Abstract
DAXX, a modulator of apoptosis and a repressor of basal transcription, was identified in a two-hybrid screen as a protein capable of interacting with a trimeric form of human heat shock factor 1 (HSF1). In human cells, DAXX interacted with HSF1 essentially only during stress, i.e., when factor trimerization occurred. Several lines of experimentation suggested that DAXX is an important mediator of HSF1 activation: (i) overexpression of DAXX enhanced basal transactivation competence of HSF1 in the absence of a stress; (ii) a DAXX fragment exerted dominant-negative effects on HSF1 activation by different types of stress; (iii) induction of heat shock or stress protein (HSP)70 by heat stress was defective in a cell line lacking functional DAXX; and (iv) RNA interference depletion of DAXX also substantially reduced heat induction of HSF1 activity and HSP70 expression. HSF1 transactivation competence is repressed by an HSP90-containing multichaperone complex that interacts with trimeric factor. Overexpressed HSF1, known to be largely trimeric, only marginally increased HSF1 activity on its own but potentiated the activating effect of DAXX overexpression. Expression of a nonnative protein capable of competing for multichaperone complex also synergistically enhanced activation of HSF1 by DAXX. These observations suggest a model in which DAXX released from its nuclear stores during stress opposes repression of HSF1 transactivation competence by multichaperone complex through its interaction with trimerized HSF1. Our identification of DAXX as a mediator of HSF1 activation raises the question whether DAXX produces some of its pleiotropic effects through modulation of HSP levels.
When cells are exposed to a physical (e.g., heat) or chemical stress, they respond by altering their program of gene expression to cope with the adverse situation. Expression of a small set of genes, so-called heat shock or stress protein (HSP) genes, is dramatically increased (referred to herein as the stress protein response), and expression of many other genes is reduced (1–4). Elevated levels of HSPs typically provide a survival advantage to cells exposed to physical or chemical stresses (2, 5). The key transcription factor that mediates the stress protein response in vertebrates is heat shock factor 1 (HSF1) (6–11). HSF1 is constitutively present and is activated in response to a stress. Activation involves two distinct steps (12–15). In the first step, HSF1 converts to a DNA-binding homotrimer, and in the second step the trimerized factor acquires transactivation competence. Both steps are repressed by a bound HSP90-containing multichaperone complex (16–19). Activating stresses cause accumulation of denatured proteins that compete with HSF1 for HSP90-containing multichaperone complexes. Unbound HSF1 rapidly becomes homotrimeric and acquires transactivation competence. It seems fairly certain that HSF1 activity is also subject to additional control mechanisms. Activation of HSF1 is accompanied by hyperphosphorylation that may enhance transcriptional competence (20–22). In certain cell types, HSF1 also becomes rapidly organized in nuclear granules, termed stress granules, upon exposure of the cells to a stress (21, 23, 24). Furthermore, HSF1 is subject to modification by ubiquitin-like protein SUMO-1 (25). This sumoylation was reported to direct the factor to stress granules (25). Finally, Ral-binding protein 1 was proposed to function as a corepressor of HSF1 oligomerization (26).
DAXX is a ubiquitously expressed vertebrate protein (of 740 residues in humans) that was assigned to a variety of cellular functions (27, 28). DAXX is a largely nuclear protein in most cell types (29–32). Within the nucleus, DAXX predominantly associates with specific domains known as promyelocytic leukemia oncogenic domains (PODs) or nuclear domains 10 (31–33). DAXX is released from the latter depots during stress (34, 35). Overexpression of DAXX was reported to enhance Fas-induced apoptosis through activation of apoptosis signal-regulating kinase 1 (27, 36, 37). Other studies reported that the ability of DAXX to enhance Fas-mediated apoptosis required localization of the protein to PODs and the presence of promyelocytic leukemia protein (32, 33). DAXX was also shown to participate in transforming growth factor β-induced apoptosis in B cell lymphomas and hepatocytes (38). Various transcription factors were found to be repressed by DAXX (30, 39, 40). Depending on circumstances, homeodomain transcription factor PAX5 was either corepressed or coactivated by DAXX (41). Somewhat unexpectedly, depletion of DAXX by genetic means or by RNA interference (RNAi) revealed DAXX to be an antiapoptotic regulator (42, 43).
This report describes a nuclear function for DAXX as an important mediator of transcriptional competence of HSF1. Our finding that HSP expression is modulated by DAXX may prove helpful for understanding the diverse roles of the protein.
Materials and Methods
DNA Constructs. Cytomegalovirus promoter-driven expression constructs for cBSA and a matched β-galactosidase control and reporter gene lexA-fluc were described in ref. 19. Reporter gene Rluc was pRL-cytomegalovirus from Promega. Expression construct for HSF1 was as described in Zuo et al. (14), except that the hsf1 gene was recloned in pcDNA3.1 (Invitrogen). Expression construct B-galactosidase (pcDNA3.1/His B/lacZ, Invitrogen) was used as a filler (for equalizing amounts of DNA in transfection mixtures) and a control protein. Expression construct LEXA1–87HSF179–529 encoded a FLAG-tagged version of the protein. The construct was prepared by ligating a PCR fragment containing the protein-coding sequence (14) to the EcoRI and XhoI sites of p31FLAG. p31FLAG contained the nucleotide sequence ATGGACTACAAGGACGACGATGACAAG between the Asp-718 and EcoRI sites of pcDNA3.1. The K298R and S326A mutations were introduced by using the QuikChange procedure (Stratagene). Mutants were verified by restriction, in vitro expression, and nucleotide sequencing across the mutated site. Expression construct DAXX was prepared by insertion of an EcoRI/SalI daxx fragment from pQE-30-DAXX (44) into the EcoR1 and XhoI sites of p31FLAG. Expression construct for DAXX625–740 was derived from construct DAXX by using the QuikChange procedure. Reporter gene hsp70-fluc was obtained by subcloning promoter and RNA leader sequences of the human hsp70B gene into a plasmid containing a firefly luciferase gene.
Cell Culture and Transfection. Human HeLa-CAT (45) and HEK293 cells were grown in DMEM (high glucose) supplemented with 10% FCS/penicillin-streptomycin-glutamine (Invitrogen) at 37°C and under 5% CO2. Note that while most results shown are from experiments with HeLa-CAT cells, many experiments were repeated with HEK293 cells and similar data were obtained. Embryonic stem (ES) cell lines were cultured in knockout DMEM supplemented with 16% knockout serum replacement/penicillin-streptomycin-glutamine/nonessential amino acids/Hepes (all from Invitrogen)/0.14 mM 2-mercaptoethanol/103 units per ml ESGRO (Chemicon) in gelatinized tissue culture plates on a primary embryo fibroblast (Cell and Molecular Technologies, Phillipsburg, NJ) feeder layer at 37°C and under 7.5% CO2. The rapid protocol of Invitrogen was followed to transfect cells in 96-well plates by using Lipofectamine 2000. Transfection of larger cultures was by Lipofectamine Plus used as suggested by the manufacturer (Invitrogen). Reporter genes hsp70-fluc or lexA-fluc were present at a 250-fold excess over Rluc in transfection mixtures. Typically, cultures were harvested on the day following transfection. When cultures were treated with heat (typically for 30 min at 43–44°C) or chemicals 1 day after transfection, treated and untreated cultures were typically harvested 6–7 h after treatment unless indicated otherwise.
Determination of Transcriptional Activity of HSF1 Forms. Reporter activity was determined by using the dual-luciferase reporter assay system according to the manufacturer's instructions (Promega). HSF1 and LEXA1–87HSF179–529 activities were calculated as ratios of firefly luciferase to Renilla luciferase activities. Activities reported in the figures are relative to the activity of the most active sample.
Depletion by RNAi. Wells of 12-well dishes were seeded with 3 × 104 cells. On the next day, wells were transfected with 2.5 μl of either buffer (mock) or 20 μM mDx2 or hDx2 oligonucleotides (Dharmacon) by using the Oligofectamine protocol of Invitrogen (for a description of oligonucleotide sequences, see ref. 43). RNAi depletion was typically for 4 days. One day before heat treatment and analyses, each well was additionally transfected with 100 ng of a mixture of hsp70-fluc and Rluc reporter constructs by using 1 μl of Lipofectamine 2000 in 50 μl of OptiMEM (Invitrogen) without subsequent medium change.
Immunoprecipitation and Western Blots. Immunoprecipitations and Western blots were performed as described (14, 19). Expression of all exogenous proteins was verified by Western blot. Antibodies were as follows: FLAG M5 and A-tubulin (Sigma), HSF1 (SPA-901) and HSP70 (SPA-812) for mouse cells, and HSP70B (SPA-756) for human cells (Stressgen Biotechnologies, Victoria, BC, Canada), DAXX (Chemicon International), RGS-His tag (Qiagen), and GRP78 (Santa Cruz Biotechnology). Mouse HSF1 antibody was a gift from R. I. Morimoto (Northwestern University, Evanston, IL). Total protein concentration of cell lysate was determined by the Bradford-based protein assay (Bio-Rad) to equalize protein amounts for Western blots.
Results
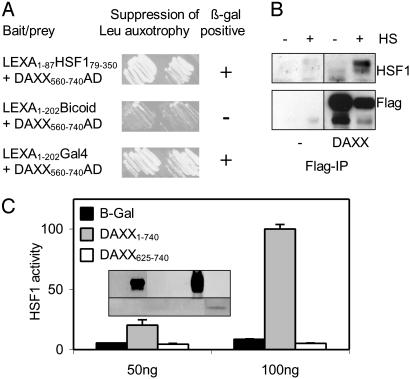
Two-Hybrid Screen-Identified Proteins Interacting with Trimeric HSF1. Chimeric transcription factor LEXA1–87HSF179–529 had been derived from human HSF1 by replacement of its DNA-binding domain (residues 1–78) with the DNA-binding domain of bacterial repressor LEXA (residues 1–87) (14). Like HSF1, LEXA1–87HSF179–529 appeared to form DNA-binding homotrimers when overexpressed in mammalian cells (ref. 14 and unpublished observations). LEXA1–87HSF179–350, which is LEXA1–87HSF179–529 minus carboxyl-terminal activation domains, was used as the bait in a two-hybrid screen of a human cDNA interaction library (46). Because monomeric LEXA does not bind DNA with high affinity, the screen specifically discovered proteins interacting with the trimerized bait; 120 positives representing 43 different prey constructs were identified. Nucleotide sequencing revealed that the majority of constructs included sequences coding for a carboxyl-terminal portion of human DAXX (residues 492–740). An experiment illustrating an interaction in yeast cells of LEXA1–87HSF179–350 and a prey protein containing DAXX residues 560–740 is shown in Fig. 1A. Full-length DAXX was also capable of associating with LEXA1–87HSF179–350 (data not shown). Thus, DAXX can interact with a trimeric form of HSF1. The above screen also identified SUMO-1-conjugating enzyme UBC9 as a potential HSF1-interacting protein.
Fig. 1.
DAXX interacts with trimeric HSF1 forms and stimulates basal HSF1 activity. (A) Interaction of HSF179–350 and DAXX560–740 sequences in yeast strain EGY48 grown on galactose-containing medium lacking leucine. AD, activation domain. (B) Interaction of DAXX and HSF1 in human cells. Two sets of cultures were transfected with 2 μg (per 60-mm dish) of expression construct for FLAG-tagged DAXX or FLAG vector. One day later, one set was heat-treated (HS). DAXX-containing complexes were immunoprecipitated from extracts by using FLAG antibody, and immunoprecipitates were analyzed by Western blot by using HSF1 antibody (Upper) or FLAG antibody (Lower). (C) Enhancement of basal HSF1 activity by overexpressed DAXX. Amounts of expression constructs cotransfected in addition to 25 ng of hsp70-fluc/Rluc reporter mixture are shown below. (Inset) Anti-FLAG Western blot signals produced by expression of DAXX (Upper) and DAXX625–740 (Lower).
To demonstrate that the HSF1–DAXX interaction could also occur in human cells, two sets of cultures of HeLa cell line HeLa-CAT were transiently transfected with expression construct for (FLAG-tagged) DAXX or vector. One set was heat-treated to induce trimerization of HSF1. DAXX complexes were immune-isolated using a FLAG antibody and were analyzed by Western blot (Fig. 1B). Exogenous DAXX could be specifically immunoprecipitated from both untreated and heat-treated cells (Fig 1B Lower). Presumably due to increased turnover of the protein during heat treatment, less DAXX was recovered from heat-treated cells. As revealed by the anti-HSF1 blot (Fig. 1B Lower), DAXX was capable of interacting with endogenous HSF1 in the human cells. This interaction predominantly occurred during heat treatment. Thus, DAXX associates primarily with trimerized HSF1 in human cells.
Overexpression of DAXX Enhanced Basal HSF1 Transactivation Competence. A firefly luciferase gene under the control of the strictly stress-regulated human hsp70B gene promoter (hsp70-fluc) was used to assess HSF1 transcriptional activity. To control for differences in transfection efficiency, cell lysis, and other generalized effects, a Renilla luciferase gene driven by a cytomegalovirus immediate-early promoter (Rluc) was included in all transfections.
Unstressed cells contain a low level of basal HSF1 activity, presumably reflecting spontaneous trimerization and activation of HSF1. To find out whether overexpression of DAXX specifically affected basal HSF1 activity, parallel cultures were co-transfected with the above reporter genes and with expression constructs for full-length DAXX1–740, target protein-binding region DAXX625–740, or control protein β-galactosidase. Expression of full-length DAXX and the short DAXX625–740 fragment was verified by Western blot (Fig. 1C Inset). Reporter assay revealed that overexpressed DAXX clearly, albeit only weakly, enhanced basal HSF1 activity (Fig. 1C). Note that the latter activity corresponded to <5% of that in severely heat-stressed cells (not shown). Target protein-binding fragment DAXX625–740, which accumulated to a considerably lower level than full-length protein, weakly inhibited rather than stimulated basal HSF1 activity (best seen at 100 ng). Thus, full-length DAXX has the ability to enhance HSF1 activity.
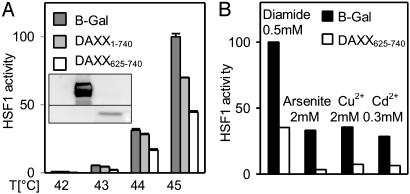
DAXX625–740 Dominantly Interfered with Stress Activation of HSF1. To begin addressing the question whether endogenous DAXX plays a critical role in activation of HSF1 by a heat stress, cells transfected with expression constructs for target protein-interacting DAXX fragment DAXX625–740 or control protein β-galactosidase were heat-treated for 30 min at different temperatures, and HSF1 activity was determined subsequently by dual luciferase assay (Fig. 2A). DAXX625–740 substantially interfered with activation of endogenous HSF1 at all temperatures (up to 70% inhibition was observed in other experiments). This finding implies that DAXX plays a positive, dominant role in the activation of HSF1 transcriptional competence by heat stress. Note that under conditions of severe heat stress and at a high level of overexpression, full-length DAXX also had a modest inhibitory effect (Fig. 2 A). This observation suggests the presence of an unknown DAXX-interacting protein that also contributes to HSF1 activation by DAXX.
Fig. 2.
Dominant-negative effect of DAXX625–740 on stress activation of HSF1. (A) Interference with activation by heat stress. Cultures were cotransfected with 150 ng of the constructs named (175 ng total DNA per well of 96-well dish). Heat treatment was for 30 min at the temperatures indicated. (Inset) Expression of DAXX625–740 and DAXX are documented in the Western blot. (B) Interference with activation by chemical stress. Cultures were cotransfected with 4 μg of DAXX625–740 or B-galactosidase (4.1 μg of total DNA per well of a six-well dish. Diamide, sodium arsenite, copper sulfate, or cadmium chloride were added to the medium at the indicated final concentrations for 1 h. Recovery started after changing the medium.
To determine whether DAXX specifically mediates activation of HSF1 by heat or whether it generically enhances stress activation of the transcription factor, interference of DAXX625–740 with activation of HSF1 by different toxicants known to trigger the stress protein response was tested (Fig. 2B). Results support the notion of a common mechanism of activation of HSF1 transcriptional competence that is triggered by toxicant stress and involves DAXX as a dominant, positively acting factor.
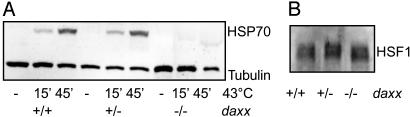
Defective Heat Induction of HSP70 in DAXX-Deficient Cells. To find out whether DAXX is a nonredundant mediator of HSF1 activation, advantage was taken of the daxx+/+, daxx+/–, and daxx–/– mouse ES cell lines established by Michaelson et al. in an earlier study (42). Parallel cultures of the different lines were either left untreated or were exposed to a moderately severe heat treatment at 43°C for 15 or 45 min. Induction of HSP70 was assessed by Western blot. A heat-induced increase in the HSP70 level was readily detected in daxx+/+ and daxx+/– cells (Fig. 3A). However, induction of HSP70 was defective in the daxx–/– cells. It is noted that the three lines expressed similar levels of HSF1 (Fig. 3B). These results strongly argued for a critical, nonredundant role for DAXX in HSF1 activation.
Fig. 3.
(A) Heat-induced HSP70 expression in ES cells homozygous (+/+) or heterozygous (+/–) for the daxx gene or lacking an intact daxx gene (–/–). Cells were exposed to a 15- or 45-min heat treatment at 43°C as indicated (HS) and were processed for Western blot after 16 h of recovery at 37°C. Tubulin Western blot served as a loading control. (B) HSF1 Western blot.
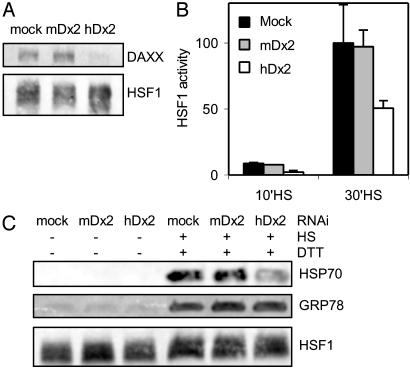
Inhibition of HSF1 Activation by Depletion of DAXX by RNAi. For an independent and more direct demonstration that DAXX is an important, nonredundant regulator of HSF1 activity, RNAi was used to specifically deplete endogenous DAXX. Parallel cultures of HeLa-CAT cells were transfected with 21-base pair double-stranded RNA oligonucleotides hDx2, mDx2 (43), or dilution buffer (mock). HDx2 contained human daxx sequence. mDx2, having the corresponding, nonidentical mouse daxx sequence, served as the control oligonucleotide. Three days after RNAi transfection, all cultures were retransfected with hsp70-fluc and Rluc reporter genes. One day later, the cultures were heat-treated at 44°C for 10 or 30 min, or were not heat-treated. Extracts were prepared 6 h later and were used to assess heat-induced HSF1 activity by dual luciferase assay and DAXX depletion by Western blot, respectively. As was expected based on the earlier study that used the same oligonucleotides (43), DAXX level was substantially reduced in the hDx2-transfected cells compared to the mDx2 or mock-transfected cells (Fig. 4A). The reporter assays revealed a substantial reduction of heat-induced HSF1 activity only in hDx2-transfected cells (Fig. 4B). In a similar experiment, RNAi-transfected cultures were heat-treated or not heat-treated on the fourth day after transfection, and HSP70 levels were determined subsequent to an 8-h recovery period (Fig. 4C). Cells transfected with hDx2 were found to have a greatly reduced heat-induced HSP70 level compared to the control-transfected cells. For ruling out the possibility that DAXX depletion could have resulted in a reduced capacity for inducible gene expression, heat-treated cultures were also exposed early during the recovery period to 15 mM DTT to induce glucose-regulated protein GRP78. As indicated by the Western blot results (Fig. 4C), DAXX depletion did not reduce induced GRP78 synthesis subsequent to heat treatment. These results further confirm that DAXX is an important mediator of stress activation of HSF1 and the stress protein response.
Fig. 4.
Depletion of DAXX by RNAi reduces heat-induced HSF1 activity and HSP70 expression in human cells. (A) DAXX Western blot of extracts from mock-treated cells or cells treated with RNAi (mDx2, hDx2) for 4 days. Extracts were equalized for protein concentration. HSF1 served as an additional control. (B) HSF1 activity in heat-stressed RNAi-treated cells. Note that cells had been transfected with hsp70-fluc and Rluc reporter genes on the third day of RNAi treatment, i.e., 1 day before heat treatment (see Materials and Methods). Heat treatment was for 10 min or 30 min at 44°C. (C) HSP70 and GRP78 induction in heat-stressed RNAi-treated cells. Extracts of similar protein concentrations were tested in the Western blots (see also the HSF1 loading control). Cells either were not heat-treated or were heat-treated for 30 min at 44°C. One hour after heat treatment, cells were also exposed to 15 mM DTT for 1 h. All cultures were harvested 8 h after heat treatment.
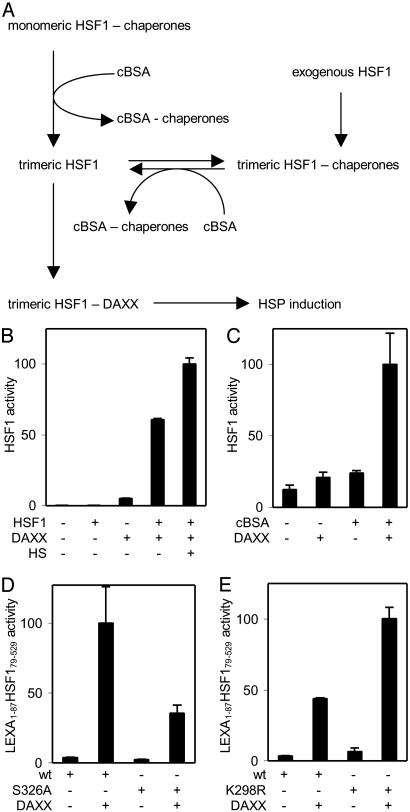
DAXX Opposes Repression of HSF1 Transactivation Competence. The coimmunoprecipitation experiment in Fig. 1B suggested that DAXX preferentially interacts with trimeric HSF1. That overexpression of DAXX enhanced basal HSF1 activity in the absence of a stress only by a small factor (Fig. 1C) suggested that DAXX was unable to activate nontrimeric HSF1 but acted on a small amount of spontaneously trimerized factor. Upon overexpression, HSF1 largely accumulates as DNA-binding trimers that have only minimal transcriptional activity (14). This repression of transactivation competence is caused by a dynamic interaction of trimeric HSF1 with an HSP90-containing multichaperone complex (ref. 19; see also the scheme in Fig. 5A). Presumably, the same mechanism also represses the small amount of endogenous HSF1 trimers present in unstressed cells. Hence, our observation that overexpression of DAXX-enhanced basal HSF1 activity also implied that exogenous DAXX was able to counteract this repression mechanism. If this were correct, cooverexpression of HSF1 to provide more trimeric factor could be expected to synergistically enhance HSF1 activity. The experiment to test this prediction not only yielded the expected result but additionally showed that DAXX was capable of inducing a level of HSF1 activity that was only increased further by a small factor (<2-fold) when the cells were also subjected to a heat treatment (Fig. 5B). Thus, overexpressed DAXX was capable to virtually completely relieve repression of HSF1 transcriptional activity by multichaperone complex. Based on these results, we hypothesized that when a cell is subjected to a stress, endogenous DAXX released from its nuclear stores may similarly enhance the activity of HSF1 by reducing the prevailing level of chaperone-mediated repression of its transcriptional competence.
Fig. 5.
DAXX counteracts repression of transcriptional competence. (A) Model. (B) Synergistic effect of DAXX and HSF1 cooverexpression, in a case in which 0.1 μg of HSF1 expression construct and 1.5 μg of DAXX expression construct were transfected (2.1 μg of total DNA per well of a 12-well dish). HS, heat treatment at 43°C for 30 min. (C) Synergistic effect of cBSA and DAXX coexpression. Expression constructs transfected were 25 ng of HSF1 and 100 ng of cBSA (150 ng of total DNA per well of a 96-well dish). (D) DAXX enhances the activities of both LEXA1–87 HSF179–529 (wt) and Ser-326 substitution S326A. Expression constructs transfected were 50 ng of DAXX, 1 ng of LEXA1–87 HSF179–529, or substitution (150 ng of total DNA per well of a 96-well dish). (E) DAXX stimulates both LEXA1–87 HSF179–529 (wt) and sumoylation-defective substitution K298R. Expression constructs transfected: 1 ng of LEXA1–87 HSF179–529 constructs and 200 ng of DAXX (250 ng of total DNA per well of a 96-well dish).
The hypothesis predicted that DAXX and a factor that specifically relieves repression of HSF1 by multichaperone complex should also synergistically enhance HSF1 activity (Fig. 5A). A previous study (19) showed that cytoplasmically expressed BSA (termed cBSA), incapable of acquiring its normal conformation in the reducing intracellular environment, is a preferential target of HSP90-containing multichaperone complex. Expression of this mutant protein (cBSA) allowed us to simulate the presence of misfolded proteins without subjecting the cells to stress conditions. In the experiment shown in Fig. 5C, cells were transfected with expression constructs for cBSA, DAXX, or both proteins. Each expression construct was transfected in an amount that only resulted in a weak enhancement of HSF1 activity. Results showed a substantial synergistic effect of cBSA and DAXX on HSF1 activation, confirming the prediction. Thus, binding of DAXX to trimeric HSF1 seems to counteract its repression by multichaperone complex. Note that even larger cooperative effects may have been masked by feedback effects in the above experiments. For example, HSP27 was reported to inhibit DAXX trafficking (47).
HSF1 transactivation competence may not only be negatively regulated by the above-discussed repression mechanism but may also be positively affected by phosphorylation and sumoylation. Because DAXX is known to interact with several protein kinases (36, 48, 49), the question was addressed whether its activating effect was solely mediated through phosphorylation of HSF1. Although HSF1 is phosphorylated on several residues during activation, we found that substitution of only one of these residues, Ser-326, substantially reduced factor activity (T.G., F.B., W. S. Lane, and R.V., unpublished results). Comparison of LEXA1–87HSF179–529 chimeras containing or lacking a Ser-326 to Ala substitution revealed that DAXX overexpression induced the activities of both, albeit to different levels (Fig. 5D).
Promyelocytic leukemia protein, the organizing protein with which DAXX associates in PODs, and DAXX, itself, are sumoylated (50, 51). HSF1 was also reported to be sumoylated (25). To find out whether SUMO-1 modification of HSF1 was required for activation by DAXX, the target lysine (Lys-298; see ref. 25) was substituted with arginine in LEXA1–87HSF179–529. Overexpressed DAXX activated both LEXA1–87HSF179–529 and its substituted derivative to comparable degrees (Fig. 5E). Note that, as was also described (52), the Lys-298 to Arg substitution mutant had an elevated basal activity. We conclude that enhancement of HSF1 transcription activity by DAXX is neither mediated solely through phosphorlyation of Ser-326 nor through sumoylation of Lys-298.
Discussion
In the present study, we demonstrated the existence of an interaction between DAXX and trimeric HSF1 during factor activation. That this interaction is of regulatory significance is supported by results from four different types of experiments. First, overexpression of DAXX enhanced basal transactivation competence of HSF1 in the absence of a stress and stimulated transcriptional activity synergistically with cooverexpressed (trimeric) HSF1. Second, overexpression of a dominant-negative form of DAXX interfered with activation of endogenous HSF1 by heat and chemical stresses. Third, cell lines containing a disrupted daxx gene displayed defective heat stress-induced HSP70 expression. Finally, depletion of DAXX by RNAi reduced heat-induced HSF1 activity and HSP70 expression. A recent study by others failed to observe a significantly reduced level of HSP70 induction in cell lines lacking functional DAXX (35). Although we do not know which difference in experimental conditions was responsible for the difference in results, we note that the previous study exposed cells to a mild heat stress, whereas we subjected cultures to moderately severe to severe heat stress. The stress protein response is known to be feedback-regulated under conditions of mild heat stress (53), which regulation may have obscured differences in induced HSP expression in the earlier study.
Our interaction assays in cells that overexpressed the protein suggested that DAXX is only capable of efficiently interacting with trimeric HSF1. This conclusion was implicitly corroborated by the observation that cooverexpression of DAXX and HSF1 synergistically induced HSF1 activity. This synergism would not have been observed if DAXX had been capable of effectively converting endogenous HSF1 to its trimeric form. In the absence of overexpression, DAXX is largely associated with PODs and is only released from this store during stress (31–35). Homotrimeric HSF1 is essentially only formed during stress. Hence, the availability of DAXX for interaction with HSF1 seems to be perfectly correlated with the availability of the trimerized form of HSF1 that is the form that interacts with DAXX.
Activation of HSF1 was previously shown to be subject to repression by HSP90-containing multichaperone complexes at both the levels of oligomerization and conversion to transcriptionally competent factor, the latter level being more resistant to stress than the former (16–19). The experiment in Fig. 5B suggested that overexpression of DAXX can almost completely relieve trimeric HSF1 from repression of its transcriptional activity by multichaperone complex. The experiment in Fig. 5C provided further support for this hypothesis by demonstrating a synergistic effect on HSF1 activity of DAXX and a nonnative protein that can counteract repression by neutralizing multichaperone complexes. The experiments showing that a DAXX fragment has a dominant-negative effect on activation of HSF1 by various stresses and that depletion of DAXX substantially reduces HSF1 activation in heat-stressed cells strongly suggested that DAXX is a driving force for the conversion of trimeric HSF1 to active transcription factor. These findings are most readily incorporated into a model of the type suggested in Fig. 5A, in which binding of DAXX to trimeric HSF1 is proposed to compete with the repressive interaction of multichaperone complex with the transcription factor. The model is also consistent with the phenotype of HSF1 mutants lacking part or all of the so-called regulatory domain (14). These mutants are incapable of associating effectively with multichaperone complex and display an elevated level of activity in the absence of a stress; i.e., when DAXX is unavailable. It is noted that DAXX may also stimulate HSF1 activity by additional mechanisms. For example, DAXX may serve as the adapter for a protein kinase that phosphorylates HSF1 on residues that enhance its transcriptional activity. This possibility is presently being investigated.
Our conclusion that DAXX comediates stress induction of HSF1 transactivation competence also raises a new question relating to the signals that result in activation of HSF1 and the stress protein response. Whereas stress-denatured protein is the main proximate signal that on its own causes relief of HSF1 from repression by chaperone complexes and in induction of HSF1 activity (7, 16–19), full activation apparently also requires an interaction of DAXX and HSF1 in the nucleoplasm. Hence, the signals that cause release of DAXX from its nuclear stores are expected to be coinducers of HSF1 activity and the stress protein response. Release of DAXX was reported to correlate with desumoylation of promyelocytic leukemia protein, which in turn may be induced by phosphorylation of the protein (35, 54, 55). Interaction of DAXX with HIPK1 also resulted in dissociation of DAXX from PODs (49). The signals that trigger the latter events remain to be discovered.
Targeted deletion as well as depletion by RNAi revealed DAXX to be an antiapoptotic protein (42, 43). However, overexpression of DAXX was found to enhance Fas-mediated apoptosis (27, 32, 37, 47). Furthermore, DAXX was reported to mediate transforming growth factor β-induced apoptosis and play a role in lymphocyte and keratinocyte apoptosis as well as in IFN-induced killing of lymphoid progenitors (33, 38, 56). Interestingly, a similar ambiguity emerged regarding the pro- and antiapoptotic functions of HSF1 and the stress protein response. On the one hand, elevated levels of HSPs were shown to inhibit stress-induced apoptosis (ref. 57 and references therein), and targeted disruption of HSF1 was found to abolish protection against heat-induced apoptosis (8). On the other hand, expression of a constitutively transactivating HSF1 mutant (a regulatory domain deletion) drastically enhanced Fas-induced apoptosis (58). Overexpression of DAXX was also found to result in a readily detectable increase in hsp70-fluc gene activity in cells of a HeLa line exposed to Fas antibody CH11 (data not shown). These observations suggest that, depending on circumstances, activation of HSF1 either protects against or promotes apoptosis. The possibility may be considered that DAXX mediates its pro- and antiapoptotic function in part or entirely through its activating effect on HSF1 and the stress protein response.
Acknowledgments
We thank J. Michaelson and P. Leder for DAXX ES cell lines; R. Brent for the cDNA library used in two-hybrid screens; J. Strauss for a full-length human DAXX cDNA; R. I. Morimoto for mouse HSF1 antibody, I. Dickerson for assistance with ES cell culture; G. Kraus for assistance with pilot experiments involving ribozymes; and A. So, P. Boehmer, and G. Maul for critical comments. This work was supported by National Institutes of Health Grant GM31125.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSF1, heat shock factor 1; HSP, heat shock or stress protein; POD, promyelocytic leukemia oncogenic domain; RNAi, RNA interference; ES, embryonic stem.
References
- 1.Welch, W. J. (1992) Physiol. Rev. 72, 1063–1081. [DOI] [PubMed] [Google Scholar]
- 2.Parsell, D. A. & Lindquist, S. (1993) Annu. Rev. Genet. 27, 437–496. [DOI] [PubMed] [Google Scholar]
- 3.Craig, E., Weissman, J. S. & Horvich, A. L. (1994) Cell 78, 365–372. [DOI] [PubMed] [Google Scholar]
- 4.Hartl, F. U. (1996) Nature 381, 571–579. [DOI] [PubMed] [Google Scholar]
- 5.Xia, W., Vilaboa, N., Martin, J. L., Mestril, R., Guo, Y. & Voellmy, R. (1999) Cell Stress Chap. 4, 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu, C. (1995) Annu. Rev. Cell Dev. Biol. 11, 441–469. [DOI] [PubMed] [Google Scholar]
- 7.Voellmy, R. (1996) in Stress-Inducible Cellular Responses, eds. Feige, U., Morimoto, R. I., Yahara, I. & Polla, B. S. (Birkhaeuser, Basel), pp. 121–137.
- 8.McMillan, D. R., Xiao, X., Shao, L., Graves, K. & Benjamin, I. J. (1998) J. Biol. Chem. 273, 7523–7528. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto, R. I. (1998) Genes Dev. 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- 10.Morano, K. A. & Thiele, D. J. (1999) Gene Expression 7, 271–282. [PMC free article] [PubMed] [Google Scholar]
- 11.Pirkkala, L., Nykaenen, P. & Sistonen, L. (2001) FASEB J. 15, 1118–1131. [DOI] [PubMed] [Google Scholar]
- 12.Jurivich, D. A., Sistonen, L., Kroes, R. A. & Morimoto, R. I. (1992) Science 255, 1243–1245. [DOI] [PubMed] [Google Scholar]
- 13.Bruce, J. L., Price, B. D., Coleman, C. N. & Calderwood, S. K. (1993) Cancer Res. 53, 12–15. [PubMed] [Google Scholar]
- 14.Zuo, J., Rungger, D. & Voellmy, R. (1995) Mol. Cell. Biol. 15, 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotto, J. J., Kline, M. & Morimoto, R. I. (1996) J. Biol. Chem. 271, 3355–3358. [DOI] [PubMed] [Google Scholar]
- 16.Ali, A., Bharadwaj, S., O'Carroll, R. & Ovsenek, N. (1998) Mol. Cell. Biol. 18, 4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou, J., Guo, Y., Guettouche, T., Smith, D. F. & Voellmy, R. (1998) Cell 94, 471–480. [DOI] [PubMed] [Google Scholar]
- 18.Bharadwaj, S., Ali, A. & Ovsenek, N. (1999) Mol. Cell. Biol. 19, 8033–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, Y., Guettouche, T., Fenna, M., Boellmann, F., Pratt, W. B., Toft, D. O., Smith, D. F. & Voellmy, R. (2001) J. Biol. Chem. 276, 45791–45799. [DOI] [PubMed] [Google Scholar]
- 20.Sorger, P. K., Lewis, M. J. & Pelham, H. R. B. (1987) Nature 329, 81–84. [DOI] [PubMed] [Google Scholar]
- 21.Sarge, K. D., Murphy, S. P. & Morimoto, R. I. (1993) Mol. Cell. Biol. 13, 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmberg, C. I., Tran, S. E., Eriksson, J. E. & Sistonen, L. (2002) Trends Biochem. Sci. 27, 619–627. [DOI] [PubMed] [Google Scholar]
- 23.Cotto, J. J., Fox, S. G. & Morimoto, R. I. (1997) J. Cell Sci. 110, 2925–2934. [DOI] [PubMed] [Google Scholar]
- 24.Jolly, C., Konecny, L., Grady, D. L., Kutsova, Y. A., Cotto, J. J., Morimoto, R. I. & Vourc'h, C. (2002) J. Cell Biol. 156, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, Y., Rogers, R., Matunis, M. J., Mayhew, C. N., Goodson, M., Park-Sarge, O. K. & Sarge, K. D. (2001) J. Biol. Chem. 276, 40263–40267. [DOI] [PubMed] [Google Scholar]
- 26.Hu, Y. & Mivechi, N. F. (2003) J. Biol. Chem. 278, 17299–17306. [DOI] [PubMed] [Google Scholar]
- 27.Yang, X., Khosravi-Far, R., Chang, H. Y. & Baltimore, D. (1997) Cell 89, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaelson, J. S. (2000) Apoptosis 5, 217–220. [DOI] [PubMed] [Google Scholar]
- 29.Pluta, A. F., Earnshaw, W. C. & Goldberg, I. G. (1998) J. Cell Sci. 111, 2029–2041. [DOI] [PubMed] [Google Scholar]
- 30.Hollenbach, A. D., Sublett, J. E., McPherson, C. J. & Grosveld, G. (1999) EMBO J. 18, 3702–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishov, A. M., Sotnikov, A. G., Negorev, D., Vladimirova, O. V., Neff, N., Kamitani, T., Yeh, E. T. H., Strauss, J. F. III, & Maul, G. G. (1999) J. Cell Biol. 147, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torii, S., Egan, D. A., Evans, R. A. & Reed, J. C. (1999) EMBO J. 18, 6037–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, S., Salomoni, P., Ronchetti, S., Guo, A., Ruggero, D. & Pandolfi, P. P. (2000) J. Exp. Med. 191, 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maul, G. G., Yu, E., Ishov, A. M. & Epstein, A. L. (1995) J. Cell. Biochem. 59, 498–513. [DOI] [PubMed] [Google Scholar]
- 35.Nefkens, I., Negorev, D. G., Ishov, A. M., Michaelson, J. S., Yeh, E. T. H., Tanguay, R. M., Mueller, W. E. G. & Maul, G. G. (2003) J. Cell Sci. 116, 513–524. [DOI] [PubMed] [Google Scholar]
- 36.Chang, H. Y., Nishitoh, H., Yang, X., Ichijo, H. & Baltimore, D. (1998) Science 281, 1860–1863. [DOI] [PubMed] [Google Scholar]
- 37.Ko, Y. G., Kang, Y. S., Park, H., Seol, W., Kim, J., Kim, T., Park, H. S., Choi, E. J. & Kim, S. (2001) J. Biol. Chem. 276, 39103–39106. [DOI] [PubMed] [Google Scholar]
- 38.Perlman, R., Schiemann, W. P., Brooks, M. W., Lodish, H. F. & Weinberg, R. A. (2001) Nat. Cell Biol. 3, 708–714. [DOI] [PubMed] [Google Scholar]
- 39.Li, H., Leo, C., Zhu, J., Wu, X., O'Neil, J., Park, E. J. & Chen, J. D. (2000) Mol. Cell. Biol. 20, 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, R., Pei, H., Watson, D. K. & Papas, T. S. (2000) Oncogene 19, 745–753. [DOI] [PubMed] [Google Scholar]
- 41.Emelyanov, A. V., Kovac, C. R., Sepulveda, M. A. & Birshtein, B. K. (2002) J. Biol. Chem. 17, 11156–11164. [DOI] [PubMed] [Google Scholar]
- 42.Michaelson, J. S., Bader, D., Kuo, F., Kozak, C. & Leder, P. (1999) Genes Dev. 13, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaelson, J. S. & Leder, P. (2003) J. Cell Sci. 116, 345–352. [DOI] [PubMed] [Google Scholar]
- 44.Kiriakidou, M., Driscoll, D. A., Lopez-Guisa, J. M. & Strauss, J. F., III (1997) DNA Cell Biol. 16, 1289–1298. [DOI] [PubMed] [Google Scholar]
- 45.Baler, R., Welch, W. J. & Voellmy, R. (1992) J. Cell Biol. 117, 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gyuris, J., Golemis, E., Chertkov, H. & Brent, R. (1993) Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- 47.Charette, S. J., Lavoie, J. N., Lambert, J. & Landry, J. (2000) Mol. Cell. Biol. 20, 7602–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rochat-Steiner, V., Becker, K., Micheau, O., Schneider, P., Burns, K. & Tschopp, J. (2000) J. Exp. Med. 192, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ecsedy, J. A., Michaelson, J. S. & Leder, P. (2003) Mol. Cell. Biol. 23, 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sternsdorf, T., Jensen, K. & Will, H. (1997) J. Cell Biol. 139, 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jang, M. S., Ryu, S. W. & Kim, E. (2002) Biochem. Biophys. Res. Commun. 295, 495–500. [DOI] [PubMed] [Google Scholar]
- 52.Knauf, U., Newton, E. M., Kyriakis, J. & Kingston, R. E. (1996) Genes Dev. 10, 2782–2793. [DOI] [PubMed] [Google Scholar]
- 53.Abravaya, K., Phillips, B. & Morimoto, R. I. (1991) Genes Dev. 5, 2117–2127. [DOI] [PubMed] [Google Scholar]
- 54.Muller, S., Matunis, M. J. & Dejean, A. (1998) EMBO J. 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Everett, R. D., Lomonte, P., Sternsdorf, T., van Driel, R. & Orr, A. (1999) J. Cell Sci. 112, 4581–4588. [DOI] [PubMed] [Google Scholar]
- 56.Gongora, R., Stephan, R. P., Zhang, Z. & Cooper, M. D. (2001) Immunity 14, 727–737. [DOI] [PubMed] [Google Scholar]
- 57.Mehlen, P., Mehlen, A., Godet, J. & Arrigo, A. P. (1997) J. Biol. Chem. 272, 31657–35665. [DOI] [PubMed] [Google Scholar]
- 58.Xia, W., Voellmy, R. & Spector, N. L. (2000) J. Cell Physiol. 183, 425–431. [DOI] [PubMed] [Google Scholar]