis of the original membranes and the reconstituted liposomes showed that no major lipid component was absent (data not shown). Measurements on reconstituted liposomes show that ER and basolateral plasma membranes increase in thickness on protein depletion to 38.9 ± 0.4 Å and 39.8 ± 0.2 Å, respectively (increases of 1.4 ± 0.8 Å and 4.2 ± 0.8 Å), whereas Golgi membranes become thinner by 4.6 ± 1.0 Å to 34.9 ± 0.6 Å (Fig. 4B). Protein-depleted liposomes of apical plasma membranes have the same average thickness as the parental membranes. The potential loss of lipid asymmetry on lipid extraction/reconstitution should not affect bilayer thickness as demonstrated schematically in Fig. 5. Thus, our results show that, in contrast to cholesterol depletion, protein depletion has a marked effect on the bilayer thickness of certain eukaryotic membranes.
Fig. 4.
Effect of protein depletion on bilayer thickness. (A) Cholesterol to phospholipid (Chol:PL) molar ratios of membranes before (gray columns) and after (open columns) protein depletion. For comparison, the cholesterol content of intact membranes (see Fig. 3) is given (filled triangles). Standard deviations from repeated measurements are indicated. BPM, basolateral plasma membrane; APM, apical plasma membrane. (B) Mean phosphate-to-phosphate (P-P) distances of protein-depleted membranes (dotted columns) of the ER and Golgi, and basolateral (BPM) and apical (APM) plasma membranes. Distances are calculated from second maxima, with error bars shown. The thickness values before protein depletion are shown for reference (filled columns).
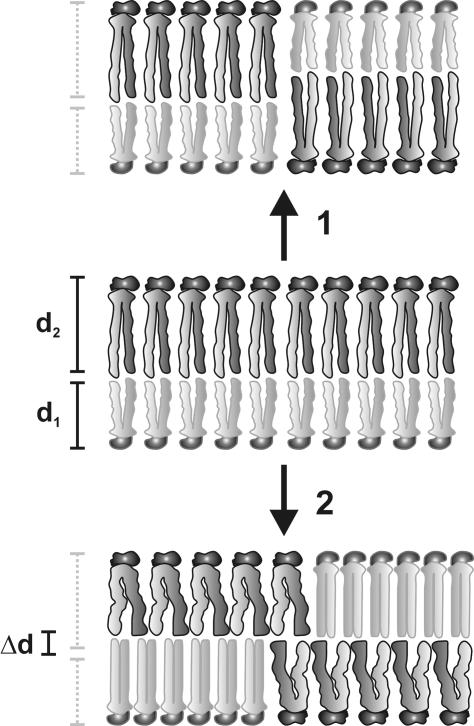
Fig. 5.
Thickness of model bilayers with symmetric or asymmetric lipid distributions. An asymmetric bilayer (Middle) with two lipid components, L1, in the inner leaflet, and L2, in the outer leaflet, with thicknesses d1 and d2, respectively, has a total thickness of d1 + d2. On loss of asymmetry, L1 and L2 are distributed equally in both leaflets. In 1, d1 and d2 are unchanged; hence, the total thickness remains d1 + d2. In 2, both lipids undergo conformational changes to hydrophobically match each other, with L1 extending by a distance Δd and L2 compressing by Δd, resulting in a total bilayer thickness of d1 + d2.
We wanted to establish the generality of this effect by repeating measurements on membranes lacking endogenous cholesterol. E. coli cytoplasmic membranes lack cholesterol and were thus used to generate samples for SXS measurements. Membranes were characterized before and after protein depletion (Table 1 and Fig. 1G), and the bilayer thicknesses of each membrane sample were measured. Scattering curves, corrected for background and intensity, were generated in triplicate, and the average bilayer thickness, d, calculated from the second peak was 37.5 ± 0.5 Å. The thickness of liposomes of extracted lipids was measured to be 33.5 ± 0.4 Å, indicating that, even in prokaryotic membranes naturally lacking endogenous cholesterol, membrane proteins modulate bilayer thickness.
Discussion
The Role of Cholesterol in Biological Membranes. Cholesterol has been shown in vitro to increase acyl chain order (41), which in turn has been demonstrated to increase bilayer thickness in simple model lipid systems (42, 43). The effect of cholesterol on the order of acyl chains has also been observed in vivo. Cholesterol increases membrane ordering in LM cells (mouse fibroblasts) (44), Chinese hamster ovary cells (45), Acholeplasma laidlawii (46), and erythrocytes (47). Analogously to simple model bilayers, acyl chain ordering in biological membranes has been extrapolated to effect an increase in membrane thickness. According to data obtained from model phospholipid/cholesterol bilayers, molar cholesterol to phospholipid ratios of 0.12 and 1 should lead to increases in bilayer thickness of 2 and 7 Å, respectively, compared with systems with no cholesterol (18–20). Thus, the predicted decrease in bilayer thickness on cholesterol depletion would have been ≈2 Å (instead of ≤1 Å) for ER and Golgi membranes and ≈5 Å (instead of ≤1 Å) for basolateral and apical plasma membranes, given cholesterol-to-phospholipid molar ratios of 0.08–0.1, 0.16–0.2, and 0.4–0.76, respectively (22, 48–51). Our data indicate that the assumption that cholesterol affects the thickness of biological membranes is not valid and suggest that the cholesterol content in cell membranes is not modulated for the purpose of controlling bilayer thickness. It should, however, be noted that our measurements provide an average bilayer thickness. Thus, the existence of domains of differing thickness, in which cholesterol could locally modulate thickness, is possible. However, because we obtain scattering curves with well defined maxima, only two types of domain distributions are probable: (i) a negligible number of domains with a large deviation in thickness from the measured average, or (ii) many domains with thicknesses close to the measured average.
Why is the modulation of bilayer thickness by cholesterol seen in model systems not observed in biological membranes? Such a discrepancy may stem from the great complexity of cell membranes, which are comprised of a large variety of lipids. Experiments on pure cholesterol/lipid systems have shown that the effect of cholesterol on the physical properties of the bilayer can vary depending on the type of other lipid components present (recently reviewed in ref. 52).
Although cholesterol may not be used in biological membranes to modulate bilayer thickness, its uses are manifold. Cholesterol can directly affect protein activity by specifically interacting with some proteins, which require specific sterols (53). Cholesterol can also indirectly affect protein/lipid function in the membrane and many important processes in the cell by modulating bulk physical properties of the membrane, such as bilayer fluidity/permeability, lateral phase separation, phase transition from the gel to the liquid crystalline state, and lateral pressure (52, 54–56).
Bilayer Thickness and the Sorting of Membrane Proteins. Our measurements on the bilayer thickness of exocytic pathway membranes provide insights into the mechanisms for the sorting of membrane proteins in eukaryotic cells. Sequence analysis has suggested that transmembrane domains of ER/Golgi residents are on average 5 amino acid residues shorter than those of plasma membrane residents (57). Experiments in unpolarized cells have shown that lengthening the transmembrane domain of Golgi resident proteins leads to their appearance on the plasma membrane (11–13). These data led to the suggestion that the sorting of membrane proteins in the exocytic pathway of eukaryotic cells is governed by hydrophobic matching of the transmembrane domains to increasingly thicker membranes along this pathway (11–13). Because in rat hepatocytes membrane proteins move from the ER to the Golgi to the basolateral plasma membrane before reaching the apical plasma membrane (58, 59), an expectation would have been that bilayer thickness would have increased along this route. We now find that the basolateral plasma membrane is the thinnest of all exocytic pathway membranes. Thus, a membrane protein in transit must cross from a relatively thick Golgi membrane through a thinner basolateral plasma membrane before it reaches the much thicker apical plasma membrane. In such a scenario, it seems unlikely that hydrophobic matching to the average bilayer thickness could be driving the localization of membrane proteins in the exocytic pathway of polarized rat hepatocytes.
Membrane Proteins, Hydrophobic Matching, and the Energy of Biological Membranes. In 1972, Singer and Nicolson (60) formulated a framework for the conceptualization of biological membrane structure that persists to this day. The fluid mosaic model posits that “the structures of the lipid in the membrane and of the lipid in isolated aqueous dispersion are closely similar” and that “hydrophobic and hydrophilic interactions are to be maximized and the lowest free energy state is to be attained for the intact membrane in an aqueous environment” (60). Thus, this prevalent view of membrane structure would have predicted that removal of resident integral membrane proteins from a naturally hydrophobically matched biological membrane would not affect bilayer thickness. We show that removal of membrane proteins from rat hepatocyte Golgi and basolateral plasma membranes, and E. coli cell membranes, can increase or decrease bilayer thickness by up to 4–5 Å, depending on the membrane studied, suggesting that membrane proteins can significantly modulate the bilayer thickness of these membranes. Supporting this possibility, Weiss et al. (61) have recently shown, using proteosomes consisting of pure phospholipid and the membrane protein gramicidin, that phospholipids can clearly modulate their bilayer thickness to match the hydrophobic thickness of gramicidin channels. Likely, transmembrane domains modulate bilayer thickness by modulating lipid acyl chain conformation and packing by means of the hydrophobic effect (19, 62). The negligible contribution of transmembrane domains to bilayer thickness in ER/apical plasma membranes is probably due to the large percentage of dolichol/sphingomyelin in these membranes, both of which may diminish the malleability of the bulk lipid to conform to the hydrophobic length of membrane protein components.
Our data suggest a modified view of the structure of biological membranes, in which component lipids and proteins may not be naturally matched. These membranes are consequently in a high-energy state because lipids and proteins strain (expend energy) to match each other hydrophobically (Fig. 6). Membranes that are not at their free-energy minimum would have altered properties, such as a lower energy of membrane deformation. We suggest that cells might take advantage of such altered membrane properties and maintain some of their membranes in a high-energy state to facilitate vital cellular and physiological processes, such as membrane fusion and protein insertion.
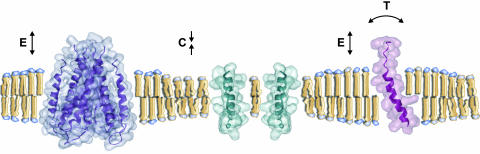
Fig. 6.
Modified view of the structure of a biological membrane. Because component lipids and proteins are not naturally matched in this membrane, they must strain (expend energy) to match each other hydrophobically, resulting in a high-energy membrane. Compensatory conformational changes include lipid acyl chain extension (E) and transmembrane helix tilting (T) when lipids surround a protein with a long transmembrane region, and lipid acyl chain compression (C) when lipids surround a protein with a short transmembrane domain.
Acknowledgments
We thank H. Wyckoff for advice on SXS instrumentation and G. Johnson and P. Pepin for assistance with SXS instrumentation, and A. Perlo for writing the SXS data processing software. K.M. was supported in part by a Postgraduate Scholarship fellowship from the Natural Science and Engineering Research Council of Canada. I.U.-B. was supported in part by a postdoctoral fellowship from the Basque Government (Spain). T.T. was supported in part by a JSPS (Japan Society for the Promotion of Science) overseas research fellowship. This work was supported by grants from the National Institutes of Health, the National Science Foundation, and the Ludwig Institute for Cancer Research.
Abbreviations: ER, endoplasmic reticulum; SXS, solution x-ray scattering.
References
- 1.Mouritsen, O. G. & Bloom, M. (1984) Biophys. J. 46, 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Killian, J. A. (1998) Biochim. Biophys. Acta 1376, 401–415. [DOI] [PubMed] [Google Scholar]
- 3.Cornelius, F. (2001) Biochemistry 40, 8842–8851. [DOI] [PubMed] [Google Scholar]
- 4.Johannsson, A., Smith, G. A. & Metcalfe, J. C. (1981) Biochim. Biophys. Acta 641, 416–421. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco, C., Smith, G. A., Dabbeni-sala, F., Johannsson, A., Galante, Y. M. & Bisson, R. (1982) FEBS Lett. 144, 145–148. [DOI] [PubMed] [Google Scholar]
- 6.Cornea, R. L. & Thomas, D. D. (1994) Biochemistry 33, 2912–2920. [DOI] [PubMed] [Google Scholar]
- 7.Lee, A. G. (1998) Biochim. Biophys. Acta 1376, 381–390. [DOI] [PubMed] [Google Scholar]
- 8.Dumas, F., Tocanne, J. F., Leblanc, G. & Lebrun, M. C. (2000) Biochemistry 39, 4846–4854. [DOI] [PubMed] [Google Scholar]
- 9.Pilot, J. D., East, J. M. & Lee, A. G. (2001) Biochemistry 40, 8188–8195. [DOI] [PubMed] [Google Scholar]
- 10.Sprong, H., van der Sluijs, P. & van Meer, G. (2001) Nat. Rev. Mol. Cell. Biol. 2, 504–513. [DOI] [PubMed] [Google Scholar]
- 11.Masibay, A. S., Balaji, P. V., Boeggeman, E. E. & Qasba, P. K. (1993) J. Biol. Chem. 268, 9908–9916. [PubMed] [Google Scholar]
- 12.Munro, S. (1991) EMBO J. 10, 3577–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro, S. (1995) EMBO J. 14, 4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman, D. M. (1971) J. Mol. Biol. 58, 153–165. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, B. A. & Engelman, D. M. (1983) J. Mol. Biol. 166, 211–217. [DOI] [PubMed] [Google Scholar]
- 16.Thurmond, R. L., Niemi, A. R., Lindblom, G., Wieslander, A. & Rilfors, L. (1994) Biochemistry 33, 13178–13188. [DOI] [PubMed] [Google Scholar]
- 17.Wilkins, M. H., Blaurock, A. E. & Engelman, D. M. (1971) Nat. New Biol. 230, 72–76. [DOI] [PubMed] [Google Scholar]
- 18.Hui, S. W. & He, N.-B. (1983) Biochemistry 22, 1159–1164. [DOI] [PubMed] [Google Scholar]
- 19.Nezil, F. A. & Bloom, M. (1992) Biophys. J. 61, 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smondyrev, A. M. & Berkovitz, M. L. (1999) Biophys. J. 77, 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergstrand, A. & Dallner, G. (1969) Anal. Biochem. 29, 351–356. [DOI] [PubMed] [Google Scholar]
- 22.Keenan, T. W. & Morre, D. J. (1970) Biochemistry 9, 19–25. [DOI] [PubMed] [Google Scholar]
- 23.Meier, P. J., Sztul, E. S., Reuben, A. & Boyer, J. L. (1984) J. Cell Biol. 98, 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orci, L., Montesano, R., Meda, P., Malaisse-Lagae, F., Brown, D., Perrelet, A. & Vassalli, P. (1981) Proc. Natl. Acad. Sci. USA 78, 293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Görlich, D., Prehn, S., Hartmann, E., Kalies, K.-U. & Rapoport, T. A. (1992) Cell 71, 489–503. [DOI] [PubMed] [Google Scholar]
- 26.Hui, N., Nakamura, N., Slusarewicz, P. & Warren, G. (1998) in Cell Biology: A Laboratory Handbook (Academic, San Diego), Vol. 2, pp. 46–55. [Google Scholar]
- 27.Bretz, R. & Staubli, W. (1977) Eur. J. Biochem. 77, 181–192. [DOI] [PubMed] [Google Scholar]
- 28.Reinhart, P. H. & Bygrave, F. L. (1981) Biochem. J. 194, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry, O. H., Rosebrough, J. N., Farr, A. L. & Randall, R. J. (1951) J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- 30.Rouser, G., Fleischer, S. & Yamamoto, A. (1969) Lipids 5, 494–496. [DOI] [PubMed] [Google Scholar]
- 31.Ishidate, K., Creeger, E. S., Zrike, J., Deb, S., Glauner, B., MacAlister, T. J. & Rothfield, L. I. (1986) J. Biol. Chem. 261, 428–443. [PubMed] [Google Scholar]
- 32.Osborn, M. J. & Munson, R. (1974) Methods Enzymol. 31, 642–653. [DOI] [PubMed] [Google Scholar]
- 33.Bligh, E. G. & Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 34.Amundson, D. M. & Zhou, M. (1999) J. Biochem. Biophys. Methods 38, 43–52. [DOI] [PubMed] [Google Scholar]
- 35.Hope, M. J., Bally, M. B., Webb, G. & Cullis, P. R. (1985) Biophys. Biochim. Acta 812, 55–65. [DOI] [PubMed] [Google Scholar]
- 36.Bu, Z., Perlo, A., Johnson, J., Olack, J., Engelman, D. M. & Wyckoff, H. W. (1998) J. Appl. Crystallogr. 31, 533–543. [Google Scholar]
- 37.Bouwstra, J. A., Gooris, G. S., Bras, W. & Talsma, H. (1993) Chem. Phys. Lipids 64, 83–98. [DOI] [PubMed] [Google Scholar]
- 38.Gidwani, A., Holowka, D. & Baird, B. (2001) Biochemistry 40, 12422–12429. [DOI] [PubMed] [Google Scholar]
- 39.Ohtani, Y., Irie, T., Uekama, K., Fukunaga, K. & Pitha, J. (1989) Eur. J. Biochem 186, 17–22. [DOI] [PubMed] [Google Scholar]
- 40.Scheiffele, P., Roth, M. G. & Simons, K. (1997) EMBO J. 16, 5501–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeagle, P. L. (1985) Biochim. Biophys. Acta 822, 267–287. [DOI] [PubMed] [Google Scholar]
- 42.Seelig, A. & Seelig, J. (1974) Biochemistry 13, 4839–4845. [DOI] [PubMed] [Google Scholar]
- 43.Ipsen, J. H., Mouritsen, O. G. & Bloom, M. (1990) Biophys. J. 57, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rintoul, D. A., Chou, S. M. & Silbert, D. F. (1979) J. Biol. Chem. 254, 10070–10077. [PubMed] [Google Scholar]
- 45.Sinensky, M., Duwe, G. & Pinkerton, F. (1979) J. Biol. Chem. 254, 4482–4486. [PubMed] [Google Scholar]
- 46.Davis, J. H., Bloom, M., Butler, K. W. & Smith, I. C. (1980) Biochim. Biophys. Acta 597, 477–491. [DOI] [PubMed] [Google Scholar]
- 47.Kelusky, E. C., Dufourc, E. J. & Smith, I. C. (1983) Biochim. Biophys. Acta 735, 302–304. [DOI] [PubMed] [Google Scholar]
- 48.van Meer, G. (1998) Trends Cell Biol. 8, 29–33. [DOI] [PubMed] [Google Scholar]
- 49.van Meer, G. (1989) Annu. Rev. Cell Biol. 5, 247–275. [DOI] [PubMed] [Google Scholar]
- 50.Colbeau, A., Nachbaur, J. & Vignais, P. M. (1971) Biochim. Biophys. Acta 249, 462–492. [DOI] [PubMed] [Google Scholar]
- 51.Zambrano, F., Fleischer, S. & Fleischer, B. (1975) Biochim. Biophys. Acta 380, 357–369. [DOI] [PubMed] [Google Scholar]
- 52.Ohvo-Rekila, H., Ramstedt, B., Leppimaki, P. & Slotte, J. P. (2002) Prog. Lipid Res. 41, 66–97. [DOI] [PubMed] [Google Scholar]
- 53.Albert, A. D., Young, J. E. & Yeagle, P. L. (1996) Biochim. Biophys. Acta 1285, 47–55. [DOI] [PubMed] [Google Scholar]
- 54.Cantor, R. S. (1999) Biophys. J. 76, 2625–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Kruyff, B., Demel, R. A. & van Deenen, L. L. (1972) Biochim. Biophys. Acta 255, 331–347. [DOI] [PubMed] [Google Scholar]
- 56.Macdonald, A. G., Wahle, K. W., Cossins, A. R. & Behan, M. K. (1988) Biochim. Biophys. Acta 938, 231–242. [DOI] [PubMed] [Google Scholar]
- 57.Bretscher, M. S. & Munro, S. (1993) Science 261, 1280–1281. [DOI] [PubMed] [Google Scholar]
- 58.Keller, P. & Simons, K. (1997) J. Cell Sci. 110, 3001–3009. [DOI] [PubMed] [Google Scholar]
- 59.Wilton, J. C. & Matthews, G. M. (1996) BioEssays 18, 229–236. [DOI] [PubMed] [Google Scholar]
- 60.Singer, S. J. & Nicolson, G. L. (1972) Science 175, 720–731. [DOI] [PubMed] [Google Scholar]
- 61.Weiss, T. M., Van Der Wel, P. C., Killian, J. A., Koeppe, R. E., 2nd, & Huang, H. W. (2003) Biophys. J. 84, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Planque, M. R., Greathouse, D. V., Koeppe, R. E., 2nd, Schafer, H., Marsh, D. & Killian, J. A. (1998) Biochemistry 37, 9333–9345. [DOI] [PubMed] [Google Scholar]