Abstract
The Prnp gene encodes the cellular prion protein PrPC. Removal of its ORF does not result in pathological phenotypes, but deletions extending into the upstream intron result in cerebellar degeneration, possibly because of ectopic cis-activation of the Prnd locus that encodes the PrPC homologue Doppel (Dpl). To test this hypothesis, we removed Prnd from Prnpo/o mice by transallelic meiotic recombination. Balanced loxP-mediated ablation yielded mice lacking both PrPC and Dpl (Prno/o), which developed normally and showed unimpaired immune functions but suffered from male infertility. However, removal of the Prnd locus abolished cerebellar degeneration, proving that this phenotype is caused by Dpl upregulation. The absence of compound pathological phenotypes in Prno/o mice suggests the existence of alternative compensatory mechanisms. Alternatively, Dpl and PrPC may exert distinct functions despite having partly overlapping expression profiles.
Keywords: prion protein, Dpl, cerebellar degeneration, transallelic meiotic recombination
Transmissible spongiform encephalopathies are fatal neurodegenerative disorders (1). The only identified protein component of the infectious agent, or prion, is PrPSc, a modified form of the cellular prion protein PrPC. Conversion of PrPC to PrPSc is a central pathogenetic event (1); mice lacking PrPC are resistant to prion inoculation (2) and do not replicate prions (3).
Prnd encodes a 179-residue protein, termed Doppel (Dpl) (4), that shows ≈25% identity with the C-proximal domain of PrPC but lacks the octapeptide repeats and hydrophobic region of PrPC. Both PrPC and Dpl are glycosyl phosphatidyl inositol-anchored membrane proteins (5, 6). The structures of Dpl and PrPC are strikingly similar (4). Prnd is transcribed strongly in testis and heart, moderately in spleen and some other organs, and transiently in developing brain, but very weakly in adult brain (7). In contrast, Prnp is highly expressed in neurons (8).
Ablation of Prnd showed that Dpl is dispensable for prion pathogenesis (9). Although Prndneo/neo mice develop normally, males are infertile; Dpl-deficient sperms are unable to fertilize in vitro unless the zona pellucida of the ovum is partially dissected (10).
Ataxia and cerebellar Purkinje cell (PC) loss were described in Prnp-ablated Ngsk (11), Rcm0 (5), and Zürich II (ZH-II) (12) mice that overexpress Dpl but not in ZH-I (2) and Edbr (13) mice with unadulterated Dpl expression. Removal of the Prnp splice acceptor may provoke neurodegeneration by yielding transcriptional control of Prnd to the Prnp promoter and, hence, inappropriately up-regulating Dpl in brain (14, 15).
Dpl overexpression in brains of PrP-deficient mice supports the notion that ectopic Dpl, rather than loss of PrPC, causes neuronal degeneration in ataxic Prnp-deficient mice (16); onset of ataxia inversely correlates with Prnd transcription (12). Dpl-overexpressing mice were rescued from neuronal degeneration by introduction of a Prnp transgene; hence, absence of PrPC is necessary for Dpl to induce cell death.
To establish the role of Dpl in the neurodegenerative phenotype of mice with extensive Prnp deletions, we disrupted Prnd in the ataxic ZH-II mice by using transallelic targeted meiotic recombination (TAMERE) (17). Offspring carrying the double deletion exhibited no abnormalities, except for male sterility, and they did not develop ataxia, proving the causal role of Dpl overexpression in ZH-II mice. Dpl does not appear to compensate for the loss of any unrecognized function of PrP, as has been suggested (4, 18).
Materials and Methods
Meiotic Recombination. Genomic rearrangements within the Prn (Prnp and Prnd) locus were produced in vivo by TAMERE (17). ZH-II mice homozygous for a Prnp allele whose exon 3 was replaced by a loxP site (12) were crossed with hemizygous synaptonemal complex protein 1 (Sycp1)-Cre mice (19). The progeny were crossed with animals homozygous for a Prnd allele containing a neomycin resistance cassette flanked by two loxP sites (10) to generate “transloxer” male mice whose meiotic cells may undergo targeted recombination. Reciprocal chromosomal translocations gave rise to deletion of the locus comprised within the loxP sites. Transloxer males were mated with wild-type females, and PCR with tail DNA was used to screen the progeny. The breakpoint was sequenced with the same primers as used for PCR: D, 5′-TGCAGGTGACTTTCTGCATTCTGG-3′; and E, 5′-TGAGTCTTGGAGGGATCAGGAGG.
Western Blot and Northern Blot Analyses. Brain homogenates (20%, wt/vol) were prepared as described (20). Samples (50 μg for PrP and 100 μg for Dpl) were electrophoresed through 12% SDS/PAGE gels and processed as described (20). PrP was detected with mAb ICSM18 (21); Dpl was probed with polyclonal anti-Dpl rabbit antiserum (12). For Northern blotting, 20 μg of total RNA was processed as described (10) with probes labeled with [α-32P] dCTP by random priming. Probes were sequenced as follows: A (PrP exons 1–2), ATCAGCAGACCGATTCTGGGCGCTGCGTCGCATCGGTGGCAGGACTCCTGAGTATATTTCAGAACTGAACCATTTCAACCGAGCTGAAGCATTCTGCCTTCCTAGTGGTA; B (PrP ORF), CCAACCTCAAGCATGTGGCAGGGGCTGCGGCAGCTGGGGCAGTAGTGGGGGGCCTTGGTGGCTACATGCTGGGGAGCGCCATGAGCAGGCCCATGATCCATTTTGGCAAC; and C contains the entire Dpl ORF.
Viral Infection. Mice were infected i.v. with 2 × 106 plaque-forming units (pfu) of vesicular stomatitis virus (VSV)-Indiana and serum was collected at the time points indicated. Serial 2-fold dilutions of 40-fold prediluted sera were preincubated with an equal volume of medium containing 500 pfu/ml VSV for 90 min, and the remaining nonneutralized infectivity was detected on Vero cell monolayers. The highest serum dilution that reduced infectivity of virus by 50% was counted as the neutralizing Ab titer. Lymphocytic choriomeningitis virus (LCMV)-specific cytotoxic T lymphocytes were detected in blood by using gp33–41:H2Db-tetramers on day 13 after infection with 200 pfu of LCMV.WE. LCMV-specific IgG Abs were measured against recombinant LCMV nucleoprotein in an ELISA.
Histopathology. Brains were fixed in 4% paraformaldehyde, paraffin-embedded, and cut into 2-μm sections. Sections were stained with hematoxylin/eosin and for glial fibrillary acidic protein, as described (12).
Fluorescence-Activated Cell Sorting (FACS) Analysis. Two- and three-color FACS analyses were performed on a FACSCalibur cytometer (Becton Dickinson) as described (22). The following anti-mouse Abs were used (Pharmingen): peridinin-chlorophyll-a-protein (PerCP)-labeled anti-B220; FITC-labeled anti-CD21; phycoerythrin (PE)-labeled anti-CD23 and anti-CD8; FITC-labeled anti-CD4 and anti-AA4.1; PE-labeled anti-Ter119 and anti-CD11b; and FITC-labeled anti-Thy1.2.
Results
Generation of Prno/o Mutant Mice. The minimal SycP1 promoter (19) drives expression of Cre during the zygotene stage of meiotic prophase I. We performed sequential crosses of SycP1-Cre mice with ZH-II mice, whose Prnp gene is replaced by a single loxP site (Prnplox1) (12), and Prndneo/neo mice, carrying a neomycin-resistance cassette flanked by loxP sites in place of the Prnd coding exon (Prndneo) (10). This breeding scheme juxta-poses loxP sites encircling the Prnp and Prnd loci (Fig. 1A) during homologous chromosome pairing in the first meiotic division. Offspring were screened for the occurrence of a fused allele lacking both Prnp and Prnd (Prn°).
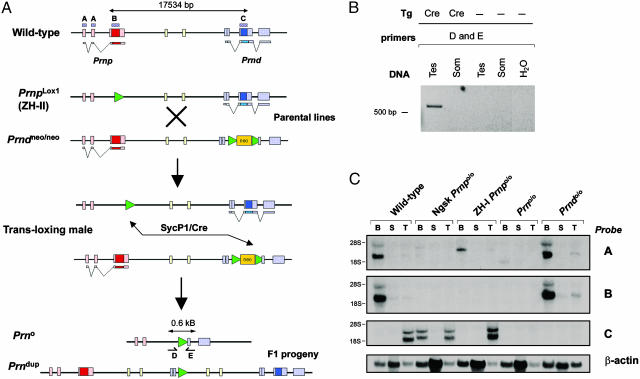
Fig. 1.
Recombination strategy. (A) Schematic representation of Cre-mediated TAMERE. In ZH-II mice, 0.26 kb of intron 2, the entire PrP ORF, and the 3′ noncoding region of exon 3 were replaced by a 34-bp loxP sequence. In Prndneo/neo mice, the Dpl ORF was replaced by a selection cassette flanked by loxP sequences. These lines were intercrossed to produce triple transgenic “transloxing” males containing the Prnplox1, Prndneo, and SycP1-Cre alleles (Fig. 1 A). After chromosome pairing and Cre expression in meiotic prophase, transallelic recombination generates two novel alleles. In the first allele (Prn°), the intergenic sequence is deleted. In the second allele (Prndup), the intergenic sequence between Prnp and Prnd is produced in the same orientation as in the original Prndup. Boxes in A–C show DNA probes used for Northern blot analysis. (B) PCR amplification of genomic DNA from testes of transloxing males, with or without the SycP1-Cre transgene. Primers D and E amplify a 0.6-kb fragment diagnostic of the deleted locus only in presence of Cre. No recombination was found in somatic transloxer tissues. (C) Northern blot analysis confirmed the absence of any transcripts in the Prno/o mice with probe B and C in different tissues, but a weak band was detected with probe A. Dup, duplication; Tes, testis; Som, somatic; B, brain; S, spleen; T, testis.
After transallelic meiotic recombination, we expected two novel alleles to segregate in haploid germ cells: one allele containing a duplication of the intergenic sequence and another allele lacking ≈16 kb encompassing Prnp and Prnd (Fig. 1 A). PCR primers D and E were designed to yield a 600-bp product after transallelic meiotic recombination; their annealing sites are separated by ≈18 kb in the wild-type locus. Genomic testis DNA from transloxing males was consistently positive for the fusion chromosome-specific PCR product (Fig. 1B). This band was not observed in control somatic liver DNA nor in testes lacking the SycP1-Cre transgene (Fig. 1B). Hence, transallelic recombination occurred in germ cells, but not in somatic cells, of transloxing males.
To obtain mice carrying fusion chromosomes with a deletion of the Prn locus, we mated transloxing males with wild-type females. We found that 1 of 482 pups (0.2%) carried the recombined allele, as judged by the appearance of the diagnostic 600-bp PCR product. Offspring were not screened for the duplicated allele. Sequencing of the floxed allele confirmed the absence of the Prnp and Prnd loci (data not shown).
Because the loss of Dpl affects male fertility, and because Cre may be genotoxic, we investigated whether the loss of both PrP and Dpl would affect spermatozoa. However, transmission of the Prnplox1, Prndneo, and SycP1-Cre alleles occurred in an undistorted Mendelian ratio, and sperms from transloxing males did not show any obvious abnormalities (data not shown).
We then performed detailed transcriptional analyses of various PrP and Dpl-related mutant mice (Fig. 1C). By using a 100-bp probe specific for Prnp exons 1 and 2 (probe A), a low-intensity 1.5-kb transcript was detectable in Prno/o brains, which may arise from intergenic splicing including Prnd exons 2b and 3, which were not removed by gene targeting. A larger Prnp transcript was present also in ZH-I brains, possibly resulting from a fused mRNA containing the cassette inserted into Prnp exon 3 (23). None of these transcripts were detected with a probe containing 100 bp of the Prnp ORF (probe B). No hybridization was detected in Prno/o and Prndo/o mice with a probe containing the Dpl ORF (probe C), but Dpl transcripts were slightly larger in ZH-I and Ngsk (11) than in wild-type testes. RT-PCR confirmed the absence of Prnp and Prnd mRNAs in homozygous mice (data not shown).
Male Prno/o Mice Develop Normally but Are Infertile. Male mice lacking Dpl are infertile (10). PrPC is also highly expressed in testis and is processed in mature sperms into a nonanchored C-terminally truncated peptide (24). However, Prnpo/o mice are fertile (23). Female Prno/o mice were fertile and yielded litter sizes similar to those of wild-type mice, whereas breeding of Prno/o males (n = 5) for >5 months did not yield any litters. Prn+/+ and Prn+/o male littermates yielded multiple litters. Histological analysis of various organs did not show gross abnormalities. Prno/o mice did not show any neurodevelopmental defects, yet their sperms had malformations similar to those of Dpl-deficient sperms (data not shown). Therefore, removal of PrP from Dpl-deficient mice did not induce any additional abnormalities and failed to accentuate the sperm phenotype. Hence, the absence of major defects in mice lacking PrPC is unlikely to result from functional compensation by Dpl.
The Immune System of Prno/o Mice. Prnd mRNA is transcribed in the spleen (7), and we detected splenic Dpl protein (Fig. 2G), suggesting a role for Dpl in lymphoid cells. However, immune functions are normal in Prndneo/neo mice (10). PrPC is also expressed in spleen (Fig. 2G) and was claimed to have functions in immune cells (25), including T cell activation (26).
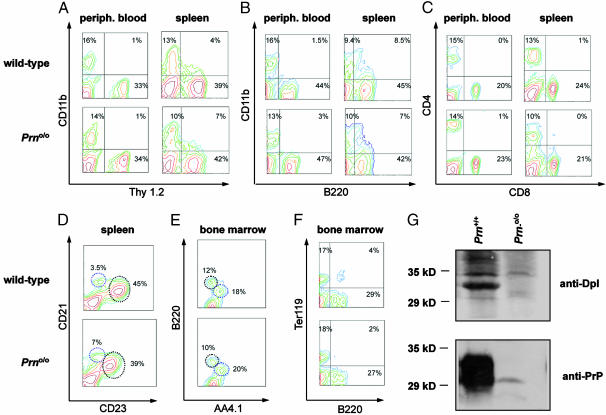
Fig. 2.
FACS analysis of immune cell subsets in Prno/o mice and wild-type littermates. Normal population of T cells (Thy1.2) (A), macrophages/monocytes (CD11b) (A and B), and B cells (B220) (B) in peripheral blood and spleen of wild-type and Prno/o mice, as indicated. (C) FACS analysis of peripheral blood and spleen revealed no alteration in CD4+ and CD8+ T cell subsets in mutant mice. (D) Similar density of B220+ splenic marginal-zone B cells (CD21high CD23neg-low) and B220+ follicular B cells (CD21med/low CD23high; circles). (E–F) Analysis of bone-marrow cells did not indicate alterations in the prevalence of AA4.1+ B220low immature B cells, AA4.1- B220high mature B cells, B220- AA4.1+ hematopoietic progenitor cells, and Ter119+ erythroblasts of Prno/o mice. Percentages of gated living cells are shown. (G) Western blot analysis of Dpl and PrPC in spleen.
If Dpl and PrP have overlapping immunological functions, Prno/o mice might exhibit immune defects. We, therefore, analyzed the prevalence of subsets of immune cells in Prno/o and littermate control mice (two 6- to 8-week-old mice for each group). B cells, T cells, and monocytes/macrophages were represented normally in peripheral blood and spleen (Fig. 2 A and B). CD4+ and CD8+ T cell subsets in spleen and peripheral blood were indistinguishable from those of controls (Fig. 2C). Spleen cell suspensions showed no abnormalities in splenic CD21highCD23neg-low marginal-zone B cells and CD21med/lowCD23high follicular B cells (Fig. 2D). There was no detectable alteration in the prevalence of immature B cells, hematopoietic progenitor cells, or erythroblasts in the bone marrow of Prno/o mice (Fig. 2 E and F).
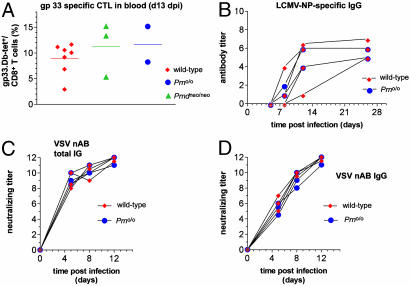
To test the functionality of the immune system as a whole, at least two 6–8 week-old Prno/o and control mice were infected with LCMV or VSV. Viability of cytotoxic CD8+ T cell immune responses was tested by immunizing wild-type, Prndneo/neo, and Prno/o mice by i.v. injection of 200 pfu of LCMV. LCMV-specific cytotoxic T lymphocytes were identified in blood by staining with gp33–41:H2Dbtetramers 13 days later. Controls and mutant mice yielded a similar high LCMV-specific responses (Fig. 3A). The LCMV-binding Ab response was similar in controls and mutant mice (Fig. 3B). The capability of mice to mount CD4+ T helper-dependent humoral immune responses was tested in the VSV-infection paradigm. On i.v. immunization of 2 × 106 pfu of VSV, wild-type and Prno/o mice did not show any significant difference in the production of neutralizing total Igs (Fig. 3C) or IgG (Fig. 3D). Therefore, loss of PrP and Dpl does not impair the efficacy of these antiviral responses.
Fig. 3.
Susceptibility of control and Prno/o mice to LCMV and VSV. (A) To assess CD8+ T cell immune responses, control mice, Prndneo/neo, and Prno/o mice were immunized by i.v. injection of 200 pfu of LCMV. LCMV-specific cytotoxic T lymphocytes (CTL) were assessed in blood by using gp33–41:H2Db-tetramers 13 days later. Controls and mutants mice yielded similar high LCMV-specific responses. (B) LCMV-neutralizing Ab responses were similar in control and mutant mice. (C and D) On i.v. immunization of 2 × 106 pfu of VSV, controls and Prno/o mice mounted a similarly strong production of neutralizing total Igs (C) or IgG (D).
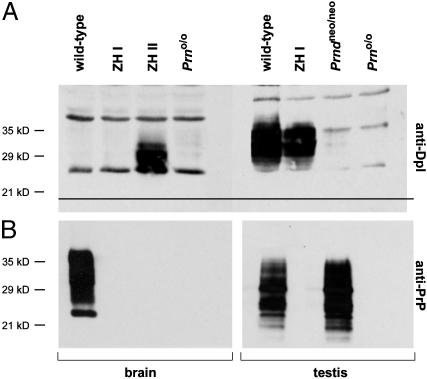
Disruption of Dpl Abrogates Cerebellar Degeneration of ZH-II Mice. At ≈6 months of age, Dpl-overexpressing ZH-II mice develop progressive tremor and unsteady gait associated with PC loss (12). This phenotype is 100% penetrant at 13 months of age. However, ZH-I mice show no ataxia or PC loss at up to at least 72 weeks (ref. 12; see Fig. 5). We, therefore, studied the cerebellar morphology of Prno/o mice. Dpl was detectable by Western blotting in ZH-II brains but not in wild-type brains, whereas removal of both Prnd alleles from ZH-II mice abrogated ectopic brain expression of Dpl (Fig. 4A). Similarly, PrP expression was absent in the brain of Prno/o mice (Fig. 4B). Testicular Western blots failed to reveal any expression of PrPC and of Dpl in testis of Prno/o mice (Fig. 4 A and B), whereas wild-type mice exhibited strong signals for both proteins. The electrophoretic mobility of testicular Dpl (≈35 kDa) was slightly slower than that of brain Dpl (≈30 kDa) possibly because of differential glycosylation.
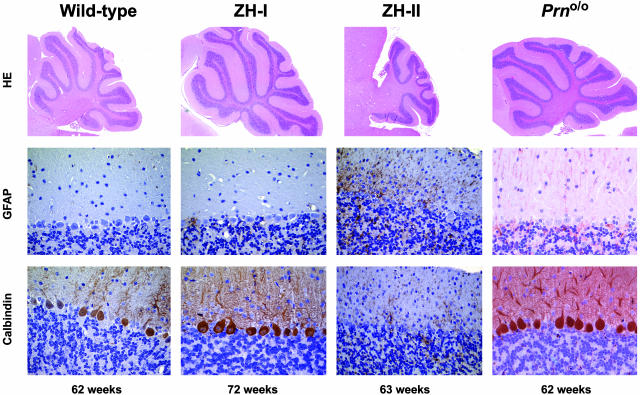
Fig. 5.
Cerebellar histopathology of Prnp and Prnd mutant mice. (Top) Parasagittal sections of wild-type, ZH-I, ZH-II, and Prno/o mice stained with hematoxylin/eosin. (Middle) Glial fibrillary acidic protein (GFAP) staining of cerebellar cortex with gliosis in ZH-II mice, but not in wild-type, ZH-I, or Prno/o mice. (Bottom) Calbindin stains (specific for PC). Wild-type, ZH-I, and Prno/o mice show an intact PC layer, whereas ZH-II mice undergo extensive cell loss.
Fig. 4.
Dpl and PrP protein levels in testes and brains of wild-type, Prndneo/neo, ZH-I, ZH-II, and Prno/o mice. (A) Western blot analysis was performed on tissue homogenates with rabbit anti-Dpl polyclonal Ab. Overexpression of Dpl in the brain of ZH-II mice is abrogated in Prno/o.(B) As expected, Western blot analysis did not reveal expression of PrPC in brain and testis of Prno/o mice. Detection was done by using anti-PrP ICSM18 Ab.
Aging Prno/o mice were scored clinically, and none exhibited tremor or unsteady gait (>62 weeks; n = 5). At 62 weeks, Prno/o mice showed intact PC layers and normally sized cerebella (Fig. 5), whereas ZH-II mice exhibited significant PC loss (12). Glial fibrillary acidic protein staining of the cerebellar cortex revealed strong astrocytosis in ZH-II mice but not in Prno/o, wild-type, or ZH-I mice (Fig. 5). These results prove that Dpl overexpression, rather than loss of Prnp or of additional cis-acting elements, causes ataxia and PC loss in ZH-II mice.
Discussion
The Prnp gene is conserved in mammals (27), and paralogues are present in turtle (28), fish (29, 30), and amphibians (31), suggesting an important function of PrPC. No natural Prnp null alleles were described despite negative evolutionary pressure (32). Mild phenotypes were found in Prnp knockout mice (23), but no molecular explanations for these observed phenotypes have come forward (18). Identifying the function of PrPC may be crucial to understanding the basis for neurodegeneration in prion diseases (33).
Dpl-deficient mice develop normally yet suffer from male infertility (10). Structural similarities and overlapping expression patterns in certain tissues suggest that Dpl may compensate for the loss of PrPC, and vice versa. Yet, neurotoxicity related to overexpression of Dpl can be counteracted by PrPC, suggesting that Dpl and PrPC may feed antagonistically into a common pathway.
We addressed the functional relationship between PrPC and Dpl by sequentially deleting Prnp and Prnd in mice. These two genes are separated by only 16 kb; hence, double-knockout mice could not be generated by Mendelian crosses. We, therefore, used TAMERE, which has been used to modify Hox gene clusters (17). The generation of Prno/o mice confirms that TAMERE is not restricted to specific loci and that it can be used broadly for producing complex “designer chromosomes.”
The low frequency of TAMERE in our experiments (0.2% deletion events) contrasts with ref. 17 (10% deletion and duplication events). Recombination frequency can vary 1–10% (34). There does not appear to be a clear-cut correlation between the genomic distance of the loxP sites and the frequency of recombination (M. Kmita and D. Duboule, personal communication). For Prn, the distance between the two loxP sites was 17,534 bp, whereas Hox-cluster deletions encompassed 0.5–22 kb.
The normal development of Prno/o mice suggests that Dpl does not transcomplement PrPC during brain development. Prno/o males suffered from infertility similarly to Prndneo/neo mice, indicating that removal of PrPC does not rescue sperm dysfunction. Although it is surprising in view of the structural homology between PrPC and Dpl (35), this finding may be due to the lack of functional overlap between PrPC and Dpl. Alternatively, further unknown homologues, which may have retained a structure similar to PrPC and Dpl despite sequence diversification (36), may provide redundancy.
Although PrPC and Dpl are expressed in lymphoid organs, Prno/o mice had normal hematopoiesis and lymphopoiesis, and they mounted normal cytotoxic and humoral immune responses against LCMV and VSV. This finding is exciting in view of the resiliency to immunization of certain C-proximal domains of both proteins. Dpl may cross-tolerize against PrPC because PrPC expression in lymphoid organs is extremely tolerogenic and the C termini of PrPC and Dpl are structured similarly. Because Prno/o mice appear to be fully immunocompetent, they may be useful for generation of Abs to novel PrP epitopes.
Dpl is overexpressed in ZH-II brains by means of intergenic splicing events that place Prnd under the control of the Prnp promoter. ZH-II mice develop ataxia and PC degeneration. Reintroduction of a single allele of Prnp abrogates degeneration. Several lines of evidence suggested that overexpression of Dpl was responsible for this neurodegenerative phenotype in the brain of ZH-II mice. (i) Dpl is highly expressed in brain of ataxic ZH-II and Ngsk mice but not in brains of ZH-I mice that never become ataxic (16); (ii) the onset of ataxia correlates inversely with Dpl levels in brains of PrPC-deficient mice (12); and (iii) targeted expression of a Dpl transgene to PC induces ataxia in Prnpo/o mice but not in Prnp+/+ mice (37).
Although the above data suggest a causal role for Dpl in neurodegeneration of Prnpo/o mice, deletion of the Prnd locus in neurodegeneration-prone ZH-II mice provides formal proof that Prnd expression, rather than loss of PrP or other cis-acting regions, causes the development of neurological syndromes and PC degeneration.
Overexpression of N-terminally truncated PrPC transgenes (ΔPrP) causes degeneration of cerebellar granule-cell layers in ZH-I Prnpo/o mice. However, reintroduction of PrPC prevents disease (38). Because the truncation deprives PrPC of regions that are absent also from Dpl, ΔPrP and Dpl may cause disease by means of similar mechanisms. The present results confirm that Dpl and PrPC feed antagonistically into a common cerebellar pathway, possibly because they bind to a common receptor (provisionally termed “LPrP” in ref. 39) or because they form heteromultimers whose neurotoxicity depends on their PrP/Dpl stoichiometry (18).
Conversion of PrPC to PrPSc may bring about a toxic alteration of its normal function, which may be ultimately responsible for prion pathology. The toxicity induced by Dpl and ΔPrP, as well as its specific suppression by PrPC, represents the only possibility currently available for understanding the function of PrPC in vivo. The Prno/o mice described here may help to elucidate the structure–function relationships of PrPC domains, which, in turn, may contribute to the understanding of prion diseases.
Acknowledgments
We thank M. Rassoulzadegan for SycP1-Cre mice and H. Naumann for technical help. This work was supported by grants from the Bundesamt für Bildung und Wissenschaft, the Swiss National Foundation, and the National Center of Competence in Research on Neural Plasticity and Repair (to A.A.). N.G. is a Koetser Foundation fellow, and G.M. is a Biotechnology and Biological Sciences Research Council Fellow.
Abbreviations: Dpl, Doppel; PC, Purkinje cell; ZH-I/II, Zürich I and II; TAMERE, targeted meiotic recombination; pfu, plaque-forming units; VSV, vesicular stomatitis virus; LCMV, lymphocytic choriomeningitis virus; Sycp1, synaptonemal complex protein 1; FACS, fluorescence-activated cell sorting; PE, phycoerythrin.
References
- 1.Aguzzi, A., Montrasio, F. & Kaeser, P. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 118-126. [DOI] [PubMed] [Google Scholar]
- 2.Büeler, H. R., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339-1347. [DOI] [PubMed] [Google Scholar]
- 3.Sailer, A., Büeler, H., Fischer, M., Aguzzi, A. & Weissmann, C. (1994) Cell 77, 967-968. [DOI] [PubMed] [Google Scholar]
- 4.Mo, H., Moore, R. C., Cohen, F. E., Westaway, D., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (2001) Proc. Natl. Acad. Sci. USA 98, 2352-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman, G. L., Qin, K., Moore, R. C., Yang, Y., Mastrangelo, P., Tremblay, P., Prusiner, S. B., Cohen, F. E. & Westaway, D. (2000) J. Biol. Chem. 275, 26834-26841. [DOI] [PubMed] [Google Scholar]
- 6.Lu, K., Wang, W., Xie, Z., Wong, B. S., Li, R., Petersen, R. B., Sy, M. S. & Chen, S. G. (2000) Biochemistry 39, 13575-13583. [DOI] [PubMed] [Google Scholar]
- 7.Li, A., Sakaguchi, S., Shigematsu, K., Atarashi, R., Roy, B. C., Nakaoke, R., Arima, K., Okimura, N., Kopacek, J. & Katamine, S. (2000) Am. J. Pathol. 157, 1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moya, K. L., Sales, N., Hassig, R., Creminon, C., Grassi, J. & Di Giamberardino, L. (2000) Microsc. Res. Tech. 50, 58-65. [DOI] [PubMed] [Google Scholar]
- 9.Behrens, A., Brandner, S., Genoud, N. & Aguzzi, A. (2001) EMBO Rep. 2, 347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behrens, A., Genoud, N., Naumann, H., Rulicke, T., Janett, F., Heppner, F. L., Ledermann, B. & Aguzzi, A. (2002) EMBO J. 21, 3652-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakaguchi, S., Katamine, S., Nishida, N., Moriuchi, R., Shigematzu, K., Sugimoto, T., Nakatni, A., Kataoka, Y., Houtani, H., Shirabe, S., et al. (1996) Nature 380, 528-531. [DOI] [PubMed] [Google Scholar]
- 12.Rossi, D., Cozzio, A., Flechsig, E., Klein, M. A., Aguzzi, A. & Weissmann, C. (2001) EMBO J. 20, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manson, J. C., Clarke, A. R., Hooper, M. L., Aitchison, L., McConnell, I. & Hope, J. (1994) Mol. Neurobiol. 8, 121-127. [DOI] [PubMed] [Google Scholar]
- 14.Moore, R. C., Lee, I. Y., Silverman, G. L., Harrison, P. M., Strome, R., Heinrich, C., Karunaratne, A., Pasternak, S. H., Chishti, M. A., Liang, Y., et al. (1999) J. Mol. Biol. 292, 797-817. [DOI] [PubMed] [Google Scholar]
- 15.Weissmann, C. & Aguzzi, A. (1999) Science 286, 914-915. [DOI] [PubMed] [Google Scholar]
- 16.Moore, R. C., Mastrangelo, P., Bouzamondo, E., Heinrich, C., Legname, G., Prusiner, S. B., Hood, L., Westaway, D., DeArmond, S. J. & Tremblay, P. (2001) Proc. Natl. Acad. Sci. USA 98, 15288-15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herault, Y., Rassoulzadegan, M., Cuzin, F. & Duboule, D. (1998) Nat. Genet. 20, 381-384. [DOI] [PubMed] [Google Scholar]
- 18.Behrens, A. & Aguzzi, A. (2002) Trends Neurosci. 25, 150-154. [DOI] [PubMed] [Google Scholar]
- 19.Vidal, F., Sage, J., Cuzin, F. & Rassoulzadegan, M. (1998) Mol. Reprod. Dev. 51, 274-280. [DOI] [PubMed] [Google Scholar]
- 20.Meier, P., Genoud, N., Prinz, M., Maissen, M., Rulicke, T., Zurbriggen, A., Raeber, A. J. & Aguzzi, A. (2003) Cell 113, 49-60. [DOI] [PubMed] [Google Scholar]
- 21.White, A. R., Enever, P., Tayebi, M., Mushens, R., Linehan, J., Brandner, S., Anstee, D., Collinge, J. & Hawke, S. (2003) Nature 422, 80-83. [DOI] [PubMed] [Google Scholar]
- 22.Heppner, F. L., Musahl, C., Arrighi, I., Klein, M. A., Rulicke, T., Oesch, B., Zinkernagel, R. M., Kalinke, U. & Aguzzi, A. (2001) Science 294, 178-182. [DOI] [PubMed] [Google Scholar]
- 23.Büeler, H. R., Fischer, M., Lang, Y., Bluethmann, H., Lipp, H. P., DeArmond, S. J., Prusiner, S. B., Aguet, M. & Weissmann, C. (1992) Nature 356, 577-582. [DOI] [PubMed] [Google Scholar]
- 24.Shaked, Y., Rosenmann, H., Talmor, G. & Gabizon, R. (1999) J. Biol. Chem. 274, 32153-32158. [DOI] [PubMed] [Google Scholar]
- 25.Cashman, N. R., Loertscher, R., Nalbantoglu, J., Shaw, I., Kascsak, R. J., Bolton, D. C. & Bendheim, P. E. (1990) Cell 61, 185-192. [DOI] [PubMed] [Google Scholar]
- 26.Mabbott, N. A., Brown, K. L., Manson, J. & Bruce, M. E. (1997) Immunology 92, 161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatzl, H. M., Da Costa, M., Taylor, L., Cohen, F. E. & Prusiner, S. B. (1995) J. Mol. Biol. 245, 362-374. [DOI] [PubMed] [Google Scholar]
- 28.Simonic, T., Duga, S., Strumbo, B., Asselta, R., Ceciliani, F. & Ronchi, S. (2000) FEBS Lett. 469, 33-38. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, T., Kurokawa, T., Hashimoto, H. & Sugiyama, M. (2002) Biochem. Biophys. Res. Commun. 294, 912-917. [DOI] [PubMed] [Google Scholar]
- 30.Rivera-Milla, E., Stuermer, C. A. & Malaga-Trillo, E. (2003) Trends Genet. 19, 72-75. [DOI] [PubMed] [Google Scholar]
- 31.Strumbo, B., Ronchi, S., Bolis, L. C. & Simonic, T. (2001) FEBS Lett. 508, 170-174. [DOI] [PubMed] [Google Scholar]
- 32.Mead, S., Stumpf, M. P., Whitfield, J., Beck, J. A., Poulter, M., Campbell, T., Uphill, J. B., Goldstein, D., Alpers, M., Fisher, E. M. & Collinge, J. (2003) Science 300, 640-643. [DOI] [PubMed] [Google Scholar]
- 33.Aguzzi, A. & Heikenwalder, M. (2003) Nature 423, 127-129. [DOI] [PubMed] [Google Scholar]
- 34.Kmita, M., Fraudeau, N., Herault, Y. & Duboule, D. (2002) Nature 420, 145-150. [DOI] [PubMed] [Google Scholar]
- 35.Luhrs, T., Riek, R., Guntert, P. & Wuthrich, K. (2003) J. Mol. Biol. 326, 1549-1557. [DOI] [PubMed] [Google Scholar]
- 36.Premzl, M., Sangiorgio, L., Strumbo, B., Marshall Graves, J. A., Simonic, T. & Gready, J. E. (2003) Gene 314, 89-102. [DOI] [PubMed] [Google Scholar]
- 37.Flechsig, E., Hegyi, I., Leimeroth, R., Zuniga, A., Rossi, D., Cozzio, A., Schwarz, P., Rulicke, T., Gotz, J., Aguzzi, A. & Weissmann, C. (2003) EMBO J. 22, 3095-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shmerling, D., Hegyi, I., Fischer, M., Blattler, T., Brandner, S., Gotz, J., Rulicke, T., Flechsig, E., Cozzio, A., von Mering, C., et al. (1998) Cell 93, 203-214. [DOI] [PubMed] [Google Scholar]