Abstract
In eukaryotes, a surveillance mechanism known as nonsense-mediated decay (NMD) degrades the mRNA when a premature-termination codon (PTC) is present. NMD requires translation to read the frame of the mRNA and detect the PTC. During pre-mRNA splicing, the exon–exon junction complex (EJC) is recruited to a region 20–24 nt upstream of the exon junction on the mature mRNA. The presence of a PTC upstream from the EJC elicits NMD. Eukaryotic initiation factor 4A (eIF4A) III is a nuclear protein that interacts physically or functionally with translation initiation factors eIF4G and eIF4B, respectively, and shares strikingly high identity with the initiation factors eIF4AI/II. Here we show that siRNA against eIF4AIII, but not against eIF4AI/II, inhibits NMD. Moreover, eIF4AIII, but not eIF4AI, is specifically recruited to the EJC during splicing. The observations that eIF4AIII is loaded onto the mRNA during splicing in the nucleus, has properties related to a translation initiation factor, and functions in NMD raises the possibility that eIF4AIII substitutes for eIF4AI/II during NMD.
Ribosome recruitment to the mRNA in eukaryotes requires the RNA helicase, eukaryotic initiation factor 4A (eIF4A), which is thought to unwind 5′ secondary structure in the mRNA and facilitate binding of the small ribosomal subunit (1). eIF4AI and eIF4AII are two similar gene products that are functionally redundant for translation initiation (2); eIF4AI accounts for the majority (≈75%) of total eIF4A in a rabbit reticulocyte lysate (3). eIF4AIII, which is ≈70% identical to eIF4AI/II (4), cannot substitute for these proteins in an in vitro mRNA ribosome binding assay (4). Nevertheless, a relationship of eIF4AIII to a translation-like mechanism is suggested by the observation that eIF4AIII requires the initiation factor eIF4B for efficient RNA unwinding activity (4). In addition, eIF4AIII interacts with another essential initiation factor, eIF4G (4). eIF4AIII can inhibit translation in a rabbit reticulocyte lysate when added in large amounts (4), whereas it may have a stimulatory effect on selective mRNA translation in Xenopus oocytes (5). Importantly, in contrast to eIF4AI/II, which are cytoplasmic, eIF4AIII is nuclear and colocalizes in speckle domains (6).
Nonsense-mediated decay (NMD) is an RNA surveillance mechanism that rids the cells of defective mRNAs bearing a premature-termination codon (PTC) (7–9). About 30% of inherited genetic disorders are the consequence of mutations that create a PTC (10). NMD is both a splicing and translation-dependent event (11). An EJC is assembled 20–24 nt upstream of the exon–exon junction on the mRNA as a result of splicing (12, 13). The exon–exon junction complex (EJC) is composed of mRNA export factors (such as Aly/REF) and factors that are required for NMD (such as RNPS1, Y14, magoh, and Upf3) (11, 14, 15). Many of these splicing-dependent mRNA-binding proteins are localized in the nucleus but shuttle between the nucleus and the cytoplasm (11, 14). The EJCs are positional markers of introns; when a termination codon is present at least 50–55 nt upstream of a spliced intron, i.e., upstream of an EJC, it is recognized as premature (16–19). In mammalian cells, intronless, PTC-bearing mRNAs are not subject to NMD, demonstrating the importance of splicing for NMD (20–22). In addition to being splicing-dependent, NMD also requires translation. Protein synthesis inhibitors (23), mutation of the initiation codon (24), hairpin structures in the 5′ UTR (25, 26), and suppressor tRNAs all inhibit NMD (24).
NMD is thought to occur during the so-called pioneer round of translation, which is defined as the first time the reading frame of the mRNA is read (27). According to a current view, the pioneer round of translation occurs when the cap and poly(A) tail are bound by the nuclear proteins CBC and PABP2, whereas bulk translation occurs when the cytoplasmic proteins eIF4E and PABP are bound (27). Because eIF4AIII is localized to the nucleus, is a homologue of a translation factor necessary for bulk translation, and interacts with translation factors (eIF4B and eIF4G), it is conceivable that it could function in the pioneer round of translation and thus in NMD (27).
Here we report that knockdown of eIF4AIII by using RNA interference (RNAi) results in inhibition of NMD, whereas knockdown of eIF4AI and II fails to inhibit NMD. We also demonstrate that eIF4AIII, but not eIF4AI, associates with mRNA during splicing at a region containing the EJC. Finally, we show that eIF4AIII is a nucleocytoplasmic shuttling protein. Together, these data indicate that eIF4AIII is loaded onto the mRNA during splicing in the nucleus and then functions during NMD.
Materials and Methods
Small Interfering RNA (siRNA) Transfections. siRNA transfections were performed in HeLa cells by using Lipofectamine Plus Reagent (Invitrogen). One day before siRNA transfection cells were trypsinized to achieve 50–60% confluency. For a six-well plate, 15 μl of siRNA duplex (20 μM annealed duplex from Dharmacon) was mixed with 100 μl of OPTI-MEM medium and 10 μl of Plus Reagent and incubated for 15 min at room temperature. In a separate tube, 20 μl of Lipofectamine (Invitrogen) was added to 100 μl of OPTI-MEM. Lipofectamine mix was added to precomplexed RNA mix and incubated for an additional 15 min. RNA and protein were harvested 48 h after transfection for use in Northern and Western blots, respectively. For double knockdown of eIF4AI and eIF4AII, eIF4AII siRNA was transfected by using conditions described above, 12 h later eIF4AI siRNA was transfected in the same manner and cells were collected 60 h after transfection.
Western Blot. Cells were washed twice and scraped in ice-cold PBS and pelleted at maximum speed for 10 min in a microcentrifuge. Cell pellets were frozen on dry ice and thawed in RIPA buffer [150 mM NaCl/50 mM Tris·HCl, pH 7.5/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS and a mixture of protease inhibitors (Roche Diagnostics)], incubated on ice for 10 min and centrifuged for 5 min at maximum speed to recover the supernatant. The amount of protein loaded is indicated in the figure legends. Proteins were separated on a SDS/10% PAGE, transferred to a nitrocellulose membrane (0.22 μM), blocked overnight in 5% nonfat milk, and incubated with antibodies [anti-eIF4AIII (4), anti-eIF4AI (4), and anti-eIF4AII (28)] prepared in 1% BSA/PBST (0.5% Tween) at a dilution of 1/1,000 (except for 4AII, which was used at 1/500 dilution) for 1 h at room temperature, washed four times for 15 min with PBST (0.5% Tween), and incubated with secondary antibody (1/5,000) for 1 h at room temperature.
NMD Assay. Cells stably expressing TCR-β wt (3C1) and TCR-β containing a PTC (7C3) were a generous gift from Miles Wilkinson and Melissa Moore (29). Total RNA was isolated by using Qiagen (Valencia, CA) RNAeasy kit according to the manufacturer's instructions. For Northern blots, 3 μg of total RNA was separated on a 1.3% agarose/formaldehyde gel, transferred to a Hybond-N membrane (Amersham Pharmacia) and probed with 32P-labeled, random-primed DNA probes: TCR-β Vβ8.1 and human actin probes were prepared from plasmids β117 and β290, respectively (23, 30). Cycloheximide treatment was performed at a final concentration of 100 μg/ml for the indicated amount of time.
Immunoprecipitation of Spliced mRNA and Oocyte Injections. Oligonucleotide-directed RNase H cleavage and immunoprecipitation was performed essentially as described (12). Briefly, endogenous RNase H digestion was accomplished by adding each cDNA oligo to a final concentration of 0.5 μM to in vitro splicing reactions and incubating the reaction for 10 min at 30°C. For no oligo controls, equivalent volumes of TE buffer was added to the splicing reactions. Immunoprecipitated RNAs were recovered by phenol/chloroform extraction and ethanol precipitation. RNA species were analyzed on 15% denaturing polyacrylamide gels and visualized by Molecular Dynamics PhosphorImager. Oocyte microinjection was carried out according to Luo et al. (31). The plasmid encoding GST-eIF4AIII was constructed by cloning the cDNA encoding human eIF4AIII protein into protein expression vector pGEX5 at BamHI and XhoI sites. GST pull-down of the in vivo complexes was done as described (32).
Heterokaryon Assay. HeLa and NIH 3T3 fusions were performed as described (33). Hemagglutinin-tagged eIF4AIII and 4E-T were reported (4, 34) and were transfected into HeLa cells by the calcium phosphate method. Cells were treated with 5 ng/ml leptomycin B (LMB, Sigma) 3 h before cell fusion. Cells were subsequently washed with 1× PBS, and cell fusions were formed by incubation for 2 min in 50% polyethylene glycol (PEG) 8000 in PBS. Cells were washed extensively with PBS and returned to fresh media containing 100 μg/ml cycloheximide and 5 ng/ml leptomycin B. After 2 h, cells were fixed and processed for immunofluorescence as described (34).
Results
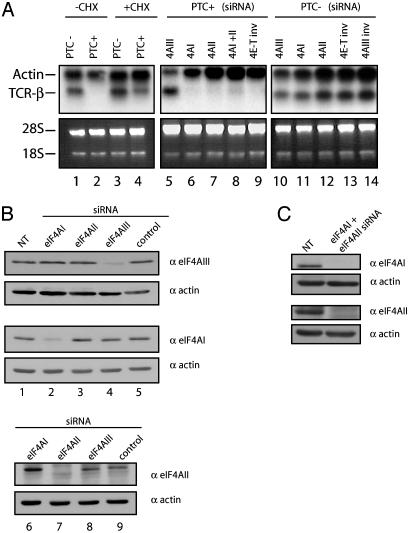
RNAi was carried out in HeLa cells to determine whether eIF4AIII plays a role in NMD. To assay for NMD, we analyzed the levels of TCR-β mRNA, a transcript that undergoes NMD in a nuclear-associated manner (24). Wild-type and PTC-containing TCR-β transcripts were analyzed (23, 24). Treatment of cells with siRNA against eIF4AIII reduced its level by 80% (Fig. 1B, lane 4). In HeLa cells expressing a PTC-containing TCR-β gene, the levels of the mutant mRNA were very low relative to the levels of the wild-type mRNA, indicating that NMD has occurred (Fig. 1A, compare lane 1 to lane 2). As expected, NMD was inhibited upon the addition of the translation inhibitor, cycloheximide (Fig. 1A, lane 4) (23). NMD was significantly inhibited in cells treated with siRNA against eIF4AIII (Fig. 1A, lane 5; siRNA against eIF4AIII is accompanied by a decrease in actin, which is presumably a consequence of increased cell death), but not in cells treated with a negative control siRNA [4E-T inverted; 4E-T inv (34); Fig. 1A, lane 9]. Furthermore, siRNA against eIF4AIII inhibits decay of PTC+ TCR-β mRNA to a similar extent as siRNA against Upf1 (35, 53). We conclude that eIF4AIII functions in NMD.
Fig. 1.
RNAi against eIF4AIII impairs NMD. (A) Northern blot analysis of poly(A) RNA hybridized with Vβ8.1 probe (23). Lanes 1 and 2 represent stable cell lines expressing wild-type TCR-β minigene and PTC-containing TCR-β minigene, respectively. Cycloheximide treatment in both cell lines was performed for 2 h at 37°C (100 μg/ml final) (lanes 3 and 4). siRNAs were transfected into stable cell lines expressing a PTC-containing TCR-β minigene (PTC+) (lanes 5–9) and wild-type TCR-β minigene (PTC–) (lanes 10–14). β-Actin served as a loading marker. The 28S and 18S ribosomal RNA serve to demonstrate equal loading for eIF4AIII siRNA treated samples because siRNA against eIF4AIII is accompanied by decreased levels of actin due to cell death (see Results). CHX, cycloheximide. (B and C) Western analysis of the levels of translation initiation factors after knockdown. Thirty micrograms of protein extract was resolved by SDS/PAGE. Proteins were quantitated against β-actin (Sigma) as a loading control and the level of protein in negative control (4E-T inv) transfected cells was normalized to 100. NT, nontransfected.
To determine whether siRNA against eIF4AI and eIF4AII affects NMD, we treated HeLa cells with siRNAs against these factors (see Table 1 for a list of sequences). The siRNA treatments resulted in a specific reduction of protein levels of 75% for eIF4AI and 95% for eIF4AII (Fig. 1B). Strikingly, and in sharp contrast to the results obtained with RNAi to eIF4AIII, knockdown of eIF4AI and II (Fig. 1B, lanes 2 and 7, respectively) failed to inhibit NMD (Fig. 1 A, lanes 6 and 7); the PTC-containing mRNA was present at levels similar to those seen in untransfected cells or cells treated with the negative control siRNA (34) (lane 9). To determine whether eIF4AII compensates for the loss of eIF4AI function or vice versa, a double knockdown was performed (Fig. 1C). No effect on NMD was observed (Fig. 1 A, compare lane 8 to lane 9). Thus, a large reduction in the levels of the cytoplasmic eIF4A translation initiation factors failed to affect NMD, whereas a similar reduction in eIF4AIII exhibited a major inhibitory effect on NMD. These data indicate that eIF4AIII, but not eIF4AI/II, functions in NMD, although we cannot rule out the possibility that the small amounts of remaining eIF4AI/II are sufficient for NMD (see Discussion). Notably, the effect of siRNA against eIF4AIII is not caused by an increase in transcription of TCR-β, as is evident in the wild-type cell line (PTC–) (Fig. 1 A, compare lane 10 to lanes 13 and 14). Furthermore, the observation that eIF4AI and eIF4AII siRNA fail to inhibit NMD, is not the result of inhibition of transcription by these siRNA (compare lanes 11 and 12 to lanes 13 and 14).
Table 1. Summary of the siRNA used for gene expression knockdown.
| Synthetic siRNA | Position in the ORF | siRNA Structure |
|---|---|---|
| eIF4AI | 226-244 | 5′- GCCCAAUCUGGGACUGGGA dTdT-3′ |
| 3′-dTdT CGGGUUAGACCCUGACCCU -5′ | ||
| eIF4AII | 122-140 | 5′- AGGAGUCUCUCCUUCGUGG dTdT-3′ |
| 3′-dTdT UCCUCAGAGAGGAAGCACC -5′ | ||
| eIF4AIII | 75-93 | 5′- AGUGGAAUUCGAGACCAGC dTdT-3′ |
| 3′-dTdT UCACCUUAAGCUCUGGUCG -5′ | ||
| 4E-T Inverted | 935-953 | 5′- CGUACCGUGGAAUAGUUCC dTdT-3′ |
| 3′-dTdT GCAUGGCACCUUAUCAAGG -5′ |
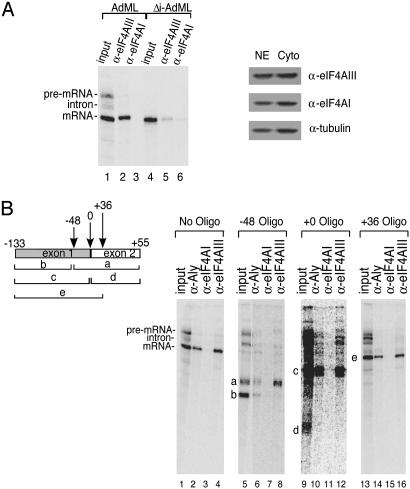
To obtain additional evidence that eIF4AI and eIF4AIII have distinct roles in NMD, we next asked whether either of these proteins associates with spliced mRNA. After splicing, the mature mRNA is packaged into a specific spliced messenger ribonucleoprotein (mRNP) complex for transport to the cytoplasm (31). To determine whether eIF4AIII and/or eIF4AI are present in the spliced mRNP, we performed in vitro splicing of AdML pre-mRNA in HeLa nuclear extract followed by immunoprecipitations (Fig. 2A Left). Significantly, the spliced mRNA is immunoprecipitated by the eIF4AIII antibody but not by the eIF4AI antibody (Fig. 2 A, compare lanes 2 and 3). As shown in Fig. 2 A Right, eIF4AI and eIF4AIII are present in both nuclear and cytoplasmic extracts. As eIF4AI is a cytoplasmic protein (36–38), this protein, like tubulin (Fig. 2 A Right), is likely to be a contaminant in the nuclear extract. Thus, the failure to immunoprecipitate the mRNA with the eIF4AI antibody is not caused by the absence of this protein from the nuclear extract. The eIF4AI antibody is functional for immunoprecipitation assays as determined by immunoprecipitation/Western blot (data not shown). Together, these data indicate that eIF4AIII, but not eIF4AI, is specifically associated with the mRNA.
Fig. 2.
eIF4AIII, but not eIF4AI, associates with the EJC on spliced mRNA. (A) eIF4AIII, but not eIF4AI, is associated with spliced mRNA but not with intronless mRNA (Left). Western analysis showing the presence of eIF4AIII, eIF4AI, and tubulin in the nuclear (NE) and cytoplasmic (Cyto) extracts (Right). (B) eIF4AIII associates with the region of the mRNA that contains the EJC. (Left) Schematic of AdML mRNA showing the locations of oligonucleotides. The numbers indicate the middle of each 12-mer oligonucleotide. (The RNA fragments containing the EJC are designated a, c, and e.) (Right) The total reaction after splicing and oligonucleotide-directed RNase H cleavage was used for the immunoprecipitations and is shown in lanes designated input. Immunoprecipitations were carried out with the indicated antibodies after cleavage with each oligonucleotide. The RNA fragments containing the EJC that were immunoprecipitated are indicated (a, c, and e). The RNA fragments that do not contain the EJC are also indicated (b and d). A darker exposure of lanes 9–12 is included to detect fragment d (55 nt) generated with the +0 oligonucleotide (lane 9). The 13-nt RNA fragment generated with the +36 oligonucleotide is not detected.
To determine whether the association of eIF4AIII with the mRNA is splicing dependent, we transcribed a construct that is identical to that encoding AdML pre-mRNA except that it is lacking the intron. This intronless mRNA (Δi-AdML) was then incubated in the nuclear extract and used for immunoprecipitations. Significantly, the Δi-AdML mRNA is immunoprecipitated very poorly by the eIF4AIII or eIF4AI antibody (Fig. 2 A, lanes 4–6). We conclude that eIF4AIII is specifically associated with mRNA in a splicing-dependent manner. This result is consistent with previous studies showing that eIF4AIII is specifically associated with highly purified spliceosomal complexes (39–42).
Proteins that function in NMD as well as proteins that function in mRNA export are localized on the spliced mRNA to a distinct region located 20–24 nt upstream of the exon–exon junction (12, 13, 15). The proteins that are bound to this region are referred to as the EJC (11–14). The EJC proteins were originally localized on the mRNA by using a combination of immunoprecipitation and oligonucleotide-directed RNase H cleavage of the mRNA (12). Thus, we next used this approach to determine where eIF4AIII is bound on the mRNA. After in vitro splicing, three different 12-mer oligos were used for RNase H digestion (see schematic, Fig. 2B). Immunoprecipitations were carried out with antibodies to eIF4AI and eIF4AIII. In addition, we used an antibody to Aly, a known EJC protein (12), as a positive control. In lanes 1–4, immunoprecipitations carried out without RNase H digestion demonstrate that eIF4AIII and Aly antibodies, but not eIF4AI antibodies, immunprecipitate spliced mRNA but not pre-mRNA in vitro. The mRNA fragments containing the exon–exon junction, bands labeled as a, c, and e, were generated by digestion with a –48 oligonucleotide (Fig. 2B, lanes 5–8), +0 oligonucleotide (Fig. 2B, lanes 9–12), and + 36 oligonucleotide (Fig. 2B, lanes 13–16), respectively. Fragments containing the exon–exon junction region are immunoprecipitated by Aly (Fig. 2B, lanes 6, 10, and 14) and eIF4AIII antibodies (Fig. 2B, lanes 8, 12, and 16), but not by eIF4AI antibodies (Fig. 2B, lanes 7, 11, and 15). The products of the digestion lacking the exon–exon junction (labeled b and d) are immunoprecipitated by neither eIF4AIII nor Aly antibodies. We note that the 3′ RNase H digestion products, which lack the 5′ cap (e.g., fragments a and d), are more readily degraded than the 5′ RNase H digestion products, which contain the cap (e.g., fragments b, c, and e). The 13-nt fragment generated with the +36 oligonucleotide is not detected at all, most likely because it is rapidly degraded as it is very small and uncapped. A long exposure of the gel shows that fragment d, which does not contain the EJC, is not immunoprecipitated by any of the antibodies (Fig. 2B, lanes 9–12). Together, these results indicate that eIF4AIII, like Aly, is present on the region of the mRNA that contains the EJC.
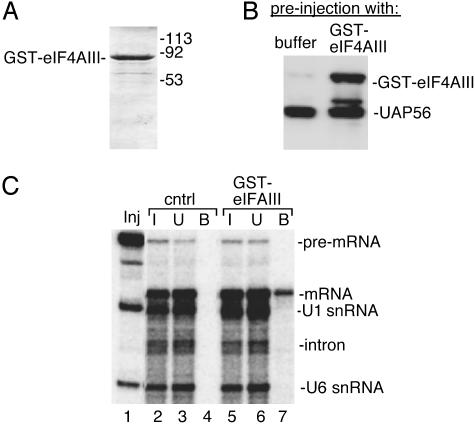
To determine whether eIF4AIII associates with spliced mRNA in vivo, GST-eIF4AIII or buffer alone (Fig. 3A) was preinjected into Xenopus oocyte cytoplasm. After incubation of the oocytes, the eIF4AIII was imported into the nucleus (Fig. 3B). UAP56, which serves as a marker for the nucleus, is detected in the nuclei of preinjected oocytes (Fig. 3B). A mixture of 32P-pre-mRNA, U1 and U6 snRNAs (Fig. 3C, lane 1) was then injected into nuclei of the preinjected oocytes and incubated to allow splicing. The nuclei were then isolated and the nuclear lysates incubated with glutathione beads. The spliced mRNA, but not the pre-mRNA or small nuclear RNAs, is bound to the beads when the oocytes contain GST-eIF4AIII (Fig. 3C, lane 7). In contrast, no RNAs bind to the beads in the buffer control (Fig. 3C, lane 4) or when other nuclear GST-fusion proteins (GST-Sir2 and GST-ΔC Aly; ref. 43) are preinjected (data not shown). We conclude that eIF4AIII is specifically associated with spliced mRNA in vivo. The observation that eIF4AIII is present in the EJC and is associated with mRNA is consistent with the possibility that eIF4AIII plays a direct role in NMD. Moreover, eIF4AIII is recruited to the mRNA during splicing, while the mRNA is still in the nucleus.
Fig. 3.
eIF4AIII is associated with spliced mRNA in vivo. (A) Coomassie-stained gel showing GST-eIF4AIII used for microinjection into oocyte cytoplasm. (B) Western analysis of Xenopus oocyte nuclei after preinjection showing the presence of UAP56, a known nuclear protein, and recombinant GST-eIF4AIII. (C) Inj indicates the total RNA before injection, and I indicates the RNA species present in the nuclear lysates before the addition of glutathione beads. Total RNA unbound (U) and bound (B) to the beads is shown.
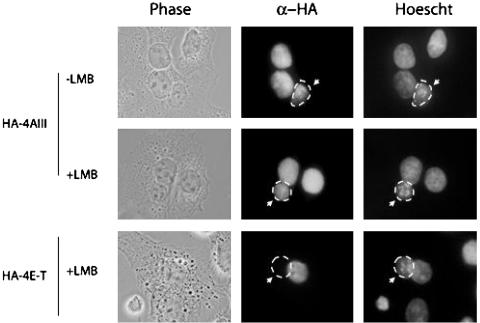
To determine whether eIF4AIII might accompany the mRNA to the cytoplasm together with other EJC components (Y14, magoh, Aly/REF), we used a heterokaryon assay (44) that measures the ability of a protein to shuttle between the nucleus and the cytoplasm. A hemagglutinin-tagged eIF4AIII expression plasmid was transfected into HeLa cells, followed by fusion to murine NIH 3T3 cells. Three hours after fusion, eIF4AIII was detected in most (>95%) of the fused murine nuclei (Fig. 4). Incubation with leptomycin B (LMB), which inhibits CRM1-dependent protein export, had no effect on eIF4AIII shuttling (Fig. 4) even after overnight incubation (data not shown). In contrast, LMB abolished export of 4E-T, a protein known to be a substrate of the CRM1 receptor (34). We conclude that eIF4AIII exits the nucleus in a CRM1-independent fashion and thus may accompany the mRNA via the TAP-mediated export pathway (45–47). The observation that eIF4AIII shuttles, suggests that it can conceivably participate in NMD in the cytoplasm and/or nucleus (37, 48, 49).
Fig. 4.
eIF4AIII is a shuttling protein. eIF4AIII and 4E-T localization was detected by staining with anti-hemagglutinin antibody (1/1,000 dilution, Covance/Babco) and Texas red-conjugated anti-mouse IgG (Molecular Probes). Human and mouse nuclei are differentiated by staining with Hoechst dye 33258 (Sigma). The mouse nucleus is indicated by an arrow. Leptomycin B (LMB, Sigma) treatments were performed at 5 ng/ml final concentration for 5 h. Images were processed with a Nikon Eclipse E800 microscope at ×60 magnification.
Discussion
Here we have shown that RNAi of eIF4AIII, a protein with striking similarity to the well known cytoplasmic translation initiation factors eIF4AI/II, inhibits NMD. In contrast, RNAi of eIF4AI/II had no effect on NMD, suggesting that these proteins do not have a function in NMD. Although we cannot rule out the possibility that small amounts of eIF4AI/II are sufficient for NMD, we also show that eIF4AIII, but not eIF4AI, is associated with the EJC, a complex known to function in NMD. Thus, both the RNAi data and the biochemical studies support a role for eIF4AIII and not its close homolog eIF4AI, in NMD. We note that eIF4AI knockdown effectively inhibits translation (>80%) of poliovirus, which is confined to the cytoplasm and is more dependent on eIF4AI for translation than other viral and cellular mRNAs (50, 51) (data not shown). Thus, eIF4AI knockdown affects its function in cytoplasmic translation.
The early recruitment of eIF4AIII to the spliced mRNA raises the possibility that the protein functions in an initial step of NMD. It was previously proposed that NMD occurs during the pioneer round of translation (27). Considering (i) the structural and functional similarities between eIF4AIII and eIF4AI/II, i.e., its interaction with the middle domain of eIF4GI and its helicase activity stimulated by eIF4B, (ii) the essential role of eIF4AIII in NMD, (iii) the presence of eIF4AIII at the EJC and its interaction with Y14 and magoh (our results and refs. 38, 53, and 54) and the recruitment of eIF4AIII to the mRNA while it is still in the nucleus, it is plausible that eIF4AIII is required for NMD because it substitutes for eIF4AI/II in the pioneer round of translation. Another, but not mutually exclusive possibility, is that eIF4AIII functions as a structural component of the EJC and that its helicase activity is required for remodeling the components of the EJC. This may be necessary for its positioning, likely through Upf2 and Upf3, in proximity to the ribosome and its associated factors to communicate the site of the PTC. In a previous study, Upf2, was shown to interact with eIF4AI (52). It is possible that this interaction occurs during a later step of NMD or functions in cytoplasmic NMD, as this interaction was shown in yeast, where NMD appears to be a purely cytoplasmic process.
It is still a matter of debate whether the first round of translation takes place in the nucleus or the cytoplasm. Given that eIF4AIII is a nucleocytoplasmic shuttling factor, it can conceivably function in either compartment. It was reported earlier that the nuclear cap binding complex, nCBC, substitutes for the cytoplasmic cap-binding protein, eIF4E, in the pioneer round of translation (27). Other potential translation initiation factors that could function uniquely in NMD include PABP2 and an eIF4G-like protein (39, 40). Similar to eIF4AIII, these proteins are present in the nucleus and are specifically recruited to spliceosomes (39, 40). eIF4G-like contains a region that is similar to the domain in eIF4G that interacts with eIF4AIII (4). By analogy to the cytoplasmic initiation complex, eIF4G-like may interact with both eIF4AIII and nCBC. The cytoplasmic translation initiation factor PABP1 interacts with eIF4G and the poly(A) tail, and PABP2 is a candidate for this protein in the nuclear initiation complex. Together, these observations raise the possibility that NMD involves a set of previously uncharacterized translation initiation factors that are loaded onto the mRNA early in its biogenesis in the nucleus.
Acknowledgments
We thank Avak Kahvejian, Hiroaki Imataka, Yuri Svitkin, Jerry Pelletier, Imed Gallouzi, and Graham Belsham for helpful discussions and critical reading of the manuscript. We thank Miles Wilkinson and Melissa Moore for providing the TCR-β stable cell lines and β117 and β290 plasmids, and Chris Proud for providing anti-eIF4AII antibody. We thank Melissa Moore and Elisa Izaurralde for communicating to us their unpublished results. We also thank Nicolas Bottari, Guylaine Roy, and Colin Lister for their assistance. This research was supported by a grant from the Canadian Institute of Health Research (CIHR) (to N.S.) and by a National Institutes of Health grant (to R.R.). N.S. is a CIHR distinguished scientist and a Howard Hughes Medical Institute International Scholar. We thank the National Cell Culture Center (Minneapolis, MN) for HeLa cells.
Abbreviations: eIF4A, eukaryotic initiation factor 4A; NMD, nonsense-mediated decay; PTC, premature termination codon; RNAi, RNA interference; siRNA, small interfering RNA; EJC, exon–exon junction complex.
References
- 1.Gingras, A. C., Raught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913–963. [DOI] [PubMed] [Google Scholar]
- 2.Hershey, J. W. B. & Merrick, W. C. (2000) in Translational Control of Gene Expression, ed. Merrick, W. C. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 33–88.
- 3.Conroy, S. C., Dever, T. E., Owens, C. L. & Merrick, W. C. (1990) Arch. Biochem. Biophys. 282, 363–371. [DOI] [PubMed] [Google Scholar]
- 4.Li, Q., Imataka, H., Morino, S., Rogers, G. W., Jr., Richter-Cook, N. J., Merrick, W. C. & Sonenberg, N. (1999) Mol. Cell. Biol. 19, 7336–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinstein, D. C., Honore, E. & Hemmati-Brivanlou, A. (1997) Development (Cambridge, U.K.) 124, 4235–4242. [DOI] [PubMed] [Google Scholar]
- 6.Holzmann, K., Gerner, C., Poltl, A., Schafer, R., Obrist, P., Ensinger, C., Grimm, R. & Sauermann, G. (2000) Biochem. Biophys. Res. Commun. 267, 339–344. [DOI] [PubMed] [Google Scholar]
- 7.Hentze, M. W. & Kulozik, A. E. (1999) Cell 96, 307–310. [DOI] [PubMed] [Google Scholar]
- 8.Wagner, E. & Lykke-Andersen, J. (2002) J. Cell Sci. 115, 3033–3038. [DOI] [PubMed] [Google Scholar]
- 9.Maquat, L. E. (2002) Curr. Biol. 12, R196–R197. [DOI] [PubMed] [Google Scholar]
- 10.Frischmeyer, P. A. & Dietz, H. C. (1999) Hum. Mol. Genet. 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell. Biol. 3, 195–205. [DOI] [PubMed] [Google Scholar]
- 12.Le Hir, H., Izaurralde, E., Maquat, L. E. & Moore, M. J. (2000) EMBO J. 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kataoka, N., Yong, J., Kim, V. N., Velazquez, F., Perkinson, R. A., Wang, F. & Dreyfuss, G. (2000) Mol. Cell 6, 673–682. [DOI] [PubMed] [Google Scholar]
- 14.Reed, R. & Hurt, E. (2002) Cell 108, 523–531. [DOI] [PubMed] [Google Scholar]
- 15.Le Hir, H., Gatfield, D., Izaurralde, E. & Moore, M. J. (2001) EMBO J. 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter, M. S., Li, S. & Wilkinson, M. F. (1996) EMBO J. 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy, E. & Maquat, L. E. (1998) Trends Biochem. Sci. 23, 198–199. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, J., Sun, X., Qian, Y. & Maquat, L. E. (1998) RNA 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, J., Sun, X., Qian, Y., LaDuca, J. P. & Maquat, L. E. (1998) Mol. Cell. Biol. 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brocke, K. S., Neu-Yilik, G., Gehring, N. H., Hentze, M. W. & Kulozik, A. E. (2002) Hum. Mol. Genet. 11, 331–335. [DOI] [PubMed] [Google Scholar]
- 21.Maquat, L. E. & Li, X. (2001) RNA 7, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neu-Yilik, G., Gehring, N. H., Thermann, R., Frede, U., Hentze, M. W. & Kulozik, A. E. (2001) EMBO J. 20, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter, M. S., Doskow, J., Morris, P., Li, S., Nhim, R. P., Sandstedt, S. & Wilkinson, M. F. (1995) J. Biol. Chem. 270, 28995–9003. [DOI] [PubMed] [Google Scholar]
- 24.Li, S., Leonard, D. & Wilkinson, M. F. (1997) J. Exp. Med. 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belgrader, P., Cheng, J. & Maquat, L. E. (1993) Proc. Natl. Acad. Sci. USA 90, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, J., Vock, V. M., Li, S., Olivas, O. R. & Wilkinson, M. F. (2002) J. Biol. Chem. 277, 18489–18493. [DOI] [PubMed] [Google Scholar]
- 27.Ishigaki, Y., Li, X., Serin, G. & Maquat, L. E. (2001) Cell 106, 607–617. [DOI] [PubMed] [Google Scholar]
- 28.Li, W., Belsham, G. J. & Proud, C. G. (2001) J. Biol. Chem. 276, 29111–29115. [DOI] [PubMed] [Google Scholar]
- 29.Muhlemann, O., Mock-Casagrande, C. S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M. F. & Moore, M. J. (2001) Mol. Cell 8, 33–43. [DOI] [PubMed] [Google Scholar]
- 30.Wang, J., Chang, Y. F., Hamilton, J. I. & Wilkinson, M. F. (2002) Mol. Cell 10, 951–957. [DOI] [PubMed] [Google Scholar]
- 31.Luo, M. J. & Reed, R. (1999) Proc. Natl. Acad. Sci. USA 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, Z., Luo, M. J., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature 407, 401–405. [DOI] [PubMed] [Google Scholar]
- 33.Fan, X. C. & Steitz, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dostie, J., Ferraiuolo, M., Pause, A., Adam, S. A. & Sonenberg, N. (2000) EMBO J. 19, 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendell, J. T., ap Rhys, C. M. & Dietz, H. C. (2002) Science 298, 419–422. [DOI] [PubMed] [Google Scholar]
- 36.Lejbkowicz, F., Goyer, C., Darveau, A., Neron, S., Lemieux, R. & Sonenberg, N. (1992) Proc. Natl. Acad. Sci. USA 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohnsack, M. T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E. & Gorlich, D. (2002) EMBO J. 21, 6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan, C. C., Dostie, J., Diem, M. D., Feng, W., Mann, M., Rappsilber, J. & Dreyfuss, G. (2004) RNA 10, 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, Z., Licklider, L. J., Gygi, S. P. & Reed, R. (2002) Nature 419, 182–185. [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. (2002) Genome Res. 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurica, M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. (2002) RNA 8, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jurica, M. S. & Moore, M. J. (2003) Mol. Cell 12, 5–14. [DOI] [PubMed] [Google Scholar]
- 43.Luo, M. L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M. & Reed, R. (2001) Nature 413, 644–647. [DOI] [PubMed] [Google Scholar]
- 44.Pinol-Roma, S. & Dreyfuss, G. (1992) Nature 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 45.Izaurralde, E. (2002) Results Probl. Cell Differ. 35, 133–150. [DOI] [PubMed] [Google Scholar]
- 46.Izaurralde, E. (2002) Eur. J. Cell Biol. 81, 577–584. [DOI] [PubMed] [Google Scholar]
- 47.Reed, R. & Magni, K. (2001) Nat. Cell Biol. 3, E201–E204. [DOI] [PubMed] [Google Scholar]
- 48.Iborra, F. J., Jackson, D. A. & Cook, P. R. (2001) Science 293, 1139–1142. [DOI] [PubMed] [Google Scholar]
- 49.Nathanson, L., Xia, T. & Deutscher, M. P. (2003) RNA 9, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svitkin, Y. V., Pause, A., Haghighat, A., Pyronnet, S., Witherell, G., Belsham, G. J. & Sonenberg, N. (2001) RNA 7, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniels-McQueen, S., Detjen, B. M., Grifo, J. A., Merrick, W. C. & Thach, R. E. (1983) J. Biol. Chem. 258, 7195–7199. [PubMed] [Google Scholar]
- 52.Mendell, J. T., Medghalchi, S. M., Lake, R. G., Noensie, E. N. & Dietz, H. C. (2000) Mol. Cell. Biol. 20, 8944–8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shibuya, T., Tange, T., Sonenberg, N. & Moore, M. J. (2004) Nat. Struct. Mol. Biol., in press. [DOI] [PubMed]
- 54.Palacios, I. M., Gatfield, D., St. Johnston, D. & Izaurralde, E. (2004) Nature 427, 753–757. [DOI] [PubMed] [Google Scholar]