Abstract
Fungi must recognize plant-specific signals to initiate subsequent morphogenetic events such as filamentation that lead to infection. Here we show that the plant hormone indoleacetic acid (IAA) induces adhesion and filamentation of Saccharomyces cerevisiae. Genome expression profiling of cells treated with IAA identified Yap1, a fungal specific transcription factor, as a key mediator of this response. Strains lacking YAP1 (yap1-1) are hypersensitive to growth on IAA because they accumulate more IAA than can wild type. Members of a family of transporters the amino acid/auxin:proton symport permeases with homology to AUX1, a putative IAA transporter from plants, are up-regulated in the yap1-1 mutant. Deletion of any one of these transporters makes yap1-1 mutants more resistant to IAA by decreasing its uptake. The permease mutants are defective in IAA perception and filamentation. The ability of a fungus to perceive a plant hormone that causes it to differentiate into an invasive form has important implications for plant–pathogen interactions.
Infection of a plant by a fungus involves mutual chemical recognition between the host and the pathogen. Plants recognize and respond to compounds such as chitin, which are unique signatures of fungal and insect pathogens (1). Reciprocally, fungi recognize chemical cues that signal the presence of the plant host and induce the subsequent morphogenetic changes that enable invasion. Fungi alight on a plant in a cellular form either as a yeast cell or spore and, after sensing the plant, proliferate into an invasive filamentous form. For example, spores of Colletotrichum gloeosporioides elaborate an invasion structure (appressorium) in response to ethylene, a plant hormone that promotes fruit ripening (2). By homing in on the plant hormone, the fungus synchronizes its invasion with a nutritionally advantageous stage in plant development.
Indoleacetic acid (IAA) is a molecule that is synthesized by plants and a few microbes (3, 4). In plants, IAA plays a key role in both root and shoot development. The hormone moves from one part of the plant to another by a designated importer (AUX1) and efflux pumps (PIN1–7) (5, 6). In peas, wounding or application of IAA induces the expression of ACS2, an IAA-responsive gene, suggesting that IAA may be present at the site of a wound (7). The presence of IAA at the wound site would make it a distinctive attractant for fungi; however, there is limited evidence for such a role. One difficulty in establishing the connection between IAA and pathogenesis is the lack of an in planta assay for the concentration of IAA at the foci of infection.
In this study, we take a different approach and ask whether the fungus Saccharomyces cerevisiae is able to perceive the phytohormone IAA. Our results show that Saccharomyces distinguishes IAA from other closely related compounds. At high concentrations, IAA inhibits growth, whereas at lower concentrations, it induces filamentation and adhesion. These responses are mediated by a family of transporters and the fungal transcription factor Yap1.
Materials and Methods
Strains and Media. Growth and media conditions were those described (8, 9). The S. cerevisiae strains are isogenic to BY4742 [the S288c background that was used for the Yeast Genome Sequencing project and the deletion project (10)]. Strains in the S288c background have a flo8 mutation, which prevents pseudohyphal growth; therefore, pseudohyphal growth and microarray studies were performed by using cognate deletions made in the Σ1278b background. Both S288c and Σ1278b are sensitive to IAA. Deletion strains were either from the Euroscarf deletion library or were constructed by replacement of the relevant ORF with the G418 resistance marker (11). The Ustilago maydis strain, (a1 b1, wild-type), used in this study was provided by S. Gold (University of Georgia, Athens).
Compounds Tested. More than 50 compounds were tested for differential growth inhibition (for a complete list, see Data Set 1, which is published as supporting information on the PNAS web site). These compounds included plant hormones (such as salicylic acid, ethylene, cytokinin, gibberellic acid, and abscisic acid), several IAA-conjugated compounds, and other indoles (such as indole, indole-3-pyruvic acid, and indole-3-acetamide), mimics and inhibitors of IAA transport (such as 2,4-(dichlorophenoxyacetic) acid and 2,3,5-triiodobenzoic acid), weak acids (sorbic acid and benzoic acid), and agents known to arrest the growth of yeast (azetidine, canavanine, and ethionine).
IAA Uptake Assay. The whole-cell assay used yeast strains grown at 30°C to an OD600 of ≈0.4 in minimal media supplemented with required amino acids. To measure IAA uptake, cells were harvested by centrifugation at 1,000 × g and were resuspended in Mes buffer at pH 4.6 with 2% glucose (12). Duplicate samples were removed after the addition of cells to medium containing 3H-labeled IAA (Amersham Pharmacia, Piscataway, NJ) 220 μCi/mmol (1 Ci = 37 GBq). Cells were collected on glass-fiber filters and were washed extensively with buffer. The filters were placed in glass scintillation vials, were treated with 350 μl of 1 M NaOH for 10 min, and the solution was neutralized with 400 μl of 1 M acetic acid. Uptake of labeled nutrients into cells was determined by using 5 ml of liquid scintillation fluid. The radioactive material extracted from cells comigrates with pure IAA in thin-layer chromatography. Uptake of IAA is not due to nonspecific binding of IAA because only live cells are capable of its transport. For 2,4-dinitrophenol (DNP) or carbonylcyanide m-chlorophenylhydrazone (CCCP) treatment, cells were exposed to 1 mM DNP or 500 μM CCCP for 45 min before the uptake assay. Tryptophan uptake was measured in the same whole cell assay, except 20 μCi/mmol 3H-tryptophan was used instead of 3H-IAA. The yap1-1 and avt yap1-1 double mutants show the same level of tryptophan uptake as wild-type cells; therefore, the yap1-1 mutation does not lead to a general elevation in the uptake of indoles (Fig. 5, which is published as supporting information on the PNAS web site).
Protein Localization. GFP was placed at the C terminus of Avt3 and Avt4 by site-directed PCR. The resulting in-frame fusion proteins, Avt3::GFP and Avt4::GFP, were shown to be functional because they suppressed growth inhibition by IAA and restored IAA uptake in avt mutants. Strains were grown on minimal media and were visualized on a Zeiss LSM 510 confocal microscope.
Results
IAA Induces Invasive Growth in S. cerevisiae. A number of plant-specific compounds were tested for their ability to inhibit either the growth of yeast or to induce filamentation (see Data Set 1 for a complete list of compounds tested). Diploid yeast cells develop into a multicellular, pseudohyphal form (invasive filaments of elongated cells) under conditions of nitrogen starvation. Haploid cells adhere to the agar surface of rich media plates (haploid invasion). Of the many compounds tested, only IAA had effects in these assays. IAA enhances both haploid invasion and diploid pseudohyphal growth (Fig. 1 B and D). The enhancement of these phenotypes by IAA is optimal when xylose is the carbon source. The recognition of IAA by yeast is specific; other plant hormones and indoles closely related in structure to IAA fail to show induction of these phenotypes.
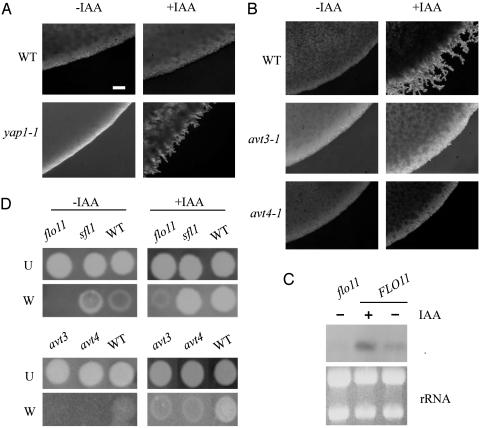
Fig. 1.
IAA enhances filamentation and surface adhesion. A and B show the edge of a yeast patch (Σ1278b) growing on filamentation-inducing media (8) with xylose as the carbon source. Plates were incubated in the dark to prevent photodegradation of IAA. For the purpose of comparing the images, A and B were taken at the same magnification. (Bar in A, 10 μm.) (A) The yap1-1 mutant filaments at low levels of IAA (2 μM), and wild type does not. (B) Wild-type filaments at higher concentrations of IAA (50 μM). IAA-enhanced filamentation is abolished in all seven avt mutants (avt3-1 and avt4-1 are shown). (C) Northern blot analysis shows that IAA induces expression of FLO11. (D) IAA enhances haploid invasive growth of S. cerevisiae. Cells were patched on media containing IAA (120 μM). U, unwashed; W, washed.
Induction of morphogenesis by IAA requires the function of Flo11, a cell-surface protein known to be required for the nutritional induction of both diploid filamentation and haploid invasive growth (13). Growth of cells in the presence of IAA leads to an increase in FLO11 mRNA (>3-fold, Fig. 1C), suggesting that IAA induces morphogenetic changes by elevating FLO11 transcription. Induction of adhesion and pseudohyphal growth by IAA is dramatically reduced by mutations that diminish FLO11 expression (flo11, ste20, ste11, and tec1; data not shown) and enhanced in mutants that have elevated levels of FLO11 (sfl1, Fig. 1D).
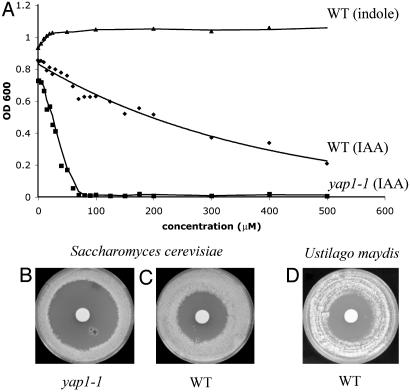
At higher concentrations, IAA, but not other indoles tested, inhibits the growth of yeast (Fig. 2A) both in liquid (EC50 = 250 μM) and on solid media (Fig. 2 B and C). IAA arrests growth in all stages of the cell cycle and, after removal from IAA, cells resume growth. Several distinct laboratory strains, including S288c and Σ1278b, were sensitive at approximately the same IAA concentrations. The inhibition by IAA is specific because other plant hormones and indolic compounds, even those structurally very similar to IAA, such as indole butyric acid and 5-hydroxy IAA, do not inhibit growth at comparable concentrations (see Data Set 1). IAA also inhibits the growth of the Ustilago maydis, a fungal pathogen of corn (Fig. 2D).
Fig. 2.
IAA growth inhibition in liquid. (A) Cells were treated with various concentrations of IAA or other compounds. The EC50 is the concentration of IAA at which growth is the half-maximum obtainable in the absence of the drug. Cells were inoculated into medium in 96-well plates. The OD at 600 nm was measured after 2 days of incubation. Each data point is the average of two samples that varied by <10%. The yap1-1 mutant is hypersensitive to IAA (EC50 = 25 μM) compared with wild type (EC50 = 250 μM). Indole and many other indolic compounds tested do not inhibit growth. (B–D) IAA growth inhibition on Petri plates. A filter disk saturated with IAA was placed on a lawn of cells. Plates were incubated for 3 days in the dark. The clear area indicates the zone of growth inhibition. The yap1-1 strain (B: yap1-1, C: wild type) is hypersensitive to IAA. Approximately 50 related compounds were tested by using this assay (see Data Set 1). Of these compounds, the yap1-1 mutant is only slightly sensitive to fluoro-IAA, indole pyruvate, and benzoic acid. (D) IAA-inhibition of Ustilago maydis growth.
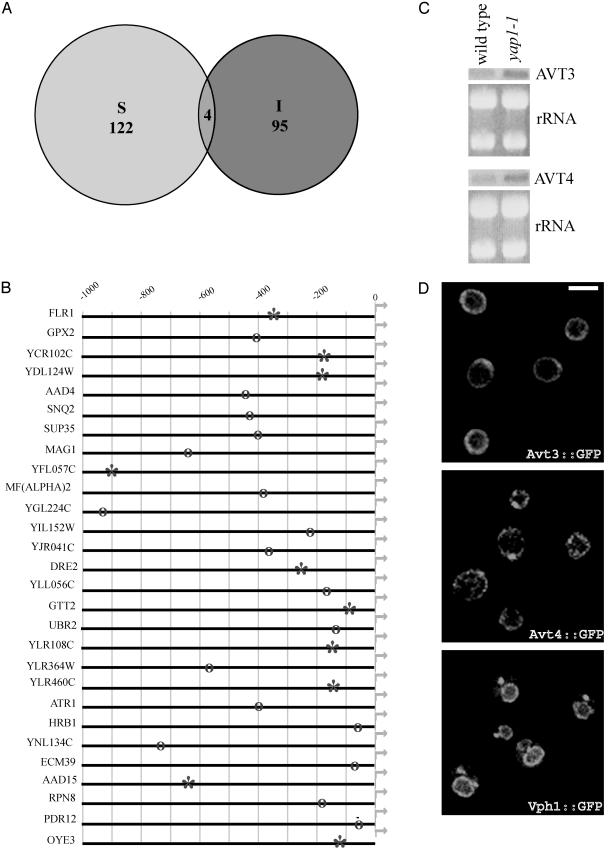
Transcription Profiling and Deletion Library Screen Implicate Yap1 in the IAA Response. Whole-genome arrays show that the transcription profile of cells growing in the presence of subtoxic levels of IAA is unique and distinct from those of cells exposed to many other diverse environmental stress conditions (Fig. 5). Of the 126 stress-induced genes identified in two independent studies (14, 15), only four are also induced after IAA exposure (Fig. 3A). Of the 99 genes whose transcripts were induced by IAA, 28 contain a consensus Yap1-binding site within 1 kb of the transcription start site (Fig. 3B). Of these genes, 10 have been shown previously to be regulated by Yap1 (asterisks, Fig. 3B, see the Saccharomyces Genome Database, which can be accessed at www.yeastgenome.org), whereas the remaining 18 genes have perfect matches to the Yap1-binding site, TTACTAA (open circles, Fig. 3B).
Fig. 3.
Analysis of IAA transcription profiles (see Fig. 5). (A) The Venn diagram compares genome-wide expression analysis under different stress conditions (S) with that of IAA treatment (I). The transcription profile of yeast treated with IAA is distinct from that of each of the published data sets of yeast genes induced in response to a large number of diverse environmental stress conditions (14, 15). Of the 126 genes induced in both data sets in response to different stress conditions, only four are also induced after IAA treatment. (B) Alignment of the upstream regulatory sequence of 99 genes significantly up-regulated after IAA exposure reveals a consensus Yap1-binding site in 28 of them. Promoter scanning of all genes in the yeast genome indicates that the Yap1-binding site is significantly enriched in this data set (P < 10–9). Asterisks represent previously documented Yap1-binding sites (Saccharomyces Genome Database at www.yeastgenome.org), and the open circles indicate perfect matches for the Yap1-binding site, TTACTAA. (C) Northern blot analysis shows that the yap1-1 strain expresses more AVT3 and AVT4 mRNA, compared with wild type; interestingly, AVT3 contains a consensus Yap1-binding site in its promoter region. (D) The pattern of localization of functional GFP fusions of Avt3 and Avt4 is different from Vph1, a known vacuolar membrane protein (24). When grown in standard minimal media (9), Avt3 and Avt4 proteins show a bias toward the periphery of the cell, as shown by immunofluorescence. The speckled appearance of Avt4 suggests that vesicles harboring Avt4 may dock at the cell periphery. (Bar, 5 μm.)
A screen of the complete yeast deletion library (10) for IAA-hypersensitive mutants identified yap1-1; no other mutant in the entire library is as IAA-sensitive as yap1-1 (EC50 = 20 μM, Fig. 2 A). In particular, mutations in paralogs of YAP1 (especially YAP2) are not hypersensitive. Furthermore, the yap1-1 mutant is induced to filament at concentrations of IAA (2 μM) that are >20-fold lower than those required to induce filamentation in wild type (50 μM) (compare Fig. 1 A and B). These data suggest that Yap1 is the key mediator of the response to IAA.
The Avt Proteins Are IAA Transporters. The specificity of IAA recognition for morphogenesis and growth inhibition suggests that yeast has transporters that recognize IAA. A search of the yeast genome for possible components of an IAA uptake system identified a family of seven genes [AVT1-7 (16)] annotated as the amino acid/auxin:proton symport permease (AAAP) family, which is part of a larger family that includes AUX1, an Arabidopsis gene implicated in IAA transport in plants (17, 18) and γ-aminobutyric acid transporters in Caenorhabditis elegans (19). Deletion of any one of the yeast AVT family members makes yap1-1 mutants more resistant to IAA (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that deletion of these putative transporters reduces uptake of this compound. Mutations in other permease genes (the general amino acid permease GAP1, the tryptophan permease TAT2, or the pleiotropic drug-resistant pump PDR12) do not alter the sensitivity of yap1-1 mutants to IAA (Fig. 4C).
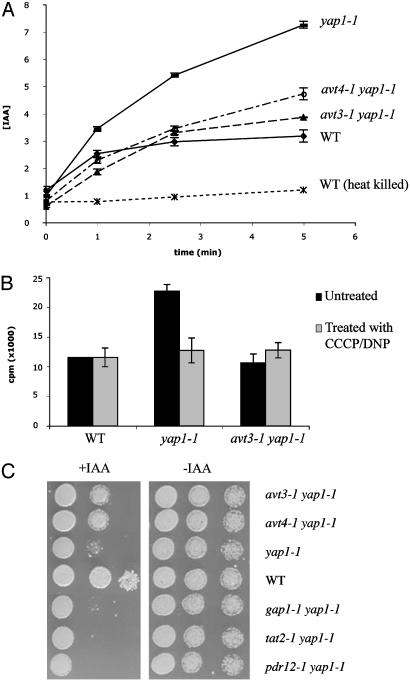
Fig. 4.
Uptake of 3H IAA into yeast cells. Cells grown to log phase in minimal media (9) were harvested and incubated in the presence of 3H-IAA for various lengths of time. Cells were filtered, and total counts retained on glass filters were monitored. (A) The yap1-1 strain accumulates more IAA than wild type; heat-killed (HK) cells show no IAA uptake. The avt3–1 yap1-1 and avt4-1 yap1-1 double mutants (dashed lines) show decreased IAA uptake. Other avt yap1-1 double mutants also reduce IAA uptake (Fig. 6A). IAA concentration is represented in μmol of IAA transported. (B) IAA accumulation depends on an intact membrane potential. Cells were treated with CCCP or DNP for 45 min at room temperature before IAA exposure. The bar graph compares IAA accumulation at 5 min with and without drug treatment. 3H-IAA transported is represented in cpm. (C) Deletion of any AVT gene makes yap1-1 mutants more resistant to IAA. Deletion of other transporters, such as the general amino acid permease (GAP1), the high-affinity tryptophan permease (TAT2), or the pleiotropic drug resistant pump (PDR12), does not alter the IAA sensitivity of the yap1-1 strain.
To determine whether the AVT genes mediate the transport of IAA, we designed an assay to measure the ability of intact yeast cells to take up IAA from the medium. In this assay, the yap1-1 mutant shows increased uptake of 3H-IAA, as compared with wild type (Fig. 4A). The yap1-1 mutation does not lead to a general elevation in the uptake of indoles (Fig. 6B). This enhanced uptake of IAA in the yap1-1 mutant is reduced to wild-type levels in the presence of DNP or CCCP, compounds that collapse the proton gradient (Fig. 4B). These data suggest that the uptake of IAA has two components, one of which requires an intact membrane potential.
The steady-state mRNA levels for AVT3 and AVT4 are elevated (>2 fold) in yap1-1 strains, as compared with wild type (Fig. 3C), which could account for the increased IAA uptake of yap1-1. As predicted by this model, the avt yap1-1 double mutant strains show a decreased rate of IAA uptake as compared with the yap1-1 mutants (Fig. 4A). It is the energy-dependent component of uptake that is missing in the avt yap1-1 strains (Fig. 4B). The reduced IAA uptake in the avt yap1-1 strains correlates with their reduced sensitivity to IAA. Consistent with their reduced uptake, the avt mutants fail to show the IAA induction of either haploid adherence (Fig. 1D) or diploid filamentation (Fig. 1B). Mutation in any one of the seven AVT genes abolishes the effects of IAA. This lack of redundancy suggests that the Avt proteins either function in the same pathway or are components of a multiprotein complex involved in some aspect of IAA transport into yeast.
The Avt Proteins Are Enriched at the Cell Periphery. Members of the Avt family in other organisms are membrane proteins that transport small molecules as diverse as γ-aminobutyric acid and amino acids (17–19). Arabidopsis orthologs of the Avt proteins have been shown to transport amino acids when expressed in S. cerevisiae (18). Previous studies in yeast have implicated some of the Avt proteins in the vacuolar transport of amino acids (16), but neither our analysis nor recent studies on the global protein localization (20) give evidence for vacuolar localization of the Avt proteins. Moreover, none of the amino acids thought to be transported by Avt proteins, can reverse the inhibition of IAA suggesting that these proteins are unlikely to transport amino acids across the plasma membrane. We find that the distribution of Avt3 and Avt4 tagged with GFP are enriched at the cell periphery (Fig. 3D). One possibility is that the intracellular distribution of the Avt proteins, like that of the Gap1 general amino acid permease, depends on environmental conditions; Gap1 shows vacuolar localization when amino acids are abundant in the growth media but localize to the plasma membrane under limiting conditions (21).
Discussion
IAA, a plant hormone, stimulates the adhesion and filamentation of yeast. Because both of these phenotypes are important for the invasion of a plant, this hormone is likely to play a role in the recognition of the plant host by fungi. Whether yeast is inhibited or stimulated by IAA depends on its concentration and the activity of Yap1, a fungal-specific transcription factor (22, 23). In the absence of Yap1, cells are more sensitive to both IAA inhibition and induction of filamentation (Fig. 1 A and D). At higher concentrations, IAA inhibits the growth of Saccharomyces and other fungi, suggesting that pathogens could use the plant defense system to cue in on the plant. These studies suggest that Yap1 and the genes it regulates could be important targets for antifungal agents.
Saccharomyces has a family of Avt proteins that mediate the uptake of IAA and its ability to inhibit growth and to stimulate morphogenesis. The yeast Avt proteins have some homology to AUX1, which is thought to mediate IAA import in Arabidopsis (5). The Arabidopsis AUX1 and Avt family of proteins are members of the larger AAAP family, which is present in most fungi that have been sequenced to date (www.broad.mit.edu). This conservation, coupled with the observation that IAA, inhibits the growth of plant fungal pathogens like Ustilago maydis (Fig. 2D) and Fusarium oxysporum (sp. var mathiolli, raphani, conglufenans; data not shown), suggests that these proteins may play a role in IAA transport in many pathogenic fungi.
Although these Avt proteins transport IAA in both fungi and plants, it is unlikely that they are devoted solely to IAA uptake. These proteins and other members of the larger AAAP family transport very diverse compounds such as amino acids and γ-aminobutyric acid (16, 19). Some of the Arabidopsis AAAP proteins appear to have a broad spectrum, capable of transporting several different amino acids (17, 18). However, the yeast Avt proteins cannot be plasma membrane amino acid transporters because, unlike their counterparts in plants, they fail to suppress the defects of yeast-specific amino acid permease mutants. Analysis of the pathway by which IAA is transported in yeast should reveal the role of the Avt proteins and the mechanism by which IAA induces these morphological transitions.
Supplementary Material
Acknowledgments
We thank the Keck Foundation for use of the Whitehead confocal microscope facility; Dr. F. Winston for his critical review of this work; Drs. A. Halme, A. Kays, and R. Wheeler for their thorough readings of the manuscript; other members of the Fink laboratory for fruitful discussions; and Drs. T. Ideker and G. Bell for consulting on computational analyses used in this study. G.R.F. is an American Cancer Society Professor of Molecular Genetics. This work was supported by National Science Foundation Grant MCB 9974451 (to G.R.F.) and a National Research Service Award Fellowship (to R.P.).
Abbreviations: IAA, indoleacetic acid; AAAP, amino acid/auxin:proton symport permeases; DNP, 2,4-dinitrophenol; CCCP, carbonylcyanide m-chlorophenylhydrazone.
References
- 1.Hammond-Kosack, K. & Jones, J. D. G. (2000) in Biochemistry and Molecular Biology of Plants, eds. Buchanan, B. B, Gruissem, W. R. & Jones, L. (Am. Soc. Plant. Physiol., Rockville, MD), Chap. 21.
- 2.Kolattukudy, P., Rogers, L., Li, D., Hwang, C. & Flaishman, M. (1995) Proc. Natl. Acad. Sci. USA 92, 4080–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basse, C. W., Lottspeich, F., Steglich, W. & Kahmann, R. (1996) Eur. J. Biochem. 242, 648–656. [DOI] [PubMed] [Google Scholar]
- 4.Yamada, T., Palm, C., Brooks, B. & Kosuge, T. (1985) Proc. Natl. Acad. Sci. USA 82, 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., Walker, A. R., Schulz, B. & Feldmann, K. A. (1996) Science 273, 948–950. [DOI] [PubMed] [Google Scholar]
- 6.Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J. & Kuhlemeier, C. (2003) Nature 426, 255–260. [DOI] [PubMed] [Google Scholar]
- 7.Peck, S. C. & Kende, H. (1998) Plant Mol. Biol. 38, 977–982. [DOI] [PubMed] [Google Scholar]
- 8.Gimeno, C., Ljungdahl, P., Styles, C. & Fink, G. (1992) Cell 68, 1077–1090. [DOI] [PubMed] [Google Scholar]
- 9.Sherman, F. (1991) in Methods in Enzymology, eds. Guthrie, C. & Fink, G. R. (Academic, San Diego), Chap. 1, Vol. 194.
- 10.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 11.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 12.Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T. & Masson, P. (1998) Proc. Natl. Acad. Sci. USA 95, 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagiano, M., van Dyk, D., Bauer, F. F., Lambrechts, M. G. & Pretorius, I. S. (1999) Mol. Microbiol. 31, 103–116. [DOI] [PubMed] [Google Scholar]
- 14.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S. & Young, R. A. (2001) Mol. Biol. Cell 12, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russnak, R., Konczal, D. & McIntire, S. L. (2001) J. Biol. Chem. 276, 23849–23857. [DOI] [PubMed] [Google Scholar]
- 17.Young, G., Jack, D., Smith, D. & Saier, M. J. (1999) Biochim. Biophys. Acta 1415, 306–322. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, W. N., Kwart, M., Hummel, S. & Frommer, W. B. (1995) J. Biol. Chem. 270, 16315–16320. [DOI] [PubMed] [Google Scholar]
- 19.McIntire, S. L., Reimer, R. J., Schuske, K., Edwards, R. H. & Jorgensen, E. M. (1997) Nature 389, 870–876. [DOI] [PubMed] [Google Scholar]
- 20.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- 21.Chen, E. J. & Kaiser, C. A. (2002) Proc. Natl. Acad. Sci. USA 99, 14837–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes, L., Rodrigues-Pousada, C. & Struhl, K. (1997) Mol. Cell. Biol. 17, 6982–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alarco, A. M. & Raymond, M. (1999) J. Bacteriol. 181, 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manolson, M. F., Proteau, D., Preston, R. A., Stenbit, A., Roberts, B. T., Hoyt, M. A., Preuss, D., Mulholland, J., Botstein, D. & Jones, E. W. (1992) J. Biol. Chem. 267, 14294–14303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.