Abstract
In eukaryotes, cyclin B-bound cyclin-dependent protein kinase 1 promotes mitotic entry but is held in check, in part, by Wee1 protein kinase. Timely mitotic entry in budding yeast requires inactivation of Swe1 (Wee1 ortholog). Perturbations of the septin collar at the bud neck lead to Swe1 stabilization, delaying the G2/M transition. Swe1 is recruited to the neck and hyperphosphorylated before ubiquitin-mediated degradation. Hsl1 kinase (Nim1 ortholog), a negative regulator of Wee1, is required for efficient Swe1 localization at the neck but seems not to phosphorylate Swe1. Here, we show that two other kinases targeted sequentially to the neck, Cla4/PAK and Cdc5/Polo, are responsible for stepwise phosphorylation and down-regulation of Swe1. This mechanism links assembly of a cellular structure to passage into mitosis.
Keywords: Cdk1 regulation, mitotic progression, G2/M transition, Saccharomyces cerevisiae
Cyclin-dependent protein kinases (Cdks) regulate cell cycle progression in all eukaryotes (1). Entry into M phase is induced by cyclin B (Clb)-bound Cdk1/Cdc2. In fission yeast and higher eukaryotes, Cdk1 is negatively regulated by phosphorylation at Tyr-15 by Wee1 (2, 3), a modification reversed by Cdc25 phosphatase (4). In budding yeast, Swe1 (Wee1 ortholog) phosphorylates Clb-bound Cdc28 (Cdk1/Cdc2 homolog) on the equivalent residue (Tyr-19) (5), a modification reversed by Mih1 (Cdc25 ortholog) (6, 7).
During a normal cell cycle, Swe1 accumulates in S phase, becomes sequentially hyperphosphorylated (8, 9), generating multiple isoforms, and undergoes ubiquitin-mediated degradation (10–12). Defects in septin filament assembly at the bud neck (9, 13, 14) result in hypophosphorylation and stabilization of Swe1 and, as a result, Swe1-dependent inhibition of Clb-Cdc28. This Swe1-imposed G2 delay leads to elongated cells because the cells fail to switch from polarized to isotropic growth during budding (15). Thus, phosphorylation and degradation of Swe1 seem critical for efficient activation of Clb-Cdc28 and timely mitotic entry.
Hsl1 (Nim1/Cdr1 ortholog) is a primary negative regulator of Swe1 (13, 16). Hsl1 activity and localization depend on septin filament assembly at the neck (13, 17, 18). Hsl1 acts in concert with Hsl7 (9, 19); absence of either protein dramatically reduces Swe1 phosphorylation in vivo and causes cell elongation. Hsl1 is required to tether Hsl7 at the neck (9), and both are required for efficient recruitment of Swe1 to the same location (14, 19). Contrary to the reported phosphorylation of Wee1 by recombinant Nim1/Cdr1 (20–22), Hsl1 seems unable to phosphorylate Swe1 in vitro (23). This observation suggests that other neck-associated kinase(s) may directly phosphorylate Swe1.
Genetic findings implicated both Cla4 (PAK homolog) (24) and Cdc5 (Polo homolog) (25, 26) in the G2/M transition, but in a manner not understood. Here, we show that the septin collar serves as an organizing platform to permit Cla4 and Cdc5 to sequentially phosphorylate Swe1 and that this cumulative multikinase-dependent modification is critical for Swe1 degradation and therefore for proper onset of mitosis.
Materials and Methods
Strain and Plasmid Construction. Yeast strains and plasmids used in this study are listed in Tables 1 and 2, which are published as supporting information on the PNAS web site. Detailed information is provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Growth Conditions and Media. Yeast cell culture and transformations were carried out by standard methods. For cell-cycle synchronization, MATa cells were arrested with 5 μg/ml α-mating pheromone (Sigma) for 2 h at 30°C. Hydroxyurea (Sigma) and nocodazole (Sigma) were used at the final concentrations of 200 mM and 15 μg/ml, respectively.
Kinase Assays and Immunoblotting. Kinase assays were carried out as described (27). For immunoblotting analyses, total cellular proteins were separated by SDS/PAGE as indicated and transferred to poly(vinylidene difluoride) membrane (Millipore). Proteins that interact with anti-Swe1 (a gift of Doug Kellogg, University of California, Santa Barbara), anti-Myc (Santa Cruz Biotechnology), and anti-Hsl7 antibodies (9) were detected by using the enhanced chemiluminescence (ECL) detection system (Pierce). To detect the phosphorylated, slow migrating, Swe1 isoforms, it was necessary to use 8% SDS/PAGE (no SDS in the separation gel).
Cell Staining and Immunofluorescence Microscopy. Indirect immunofluorescence was performed as described (27). Briefly, cells were fixed with 3.7% formaldehyde, and microtubules were visualized by using YOL1/34 rat anti-tubulin antibody (Accurate Chemical, Westbury, NY) and goat anti-rat CY3 antibody (Jackson ImmunoResearch).
Results and Discussion
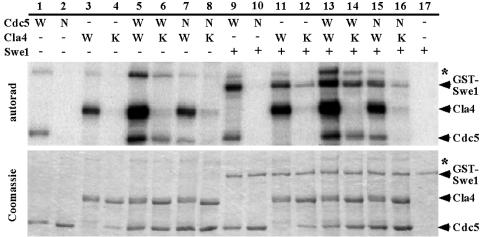
Cdc5 and Cla4 Phosphorylate Swe1 in Vitro. We tested six protein kinases known to associate with (and/or to contribute to assembly of) septin filaments at the bud neck (Cdc5, Cla4, Elm1, Gin4, Hsl1, and Kcc4) for direct phosphorylation of Swe1. The enzymes were expressed and purified from Sf9 cells, incubated with catalytically inactive GST-Swe1(K473A) (23) and [γ-32P]ATP. None phosphorylated GST alone (data not shown), and GST-Swe1(K473A) was not phosphorylated in the absence of added kinase (Fig. 1, lane 17). Hsl1, Gin4, and Kcc4 (the three Nim1-related kinases in S. cerevisiae) and Elm1 catalyzed no more than trace incorporation into GST-Swe1(K473A) (data not shown), although all these preparations were active (as judged by their autophosphorylation). In contrast, Cdc5 and Cla4 exhibited robust phosphorylation of GST-Swe1(K473A) (Fig. 1, lanes 9 and 11). Catalytically inactive Cdc5(N209A) (28) (Fig. 1, lane 2) did not phosphorylate GST-Swe1(K473A) (Fig. 1, lane 10). Similarly, catalytically defective (but not completely inactive) Cla4(K594A) (24) (Fig. 1, lane 4) displayed significantly reduced incorporation into GST-Swe1(K473A) (Fig. 1, lane 12). Cdc5 and Cla4 cross-phosphorylated each other weakly (Fig. 1, lanes 5–8), but Cdc5 and Cla4 did not seem to phosphorylate GST-Swe1(K473A) synergistically (Fig. 1 A, lanes 13–16).
Fig. 1.
Cdc5 and Cla4 phosphorylate Swe1 in an additive manner. Recombinant HA-Cdc5-Flag, HA-Cdc5(N209A)-Flag, HA-Cla4-Flag, and HA-Cla4(K594A)-Flag were immunoprecipitated from Sf9 cells by using an anti-Flag antibody cross-linked to agarose beads (Sigma). The reactions were carried out with or without kinase-inactive GST-Swe1(K473A) in the presence of 5 μM ATP (10 μCi of [γ-32P]ATP; 1 Ci = 37 GBq) for 30 min at 30°C, separated in 10% SDS/PAGE, stained with Coomassie blue (Lower), and then subjected to autoradiography (Upper). Asterisks indicate a contaminating protein that copurified with Cdc5 and Cla4.
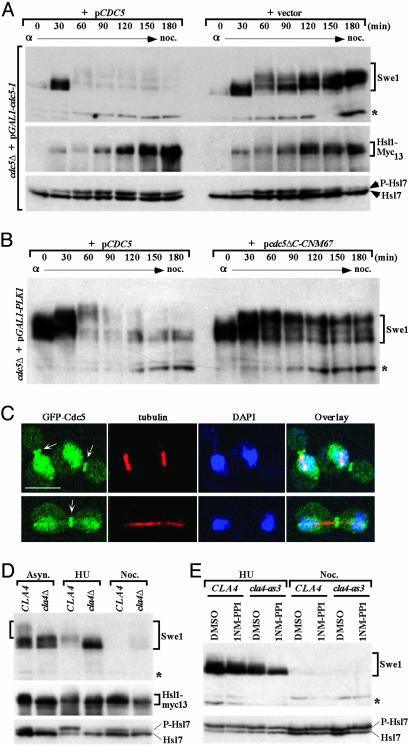
Cdc5 Is Required for Swe1 Phosphorylation in Vivo. If Cdc5-mediated phosphorylation contributes to Swe1 down-regulation, then absence of Cdc5 should prevent efficient Swe1 degradation. To test this prediction, and because CDC5 is an essential gene, a cdc5Δ strain expressing a weakly functional allele, cdc5-1, under control of the GAL1 promoter was used; these cells are viable on galactose-containing medium but are inviable when shifted to glucose medium, which represses the GAL1 promoter. To compare Cdc5-deficient to Cdc5-containing cells, the strain was transformed with either an empty centromeric (CEN) vector or the same vector expressing wild-type CDC5 from its native promoter (pCDC5). To synchronize cells in G1, cultures were treated with α-factor mating pheromone and at the same time were shifted to glucose medium to prevent expression of the Cdc5-1 mutant protein. After 2 h, cells were washed and released into glucose medium containing nocodazole (to permit passage into, but not beyond, M phase). Samples were taken at various times after release from the α-factor block and Swe1 was examined by immunoblotting of cell extracts. When Cdc5 was supplied from pCDC5, Swe1 accumulated during the first 30 min after release and was only moderately phosphorylated; by 60 min after release, however, Swe1 was almost completely absent and the most hyperphosphorylated species (the most slowly migrating bands) were barely detectable (Fig. 2A Top, left side). In contrast, in the Cdc5-deficient cells, Swe1 persisted for at least 180 min and the moderately phosphorylated species (as opposed to the most hyperphosphorylated isoforms) accumulated (Fig. 2 A Top, right side). Stabilization of Swe1 and reduction in the most hyperphosphorylated species were not due to any delay in cell cycle progression because the Cdc5-deficient cells proceeded through the cell cycle at a rate indistinguishable from cells expressing Cdc5 (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Requirement of Cdc5 and Cla4 for proper phosphorylation and degradation of Swe1. (A) Strain KLY4864 (cdc5Δ HSL1-Myc13 + YCplac33-GAL1-cdc5-1) transformed with either a centromeric CDC5 (pCDC5) (KLY4900) or a control vector (KLY4899) was arrested in G1 by α-factor treatment immediately after transferring to yeast extract/peptone (YEP)-glucose medium, then released into nocodazole-containing medium. Total cellular proteins were prepared at the indicated times and separated by 8% SDS/PAGE (without SDS in the separation gel to detect the phosphorylated Swe1 isoforms). Immunoblotting analyses were carried out by using antibodies against Swe1, c-Myc epitope (for Hsl1-Myc13), and Hsl7. P-Hsl7, phosphorylated Hsl7. Asterisks in A, B, D, and E are the same nonspecific protein cross-reacting with anti-Swe1 antibody. (B) Strain KLY4497 (cdc5Δ HSL1-Myc13 + YCplac33-GAL1-PLK1) transformed with either a centromeric pCDC5 (KLY4532) or a centromeric pCDC5ΔC-CNM67 (KLY4533) were cultured overnight in YEP-glucose medium, arrested with α-factor, and then released into YEP-glucose medium containing nocodazole. Samples were analyzed as in A. (C) To determine the timing of Cdc5 localization to the bud-neck, asynchronously growing KLY4816 cells, which express EGFP-CDC5 under endogenous CDC5 promoter control, were cultured overnight, fixed, and subjected to immunostaining with anti-tubulin antibody. Arrows indicate the bud-neck localized Cdc5. (Scale bar, 5μm.) (D) Strains KLY4071 (CLA4) and KLY4536 (cla4Δ) were grown either asynchronously or arrested with hydroxyurea (HU) or nocodazole for 4 h before the preparation of total cellular lysates. (E) Strains KLY4913 (cla4Δ + pCLA4) and KLY4914 (cla4Δ + pcla4-as3) were arrested with either hydroxyurea or nocodazole for 2 h before the addition of either 25 μM of 1-NM-PP1 or control DMSO into the medium. Cells were cultured for additional 2 h before harvesting the samples.
Because Hsl1 is critical for phosphorylation and localization of Hsl7 to the bud neck (9, 18, 23) and both proteins are necessary for efficient neck recruitment, phosphorylation, and degradation of Swe1, perturbation of the activity or localization of Hsl1 or Hsl7 in the absence of Cdc5 might account for the observed Swe1 stabilization. However, depletion of Cdc5 did not alter Hsl1 activity (Fig. 2 A Middle), Hsl7 phosphorylation (Fig. 2 A Bottom), or targeting of GFP-Hsl7 to the neck (Fig. 7, which is published as supporting information on the PNAS web site).
Swe1 Is Stabilized When Cdc5 Is Not Located at the Bud Neck. Cdc5 localizes to spindle pole bodies (SPBs) and then to the neck (27, 29). If Cdc5 at the neck is crucial for Swe1 down-regulation, preventing Cdc5 localization to that site should stabilize Swe1. Hence, a catalytically active (but localization-defective) Cdc5 truncation, Cdc5ΔC, which cannot by itself complement the inviability of cdc5Δ cells (data not shown), was fused to Cnm67, a component of the outer plaque of the SPB (30). This fusion, expressed from the native CDC5 promoter, was introduced into a cdc5Δ strain in which viability was maintained by GAL1-driven expression of a mammalian Polo kinase (PLK1) cDNA. The Cdc5ΔC-Cnm67 chimera complemented the inviability of cdc5Δ cells on glucose medium and localized to the SPB but not the bud neck (J.-E.P. and K.S.L., unpublished data). However, the resulting cells displayed an elongated bud morphology, like that of other mutants defective in Swe1 down-regulation (9, 16). To examine Swe1 stability, cdc5Δ cells carrying pCDC5 or expressing CDC5ΔC-CNM67 from the same vector were synchronized in G1 with α-factor and released into nocodazole-containing medium, as before. In cells with wild-type Cdc5, Swe1 was progressively phosphorylated and then disappeared (Fig. 2B, left side). In contrast, in cells containing just Cdc5ΔC-Cnm67, Swe1 was markedly stabilized, and isoforms phosphorylated at an intermediate level (as opposed to the most hyperphosphorylated species) accumulated (Fig. 2B, right side). These differences were not attributable to any difference in the rate of cell cycle progression (Fig. 8, which is published as supporting information on the PNAS web site) or to any perturbation in GFP-Hsl7 localization to the neck (data not shown). Thus, only Cdc5 molecules at the neck contribute to efficient Swe1 down-regulation.
This observation also predicts that Cdc5 should normally localize to the neck before onset of the G2/M transition. Therefore, timing of localization of GFP-Cdc5 expressed at its endogenous level, which is fully functional, was examined. Structure of the spindle was monitored as an internal marker for cell cycle position (Fig. 2C). Cdc5 localization to the bud neck was detectable by the time cells formed a short spindle, and this Cdc5 population remained at the neck until spindles were fully elongated (Fig. 2C). Cdc5 is thus present at the neck by G2 phase and available early enough to contribute to Swe1 modification before the onset of M phase.
Cla4 Is Required for Phosphorylation of Swe1 in Vivo. In cells lacking Cdc5 or in which Cdc5 cannot be localized to the neck, the most hyperphosphorylated Swe1 forms are eliminated and Swe1 is greatly stabilized. However, Swe1 is phosphorylated to an intermediate level. Hence, some other kinase(s) is responsible for the early phase of Swe1 phosphorylation. Cla4 is important for septin collar assembly (31) and associates tightly with septins (32). Because Cla4 was the only other neck-associated kinase able to phosphorylate Swe1 in vitro, its contribution to Swe1 phosphorylation and down-regulation in vivo was tested.
In asynchronous cultures, cla4Δ cells lack the most hyperphosphorylated Swe1 species seen in parental CLA4+ cells (Fig. 2D Top, Asyn., left bracket). Moreover, in S phase-arrested (hydroxyurea treated) cells, Swe1 accumulated predominantly in hypophosphorylated form in the cla4Δ mutant, whereas in control CLA4+ cells the bulk of Swe1 was absent (Fig. 2D). Even in M phase-arrested (nocodazole treated) cells, a small amount of Swe1 was still detectable in cla4Δ cells that was not seen in CLA4+ cells. Because cells lacking Cla4 are defective in septin filament assembly (31, 32), as indicated by diminished Hsl1 autophosphorylation (Fig. 2D Middle) and reduced Hsl7 phosphorylation (Fig. 2D Bottom), the stabilization and decrease in phosphorylation of Swe1 in Cla4-deficient cells might arise indirectly.
To confirm that Cla4 directly contributes to Swe1 phosphorylation and down-regulation, we used an analog-sensitive cla4as3 allele (33) that is inhibitable by the cell-permeable compound 1NM-PP1 (4-amino-1-tert-butyl-3-(1-naphthylmethyl) pyrazolo[3,4-d]pyrimidine) (34). A CLA4+ strain or an otherwise isogenic cla4-as3 mutant were grown to mid-exponential phase, arrested in either S phase (with hydroxyurea) or M phase (with nocodazole), and treated for2hwith solvent only (DMSO) as a control or 25 μM 1NM-PP1 to acutely inhibit Cla4 function. As observed in S phase-arrested cla4Δ cells, Swe1 was present almost exclusively in hypophosphorylated form when S phase-arrested cla4-as3 cells were treated with the analog, but not when treated with solvent alone (Fig. 2E Upper). The analog had no effect on Swe1 phosphorylation in CLA4+ cells. As expected if septin filament assembly was not perturbed under these conditions, Hsl7 was phosphorylated normally (Fig. 2E Lower) and GFP-Hsl7 localized efficiently to the neck (Fig. 9, which is published as supporting information on the PNAS web site) in both cla4-as3 and CLA4 cells with or without the analog. Thus, Cla4 activity does contribute directly to Swe1 modification in vivo, consistent with its ability to phosphorylate Swe1 in vitro (Fig. 1 A). However, as seen in cla4Δ cells (Fig. 2D), inactivation of Cla4 failed to cause Swe1 accumulation in nocodazole-arrested cells (Fig. 2E). Taken together, these observations suggest that Cla4 contributes to Swe1 phosphorylation in S phase, but Cdc5-dependent phosphorylation is able to mediate Swe1 down-regulation by the onset of M phase.
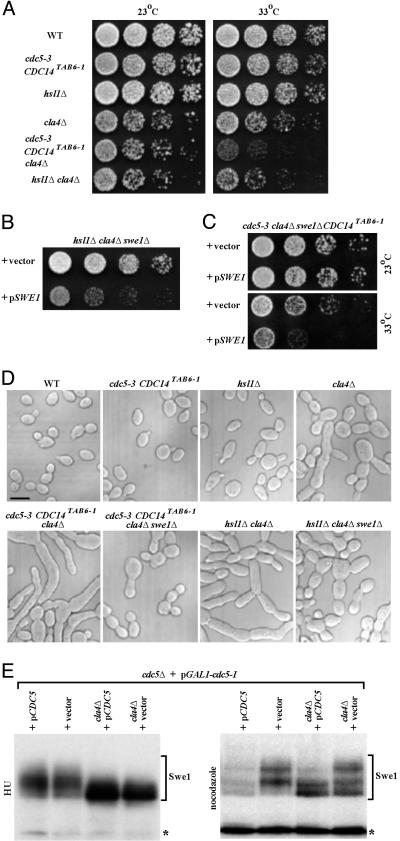
Kinase Collaboration in Swe1 Inactivation. Prior genetic results indicated that Hsl1 and Cdc5 act in parallel and upstream of Swe1 to promote entry into mitosis (26). As an independent means to assess whether Cla4 contributes in a physiologically relevant way to Swe1 down-regulation along with Hsl1 and Cdc5, we constructed double mutants. We combined a cla4Δ mutation with either an hsl1Δ mutation or the temperature-sensitive cdc5-3 allele. For the latter, we included the CDC14TAB6-1 mutation (35), which compensates for the mitotic exit defect, but not the G2/M delay, of cdc5-3 mutant cells (26). When compared to either cla4Δ or hsl1Δ single mutants, a cla4Δ hsl1Δ double mutant exhibited a more pronounced growth defect (Fig. 3A); in other strain backgrounds, cla4Δ hsl1Δ cells are inviable (36). More strikingly, the growth defect of cdc5-3 CDC14TAB6-1 cells, which are already somewhat defective in Swe1 down-regulation at semirestrictive temperature (33°C) (26), was strongly exacerbated by absence of Cla4 (Fig. 3A). As expected if the primary problem in these strains is lack of efficient down-regulation of Swe1, introduction of a swe1Δ mutation ameliorated the growth and morphology defects of the cla4Δ hsl1Δ double mutant and the cdc5-3 CDC14TAB6-1 cla4Δ cells (Fig. 3 B–D). Conversely, reintroduction of SWE1 on a CEN plasmid reimposed obvious growth defects (Fig. 3 B and C) and marked morphology defects (Fig. 10, which is published as supporting information on the PNAS web site) in both hsl1Δ cla4Δ swe1Δ cells and cdc5-3 CDC14TAB6-1 cla4Δ swe1Δ cells. This genetic synergy supports the view that Cdc5, Cla4, and Hs11 all contribute to Swe1 down-regulation.
Fig. 3.
Cdc5, Cla4, and Hsl1 function in pathways distinct from one another. (A) Strains KLY1546 (wild type), KLY4651 (cdc5-3 CDC14TAB6-1), KLY2869 (hsl1Δ), KLY4649 (cla4Δ), KLY4653 (cdc5-3 CDC14TAB6-1 cla4Δ), and KLY5098 (hsl1Δ cla4Δ) were cultured overnight. These cultures were serially diluted, spotted on YEP-glucose, and incubated at the indicated temperature. (B and C) Strain KLY5097 (hsl1Δ cla4Δ swe1Δ) (B) or strain KLY4756 (cdc5-3 CDC14TAB6-1 cla4Δ swe1Δ) (C) was transformed with either a centromeric SWE1 (pSWE1) or control vector. The resulting transformants were cultured overnight, serially diluted, and spotted on synthetic minimal plate (SDM) lacking histidine (B) or uracil (C) to select for the presence of the introduced plasmid. (D) To examine the morphologies of the strains used in A–C, cells cultured overnight at 23°C were fixed and then examined by differential interference contrast (DIC) microscopy. The synthetically elongated bud morphologies were manifest among cdc5-3 CDC14TAB6-1, cla4Δ, and hsl1Δ mutants even at 23°C. (Scale bar, 5 μm.) (E) To examine whether Cdc5 and Cla4 cooperate to regulate the Swe1 stability, cla4Δ was introduced into strains KLY4714 (cdc5Δ + pGAL1-cdc5-1 + YCplac22) and KLY4715 (cdc5Δ + pGAL1-cdc5-1 + YCplac22-CDC5), generating KLY4811 and KLY4813, respectively. All four strains were cultured overnight in YEP-galactose medium, shifted to YEP-glucose medium containing either HU or nocodazole, and cultured for an additional 4 h. Longer exposure time was required to detect Swe1 from nocodazole-arrested samples (Right). Asterisks, a cross-reacting protein with the anti-Swe1 antibody.
Biochemical criteria also support the conclusion that Cdc5 and Cla4 collaborate (albeit at different stages of the cell cycle) to regulate Swe1 modification and stability in vivo. In S phase-arrested cla4Δ cells, Swe1 accumulated in hypophosphorylated form regardless of whether Cdc5 was present or not (Fig. 3E Left, lanes 3 and 4), presumably because Cdc5 does not translocate to the neck until early G2 (Fig. 2C). When Cla4 is present, moderately phosphorylated forms of Swe1 are observed and appear to be stable (Fig. 3E Left, lanes 1 and 2), presumably because of lack of subsequent modification by Cdc5. Thus, Cla4 contributes to the early phase of Swe1 phosphorylation in a Cdc5-independent manner. In M phase-arrested cells, after Cdc5 has relocated to the neck, Swe1 is barely detectable when both Cdc5 and Cla4 are present (Fig. 3E Right, lane 1), whereas when only Cdc5 is absent, the moderately phosphorylated Swe1 species that largely depend on Cla4 for their formation accumulate (Fig. 3E Right, lane 2). In the absence of Cla4, the hypophosphorylated form of Swe1 accumulates whether or not Cdc5 is present; however, presence of Cdc5 seems to be able to drive further phosphorylation and degradation of some of the Swe1 (Fig. 3E Right, compare lanes 3 and 4). Taken together, our observations suggest Cla4 functions early and Cdc5 functions later during cell cycle progression, and that their combined activities are necessary for proper phosphorylation and efficient degradation of Swe1. Swe1 is still phosphorylated significantly in the absence of both Cla4 and Cdc5 (Fig. 3E, lane 4), suggesting that an additional kinase(s), e.g., Clb-Cdc28 (see below), also contributes to Swe1 phosphorylation.
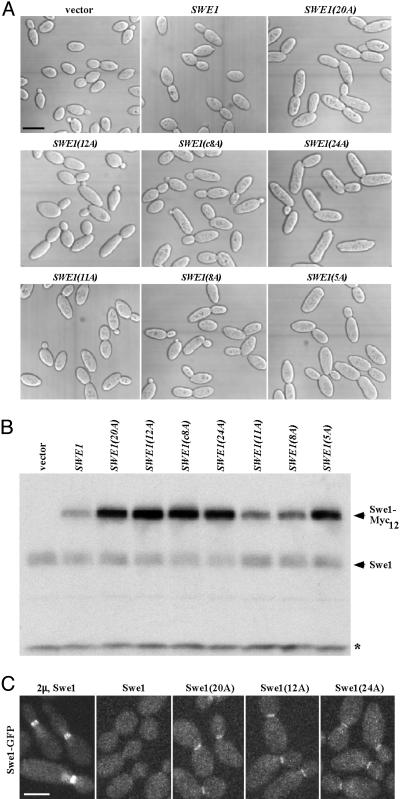
Absence of Cdc5 or Cla4 Phosphorylation Sites Stabilizes Swe1. To confirm the physiological significance of Cdc5- and Cla4-dependent phosphorylation of Swe1, we phosphorylated GST-Swe1(K473A) with each enzyme in vitro with excess ATP, and the sites phosphorylated were mapped within tryptic peptides by using mass spectrometry. Sites not definitively determined were predicted by using netphos 2.0 (37) and the known consensus phosphoacceptor motifs for Polo D/E-X-S/T-Φ-X-(D/E) (38) and for yeast PAKs (R-X-S/T-X-Φ; see refs. 32 and 39), where X is any residue and Φ is a hydrophobic residue. Seventeen definitive and four potential sites for Cdc5, and seven definitive and five potential sites for Cla4, were identified in Swe1 (Tables 3 and 4, which are published as supporting information on the PNAS web site). Eight tryptic peptides contain apparent “common” sites for both enzymes. The number of Swe1 isoforms seen in immunoblots of yeast extracts in this study and in prior work (8, 9) verify that Swe1 is phosphorylated at many sites. To date, only three sites have been determined in Swe1 recovered from yeast cells; but reassuringly, these are identical to in vitro sites we mapped (see legend of Table 3). Site-directed mutagenesis was used to introduce mutations (S to A or T to A) at individual sites in a functional (c-Myc)12-tagged Swe1 (12). The resulting constructs were integrated at the URA3 locus in a strain carrying an mih1Δ mutation, which potentiates Swe1-dependent inhibition of Clb-Cdc28 (19). Aside from one mutation (S118A) that unexpectedly destabilized Swe1 for unknown reasons (data not shown), no single point mutation detectably affected Swe1 stability (as judged by immunoblotting with anti-Myc mAb) or caused elongated cell morphology diagnostic of a G2/M delay (data not shown). Therefore, we generated Swe1 derivatives with mutations in (i) all 20 of the apparent Cdc5 phosphorylation sites (except S118), designated Swe1(20A); (ii) all 12 of the apparent Cla4 sites, dubbed Swe1(12A); (iii) the “common” sites (namely, S111A, S288A, S379A, S395A, S438A, T629A, S682A, and T688A), termed Swe1(c8A); (iv) the 24 combined Cdc5 and Cla4 sites, designated Swe1(24A); (v) just the 11 most N-terminal sites (S36A, T66A, S102A, S111A, T131A, S136A, S156A, S169A, S185A, S223A, and S225A), called Swe1(11A); (vi) just the 8 internal sites (S254A, T280A, S288A, S312A, S379A, S395A, S438A, and T501A), termed Swe1(8A); and (vii) just the 5 most C-terminal sites (S610A, T629A, T676A, S682A, and T688A), designated Swe1(5A). These multiply mutated constructs were integrated in epitope-tagged form at the URA3 locus in the mih1Δ strain and examined, as above. Compared to parental cells expressing endogenous SWE1, cells expressing the additional Swe1-Myc12 copy displayed only a slight increase in average cell length (only mother cells were measured, to avoid the inherently greater heterogeneity of buds) (Figs. 4A and 11, which is published as supporting information on the PNAS web site). Indeed, Swe1-Myc12 appears to be as unstable as endogenous Swe1, based on their steady-state level of expression in the same cell (Fig. 4B, lanes 1 and 2). In contrast, Swe1(20A) and Swe1(12A) caused pronounced cell elongation (Figs. 4A and 11). The greatest increase in cell elongation was observed in cells expressing Swe1(24A), in which all of the Cdc5 and Cla4 phosphorylation sites were mutated (Figs. 4A and 11), further supporting the conclusion that Cdc5 and Cla4 act in concert to promote Swe1 down-regulation. All of the remaining mutants [Swe1(c8A), Swe1(11A), Swe1(8A), and Swe1(5A)] caused similar degrees of cell elongation (Figs. 4A and 11). Thus, no one region within Swe1 seems uniquely responsible, when phosphorylated, for conferring instability (although absence of the C-terminal phosphorylation sites had a somewhat more pronounced effect).
Fig. 4.
Mutations in the Cdc5- and Cla4-dependent phosphorylation sites onto Swe1 induce elongated bud morphology as a result of Swe1 stabilization at the bud-neck. (A) An mih1Δ strain (M-600) was integrated with control vector or various forms of SWE1-Myc12 at the URA3 locus. The resulting strains (KLY4802–KLY4810) were cultured overnight, fixed, and subjected to DIC microscopy (A) to measure the cell size (Fig. 11). (Scale bar, 5 μm.) (B) To determine the steady-state level of various Swe1-Myc12 proteins, total cellular lysates were prepared from the asynchronously growing strains used in A. Samples were separated in 10% SDS/PAGE and then subjected to immunoblotting with anti-Swe1 antibody. Asterisk indicates a nonspecific band. (C)To examine the localization of various Swe1 proteins, the wild-type and mutant forms of SWE1 in the strains used in A were C-terminally tagged with three copies of GFP by replacing the Myc12 epitope. The resulting strains were then cultured at 23°C, arrested with hydroxyurea for 3 h to enrich the Swe1 protein, and subjected to confocal microscopy. (Scale bar, 5 μm.)
In keeping with their phenotype, all of the multiple phosphorylation site mutants were more stable than Swe1-Myc12 or endogenous Swe1 present in the same cell, as judged by immunoblotting of asynchronous cultures (Fig. 4B, lanes 3–9). Because all of these Swe1 derivatives caused cell cycle delay and cell elongation, they must all be catalytically active. Moreover, their observed accumulation cannot be due to some indirect effect because endogenous Swe1 in the same cells did not accumulate (Fig. 4B). Furthermore, GFP-tagged versions of these variants are recruited to the bud neck. Only when highly overexpressed from a multicopy vector to overcome its instability could wild-type Swe1-GFP be detected at the neck in some (47%) cells (n = 87); when expressed at a near endogenous level from a low-copy-number (CEN) plasmid, Swe1-GFP was undetectable at the neck (Fig. 4C). In contrast, when expressed from the same CEN plasmid, three different multiple phosphorylation site mutants, Swe1(20A)-GFP, Swe1(12A)-GFP, and Swe1(24A)-GFP, all displayed accumulation at the neck in >60% of the cells (n > 120 for all three mutants) (Fig. 4C). Thus, absence of Cdc5 and Cla4 phosphorylation does not perturb intracellular targeting but greatly stabilizes Swe1 in vivo.
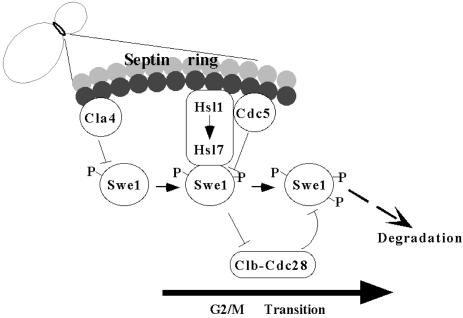
A Novel Mechanism Integrates Multiple Signals to License Passage into Mitosis. Our data demonstrate that septin collar formation establishes a platform that allows for cumulative phosphorylation of Swe1 by multiple kinases (Fig. 5). Sequential multistep phosphorylation by Cla4 and Cdc5 makes Swe1 susceptible to subsequent degradation. Cla4-mediated Swe1 modification occurs in S phase, whereas Cdc5-mediated phosphorylation happens before entry into mitosis. Consistent with this view, Cla4 binds tightly to and phosphorylates septins and promotes septin filament assembly (32). Also compatible with this proposed order, as documented here, Cdc5 translocates to the neck before the onset of M phase, and this localization is critical for Swe1 phosphorylation and down-regulation. It has been demonstrated amply that tethering of Swe1 to septin filaments at the neck requires Hsl1 and Hsl7 (9, 14, 19), and there is some evidence that Hsl1 and Hsl7 also are required for efficient recruitment of Cdc5 to this location (C. F. Hardy, personal communication). Thus, cumulative phosphorylation of Swe1 at the neck could represents a means to integrate, in one protein, stepwise signals that indicate completion of prior cell cycle events before entry into mitosis is allowed.
Fig. 5.
Model illustrating the Swe1 phosphorylation and degradation events during the cell cycle (see text for details).
Presence of Cla4 at the neck may serve as a signal that proper morphogenesis is underway and initiates Swe1 modification. Subsequent recruitment of Cdc5 to the neck after short spindles have formed may serve as a signal that septin filament assembly has been completed and further promotes Swe1 phosphorylation. Finally, given that Clb-Cdc28 also associates with and phosphorylates Swe1 (refs. 10 and 12; D. Kellogg, personal communication), accumulation of Clb-Cdc28 complexes may serve as a signal that adequate supplies of the enzyme are present to efficiently promote mitosis and, hence, Clb-Cdc28-mediated phosphorylation of Swe1 could act, in a positive feedback mechanism, as the final step of Swe1 inactivation (Fig. 5). Once unleashed from negative regulation by Swe1, Clb-Cdc28 can induce mitotic entry unimpeded.
Our findings show that phosphorylation at multiple sites scattered throughout the protein are necessary to dictate efficient Swe1 degradation. This cumulative phosphorylation by the Cla4, Cdc5, and Clb-Cdc28 kinases must establish landmarks in the protein or induce a conformational change (or both) that makes Swe1 susceptible to subsequent ubiquitination. There is evidence that the APC may be a ubiquitin ligase (E3) that contributes to Swe1 ubiquitination (ref. 40; C. F. Hardy, personal communication), but there is also evidence that a SCF-like E3 is involved in Wee1 destruction (ref. 41; T. Hunter, personal communication). Cumulative multisite phosphorylation by a single kinase is responsible for efficient degradation of the Cdk inhibitor, Sic1 (42). However, action of multiple kinases on a single protein to promote its degradation has not been seen previously.
In summary, because septin filament assembly requires Cla4, and septin filament assembly is required for localization and function of Hsl1 and Hsl7, which are, in turn, required to ensure proximity between Cdc5 and Swe1, the phosphorylation of Swe1 by neck-localized kinases links morphogenetic events with proper spatial and temporal control of mitotic entry.
Supplementary Material
Acknowledgments
We thank F. J. Gonzalez, P. Supavilai, and A. Thithapandha for their support, and R. Deshaies, D. Kellogg, D. Lew, C. F. Hardy, D. Drubin, M. Longtine, and K. Shokat for research materials, helpful comments, or communication of unpublished results. Some of these studies were conducted in partial fulfillment of requirements for the Ph.D. degree of K.S. from the Department of Pharmacology, Mahidol University, Bangkok, Thailand. This work was supported by the Royal Golden Jubilee Scholarship Fund (K.S.), funds from the Berkeley campus Cancer Research Laboratory, and National Institutes of Health Grant GM21841 (to J.T.).
Abbreviation: Clb, cyclin B.
References
- 1.Harper, J. W. & Adams, P. D. (2001) Chem. Rev. 101, 2511–2526. [DOI] [PubMed] [Google Scholar]
- 2.Russell, P. & Nurse, P. (1987) Cell 49, 559–567. [DOI] [PubMed] [Google Scholar]
- 3.Parker, L. L., Atherton-Fessler, S. & Piwnica-Worms, H. (1992) Proc. Natl. Acad. Sci. USA 89, 2917–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draetta, G. & Eckstein, J. (1997) Biochim. Biophys. A 1332, M53–M63. [DOI] [PubMed] [Google Scholar]
- 5.Booher, R. N., Deshaies, R. J. & Kirschner, M. W. (1993) EMBO J. 12, 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell, P., Moreno, S. & Reed, S. I. (1989) Cell 57, 295–303. [DOI] [PubMed] [Google Scholar]
- 7.Sia, R. A., Herald, H. A. & Lew, D. J. (1996) Mol. Biol. Cell 7, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sreenivasan, A. & Kellogg, D. (1999) Mol. Cell. Biol. 19, 7983–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulewitz, M. J., Inouye, C. J. & Thorner, J. (1999) Mol. Cell. Biol. 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sia, R. A., Bardes, E. S. & Lew, D. J. (1998) EMBO J. 17, 6678–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser, P., Sia, R. A. L., Bardes, E. G. S., Lew, D. J. & Reed, S. I. (1998) Genes Dev. 12, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMillan, J. N., Theesfeld, C. L., Harrison, J. C., Bardes, E. S. & Lew, D. J. (2002) Mol. Biol. Cell 13, 3560–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barral, Y., Parra, M., Bidlingmaier, S. & Snyder, M. (1999) Genes Dev. 13, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMillan, J. N., Longtine, M. S., Sia, R. A., Theesfeld, C. L., Bardes, E. S., Pringle, J. R. & Lew, D. J. (1999) Mol. Cell. Biol. 19, 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lew, D. J. (2003) Curr. Opin. Cell Biol. 15, 648–653. [DOI] [PubMed] [Google Scholar]
- 16.Ma, X.-J., Lu, Q. & Grunstein, M. (1996) Genes Dev. 10, 1327–1340. [DOI] [PubMed] [Google Scholar]
- 17.Hanrahan, J. & Snyder, M. (2003) Mol. Cell 12, 663–673. [DOI] [PubMed] [Google Scholar]
- 18.Theesfeld, C. L., Zyla, T. R. & Bardes, E. G. (2003) Mol. Biol. Cell 14, 3280–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longtine, M. S., Theesfeld, C. L., McMillan, J. N., Weaver, E., Pringle, J. R. & Lew, D. J. (2000) Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell, P. & Nurse, P. (1987) Cell 49, 569–576. [DOI] [PubMed] [Google Scholar]
- 21.Coleman, T. R., Tang, Z. & Dunphy, W. G. (1993) Cell 72, 919–929. [DOI] [PubMed] [Google Scholar]
- 22.Parker, L. L., Walter, S. A., Young, P. G. & Piwnica-Worms, H. (1993) Nature 363, 736–738. [DOI] [PubMed] [Google Scholar]
- 23.Cid, V. J., Shulewitz, M. J., McDonald, K. L. & Thorner, J. (2001) Mol. Biol. Cell 12, 1645–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell, D. A. & Sprague, G. F., Jr. (2001) Mol. Cell. Biol. 21, 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartholomew, C. R., Woo, S. H., Chung, Y. S., Jones, C. & Hardy, C. F. (2001) Mol. Cell. Biol. 21, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, C. J., Song, S., Lee, P. R., Shou, W., Deshaies, R. J. & Lee, K. S. (2003) Genetics 163, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, S., Grenfell, T. Z., Garfield, S., Erikson, R. L. & Lee, K. S. (2000) Mol. Cell. Biol. 20, 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy, C. F. & Pautz, A. (1996) Mol. Cell. Biol. 16, 6775–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirayama, M., Zachariae, W., Ciosk, R. & Nasmyth, K. (1998) EMBO J. 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaerer, F., Morgan, G., Winey, M. & Philippsen, P. (2001) Mol. Biol. Cell 12, 2519–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cvrckova, F., De Virgilio, C., Manser, E., Pringle, J. R. & Nasmyth, K. (1995) Genes Dev. 9, 1817–1830. [DOI] [PubMed] [Google Scholar]
- 32.Versele, M. & Thorner, J. (2004) J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]
- 33.Weiss, E. L., Bishop, A., Shokat, K. M. & Drubin, D. G. (2000) Nat. Cell Biol. 2, 677–685. [DOI] [PubMed] [Google Scholar]
- 34.Bishop, A. C., Shah, K., Liu, Y., Witucki, L., Kung, C. & Shokat, K. M. (1998) Curr. Biol. 8, 257–266. [DOI] [PubMed] [Google Scholar]
- 35.Shou, A., Sakamoto, K. M., Keener, J., Morimoto, K. W., Traverso, E. E., Azzam, R., Hoppe, G. J., Feldman, R. M., DeModena, J., Moazed, D., et al. (2001) Mol. Cell 8, 45–55. [DOI] [PubMed] [Google Scholar]
- 36.Shulewitz, M. J. (2000) Dissertation (Univ. of California, Berkeley).
- 37.Blom, N., Gammeltoft, S. & Brunak, S. (1999) J. Mol. Biol. 294, 1351–1361. [DOI] [PubMed] [Google Scholar]
- 38.Nakajima, H., Toyoshima-Morimoto, F., Taniguchi, E. & Nishida, E. (2003) J. Biol. Chem. 278, 25277–25280. [DOI] [PubMed] [Google Scholar]
- 39.Wu, C., Lytvyn, V., Thomas, D. Y. & Leberer, E. (1997) J. Biol. Chem. 272, 30623–30626. [DOI] [PubMed] [Google Scholar]
- 40.Thornton, B. & Toczyski, D. P. (2003) Nat. Cell Biol., in press. [DOI] [PubMed]
- 41.Ayad, N. G., Rankin, S., Murakami, M., Jebanathirajah, J., Gygi, S. & Kirschner, M. W. (2003) Cell 113, 101–113. [DOI] [PubMed] [Google Scholar]
- 42.Nash, P., Tang, X., Orlicky, S., Chen, S., Gertler, F. B., Mendenhall, M. D., Sicheri, F., Pawson, T. & Tyers, M. R. (2001) Nature 414, 514–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.