Abstract
The notorious resistance of melanoma cells to drug treatment can be overcome by expression of a 50-aa peptide derived from activating transcription factor 2 (ATF250–100). Here we demonstrate that ATF250–100 induced apoptosis by sequestering ATF2 to the cytoplasm, thereby inhibiting its transcriptional activities. Furthermore, ATF250–100 binds to c-Jun N-terminal kinase (JNK) and increases its activity. Mutation within ATF250–100 that impairs association with JNK and the inhibition of JNK or c-Jun expression by RNA interference (RNAi) reduces the degree of ATF250–100-induced apoptosis. In contrast, TAM67, a dominant negative of the Jun family of transcription factors, or JunD RNAi attenuates sensitization of melanoma cells expressing ATF250–100 to apoptosis after treatment with anisomycin, which is used as a model drug. Mutations within the JNK binding region of ATF250–100 or expression of TAM67 or JunD RNAi attenuates inhibition of melanoma's tumorigenicity by ATF250–100. We conclude that inhibition of ATF2 in concert with increased JNK/Jun and JunD activities is central for the sensitization of melanoma cells to apoptosis and inhibition of their tumorigenicity.
Response of melanomas to treatment and inhibition of their notorious metastatic potential are key for effective therapy (1). Common to advanced melanomas are impairments of their death cascade pathways (2, 3). Such impairments include down-regulation of FAS receptor through transcriptional silencing (4) or aberrant trafficking (5), poor expression of apoptotic protease activating factor 1 (Apaf1; ref. 6), expression of decoy tumor necrosis factorrelated apoptosis-inducing ligand receptors (TRAIL; ref. 7), or overexpression of tumor necrosis factor receptor-associated factor 2 (TRAF2; ref. 8). Over 60% of melanomas contain activating mutations within B-Raf (9), whereas 15% harbor N-Ras mutation, resulting in constitutive activation of the mitogen-activated protein kinase (MAPK) pathway (10). MAPK has been implicated in transcriptional activation of cAMP-response-element binding protein (11), c-Fos (12) and the melanocyte transcription factor microphthalmia (13), which affects melanoma cell pigmentation and viability (14). Furthermore, MAPK/extracellular response kinase has also been implicated in the activation of matrix metalloproteinase-9 (15), β-3 integrins (16), and Janus kinase (JAK)/signal transducers and activators of transcription (17).
Activating transcription factor 2 (ATF2) is phosphorylated by the stress kinases and MAPK/extracellular response kinase and is involved in the cellular response to stress (18–19). Transcriptionally active ATF2 binds to ATF/cAMP response element target sequences as a heterodimer with c-Jun (20) and activates a large set of genes including tumor necrosis factor α, transforming growth factor β, IL-6, IFN-γ, IFN-β, cyclin A, and E-selectin (21–24).
Our earlier studies indicated that ATF2 has a key role in melanoma's acquisition of resistance to apoptosis after treatment with various DNA-damaging agents (24, 25). Interference with ATF2 function via the use of dominant negative forms, or by overexpression of a short ATF2-derived peptide (ATF250–100), resulted in the sensitization of this resistant tumor to a variety of chemical and physical treatments (26, 27). Expression of the ATF250–100 inhibits endogenous ATF2 and activates c-Jun, resulting in inhibition of melanoma tumorigenicity and metastatic potential in vivo (27). Important support for the role of ATF2 in melanoma progression comes from tissue array analysis, showing a significant correlation between nuclear localization of ATF2 in melanoma tumors and poor prognosis (28).
Materials and Methods
Cell Culture and Derivation of Stable Cell Line. The mouse melanoma cell line SW1 and 293T human embryo kidney cells were maintained as described in ref. 27.
Constructs. ATF250–100 was cloned in frame into an hemagglutinin (HA)-penetratin pcDNA3 vector as described (26). The 5×Jun2-Luc and 5×12-o-tetradecanoylphorbol 13-acetate response element (TRE)-Luc constructs are described elsewhere (27). Substituting mutations in ATF250–100 amino acids MT (to AA at positions 51 and 52), RNDS (to AAAA at amino acids 59–62), or VIVA (to RRRR at amino acids 63–66) were generated by oligonucleotidedirected mutagenesis (QuikChange, Stratagene). Wild-type and mutant forms of ATF250–100 were cloned into BamHI/NotIsitesof pGEX-4T vector, resulting in GST fusion constructs. Cloning of JNK into the BamHI/NotI sites on the pEF-Flag generated pEF-Flag JNK and its mutant counterpart pEF-Flag JNK-APF. The production of ATP-pocket mutant pcDNA HA-JNK-aS3; JNK-DLD was described in ref. 29. DNA sequencing combined with Western blotting confirmed the integrity and expression of each construct. Transfections were carried out by using Lipofectamine Plus (Invitrogen), and stably expressing clones were selected as described in ref. 27.
RNAi in SW1 Cells. Nineteen-mer oligonucleotides based on sequences derived from the corresponding target transcripts were used to mediate suppression of gene transcription through RNAi as follows: murine JNK1/2 (accession nos. AB005663 and AAD22579, respectively) within nucleotides 379–397 AGAATGTCCTACCTTCTCT, murine c-Jun (accession no. BC002081) within nucleotides 983-1002 CGCAGCAGTTGCAAACGTT, and murine JunD (accession no. X15358) within nucleotides 984-1003 GTCCTCAGCCACGTCAACA. Each of these oligos was cloned into the BglII/HindIII sites of the pRETRO-SUPER (pRS) vector (30).
Protein Half-Life Measurements. SW1 cells that stably express empty vector (SW1 pcDNA) or pcDNA-ATF250–100 (SW1-ATF250–100) were treated with cycloheximide (60 mg/ml) for the indicated time periods.
Transcriptional Analysis. Reporter constructs and luciferase assays were carried out as described in ref. 27.
Apoptosis. Cells were treated with 10 μg/ml anisomycin for 36 h, and apoptosis was measured as described in ref. 4.
Retroviral Infection. Packaging of retroviral constructs (pRS containing RNAi oligos) was carried out in human 293T cells as described in ref. 31. SW1 cells (1.5 × 105) were infected for 12 h in the presence of 4 μg/ml polybrene.
In Vitro Protein-Binding Assay. 293T cells transfected with the respective JNK plasmids were lysed in binding buffer (20 mM Tris, pH7.5/150 mM NaCl/1 mM EDTA/1 mM EGTA/0.5% Nonidet P-40/1 mM NaVO4/1 mM DTT supplemented with protease inhibitors) and then incubated (500 μg per assay) in bacterially expressed and purified GST-ATF2 wild-type and mutant forms bound to glutathione beads for 2 h at 4°C. The glutathione beads were blocked with BSA (3 mg/ml in PBS) for 2 h before use. Bead-bound material was then washed (three times with 0.5 M LiCl), eluted, and separated on SDS/PAGE, followed by immunoblotting.
Western Blot and Immunohistochemistry Analysis. Cell lysates (50–150 μg) were resolved on 10% SDS/PAGE, transferred to nitrocellulose, and blotted according to standard protocols. The antibodies used were polyclonal anti-ATF2 directed to the C-terminal peptide (C-19) and anti c-Jun (SC-45) (both from Santa Cruz Biotechnology), anti-HA (Babco, San Francisco), anti-JNK (Pharmingen), and anti-JunD (Affinity BioReagents, Golden, CO). Antibody–antigen complexes were detected with the ECL system (Amersham Pharmacia). Membranes were also scanned with Bio-Rad PhosphorImager using bioanalyst software for quantification. When applicable, the x-ray films were scanned and relative band intensity was determined with a GS-800 calibrated densitometer (Bio-Rad). Immunohistochemistry analysis and confocal microscopy were carried out as described in refs. 27 and 32.
Tumorigenesis. SW1 cells expressing wild-type or mutant forms of the ATF2 peptide alone or in combination with TAM67, JunD RNAi, c-Jun RNAi, or control pRS were trypsinized, resuspended in PBS, and injected s.c. into 6- to 7-wk-old mice in the lower flank. Tumor growth was monitored as described (27).
Immunokinase Assays. Purified GST or GST fusion proteins were incubated in buffer (20 mM Mops, pH 7.2/25 mM β-glycerophosphate/5 mM EGTA/1 mM sodium orthovanadate/1 mM DTT) supplemented with 75 mM MgCl2/0.5 mM cold ATP/10 mCi (1 Ci = 37 GBq) [γ-32P]ATP at 30°C for 15 min. Samples were separated on SDS/PAGE, transferred to a membrane that was then stained with Ponceau, and autoradiographed.
Results
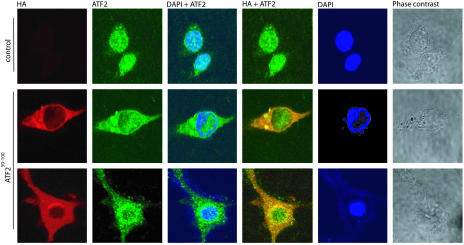
Expression of ATF250–100 Increases Cytoplasmic Localization of ATF2. Melanoma cells that express ATF250–100 (SW1-ATF250–100) exhibit a marked decrease in ATF2-dependent transcription as shown in refs. 26 and 27 (see Fig. 4b). Under normal conditions, ATF2 localizes in the nucleus of advanced human or mouse melanoma cells as shown in ref. 28 and Fig. 1. However, expression of ATF250–100 caused a substantial increase in cytoplasmic localization of ATF2 (Fig. 1 and Fig. 6, which is published as supporting information on the PNAS web site) due to inhibition of nuclear import, because addition of an inhibitor of nuclear export did not affect the level of ATF2 localization (data not shown). Minimal mutations within ATF250–100, MT, and RNDS designed to abolish JNK binding and p38 binding, respectively, failed to inhibit ATF2 nuclear localization to the degree seen with wild-type ATF250–100 (Fig. 6). This finding suggests that ATF250–100 association with JNK/p38 is important for retention of ATF2 within the cytoplasm, which consequently impairs ATF2 transcriptional activities. SW1-ATF250–100 melanoma cells also exhibited accumulation of c-Jun within the cytoplasm (Fig. 7, which is published as supporting information on the PNAS web site). In both cases, the ATF2 peptide appears to change also the morphology of the cells. Changes in the cellular distribution of ATF2 in SW1-ATF250–100 melanoma cells provide important insight into mechanisms underlying the changes observed in ATF2 and c-Jun transcriptional activities.
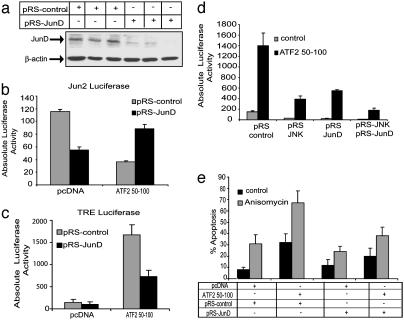
Fig. 4.
JunD is required for TRE-dependent transcription and for sensitization of melanoma cells that express ATF250–100 to apoptosis. (a) Infection of SW1 cells with pRS-JunD results in efficient inhibition of JunD expression. SW1 cells that constitutively express control or wild-type ATF250–100 were infected with control or pRS-JunD constructs. Proteins were prepared 48 h after infection, and Western blot analysis was performed with antibodies to JunD. β-actin was used as a loading control. (b and c) Inhibition of JunD expression increases Jun2-mediated transcription and inhibits TRE-dependent activities in cells expressing ATF250–100. SW1 cells that constitutively express control or ATF250–100 were infected with pRS control or pRS-JunD. Fortyeight hours after infection, the cells were transfected with Jun2-luc or TRE-Luc. Proteins were prepared 20 h later and assayed for luciferase activity. (d) Inhibition of TRE-mediated transcription is enhanced after inhibition of both JNK and JunD expression. SW1 cells that constitutively express control or ATF250–100 were infected with pRS-JunD or pRS-JNK or with both. Cells were transfected with TRE-Luc 48 h after infection, and proteins were prepared for luciferase assays after an additional 20 h. A portion of the same extracts was used to verify inhibition of JNK and JunD expression by Western blots. (e) Suppression of JunD transcription inhibits sensitization of the ATF2 peptide-expressing melanoma to apoptosis after anisomycin treatment. SW1 cells were infected with pRS-JunD or control pRS followed by transfection of the ATF2 peptide or control vectors 48 h later. Twenty hours later, cells were subjected to anisomycin treatment, and the degree of apoptosis was monitored by fluorescence-activated cell sorter analysis after an additional 24 h.
Fig. 1.
ATF250–100 promotes cytoplasmic localization of ATF2 in advanced melanomas. SW1 pcDNA and SW1-ATF250–100 wild-type were immunostained for endogenous ATF2 with ATF2 antibodies (green fluorescence) or for ATF250–100 with HA antibodies (red fluorescence). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Data shown represent multiple fields in over five experiments scoring >300 cells per experiment. Over 80% of the cells exhibited change in ATF2 localization. Shown is confocal microscopy. Phase contrast reflects cell shape.
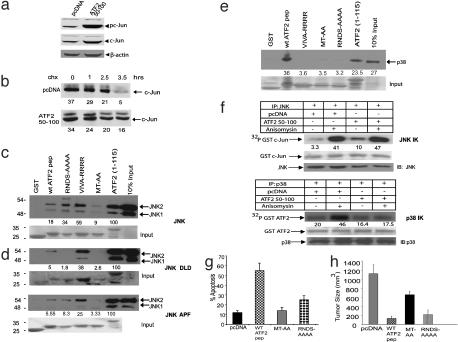
ATF250–100 Association with JNK Is Required for Sensitization of Melanoma Cells to Apoptosis. c-Jun expression is increased in SW1-ATF250–100 melanoma cells as shown in ref. 27 and Fig. 2a, because of an increase in the c-Jun half-life from 150 min to 210 min (Fig. 2b). Because JNK is implicated in the regulation of c-Jun stability (33) and ATF250–100 contains the consensus amino acid domain required for JNK binding (N.J., unpublished work), we tested whether JNK plays a role in mediating ATF250–100 effects. In vitro binding assays revealed specific association between ATF250–100 and endogenous JNK, employing ATF21–115 as a control (Fig. 2c). Mutant ATF250–100 that was altered in the two amino acids important for JNK association (MT-AA) was deficient in JNK binding, whereas the mutant deficient in p38 binding (RNDS-AAAA) or in β-sheets formation (VIVA-RRRR, expected to disturb ATF250–100 proper folding) had no significant effect on JNK association (Fig. 2c). Expression of any of the different mutant ATF250–100 listed above also failed to increase the expression of c-Jun (data not shown), suggesting that association with JNK is required for stabilization of c-Jun.
Fig. 2.
ATF250–100 associates with JNK and requires its binding to sensitize melanoma to apoptosis and inhibit tumorigenicity. (a) Expression of the ATF2 peptide increases expression of c-Jun. Protein extracts prepared from SW1 cells that stably express empty vector (control) or wild-type HA-ATF250–100 peptide (ATF2 peptide) were subjected to immunoblot analysis by using antibodies to phosphorylated c-Jun followed by reprobing with control c-Jun antibodies. The membrane was reprobed with antibodies to β-actin. (b) The half-life of c-Jun increased in melanoma cells expressing ATF250–100. The half-life of c-Jun was monitored with cycloheximide, which was added to control or ATF250–100-expressing cells for increasing time periods. Proteins were then subjected to immunoblot analysis by using antibodies to c-Jun. Numbers on the bottom of each image indicate relative change in level of expression based on a densitometry analysis normalized to background control with the quantify one program. Values represent the absolute densitometric values for each band. (c) JNK2 associates with ATF250–100. In vitro binding assays were carried with bacterially expressed and purified GST-ATF250–100 wild-type or mutant peptide incubated with extracts of 293T cells expressing Flag-tagged JNK2 for 2 h at 4°C. The beads were washed, and eluted material was subjected to immunoblot analysis with antibodies to Flag. (Lower) Coommassie blue staining reflecting the quantity of proteins used for the reaction. Relative change was quantified and normalized per binding to ATF21–115, a commonly used substrate for JNK/p38, based on densitometry. (d) Mutation within either the phosphoacceptor sites (APF) or the ATP pocket (DLD) of JNK reduces JNK2 association with ATF250–100.An in vitro binding reaction was carried out as indicated above, except that the GST peptides were incubated with extracts from 293T cells expressing Flag-tagged JNK2 APF or HA-tagged JNK2 DLD. (Lower) Quantity of proteins used for analysis. Quantifications were carried out as outlined in c.(e) Binding of ATF250–100 to p38. p38 was immunoprecipitated from cells followed by incubation with wild-type or mutant forms of the ATF250–100 or with the N-terminal region of ATF2 (1–115) as indicated in c. Quantification reflects relative binding to the corresponding proteins. (f) ATF250–100 increases basal JNK activity and reduces p38 activation after stress. Immunokinase (IK) reactions were carried out with either JNK or p38 immunoprecipitated (IP) from control or anisomycin-treated SW1 cells or SW1 cells that express ATF250–100.(Lower) Amount of substrate used (Ponceau staining) and the amount of kinase (IB). Numbers reflect quantification of phosphorylation based on PhosphorImager analysis. (g) JNK binding and, to a lesser degree, p38 binding are required for sensitization of melanoma cells to apoptosis by ATF250–100. SW1 cells expressing the wild-type or mutant forms of ATF250–100 were analyzed to determine the basal degree of apoptosis by using fluorescence-activated cell sorter analysis. (h) JNK but not p38 is required for the ability of ATF250–100 to inhibit melanoma growth in vivo. SW1 cells expressing ATF250–100 in its wild-type or mutant forms were injected s.c. into C3H mice (groups of six mice per experimental condition), and tumors were excised and analyzed to determine size and weight after 21 d.
Mutations within the ATP pocket of JNK (DLD) or its phosphoacceptor sites (APF at amino acids 183–185) were sufficient to cause a 3-fold decrease in JNK association with ATF250–100, suggesting that the active or conformationally intact forms of JNK associate with the peptide (Fig. 2d). ATF250–100 also binds p38 even more robustly than with JNK in a conformation-dependent manner, because association was abolished by each of the mutations described above (Fig. 2e).
Immunokinase assays with c-Jun as a substrate revealed that ATF250–100 increased basal JNK but not p38 kinase activity (Fig. 2f). ATF250–100 did not alter JNK activity after anisomycin treatment, but it attenuated activation of p38 kinase (Fig. 2f). These data suggest that the increase in c-Jun expression and activities in response to ATF250–100 could be attributed to changes in JNK, whereas decreased ATF2 activity could be attributed, in part, to decreases in p38 activity after treatment with anisomycin.
Mutation in ATF250–100 within the JNK binding site (MT-AA) abolished the peptide's ability to sensitize melanoma cells to apoptosis (Fig. 2g), suggesting that JNK association is required for this sensitization. Mutation within the p38-binding site (RNDSAAAA) also reduced the sensitization of the cells to apoptosis, albeit to a lesser degree compared with the mutation within the JNK-binding site (Fig. 2g). These data suggest that the association with JNK and, to a lesser degree, p38 is required for the sensitization of melanoma cells to ATF250–100-induced apoptosis.
Although SW1 cells produce aggressive tumors in mice (27, 34), a substantial inhibition of tumor growth was seen during expression of wild-type and p38 mutant ATF250–100. In contrast, mutation within the ATF250–100 JNK-binding site decreased the degree of tumor suppression by 50% (Fig. 2h). These data suggest that the association of ATF250–100 with JNK but not with p38 is required for inhibition of melanoma growth in vivo.
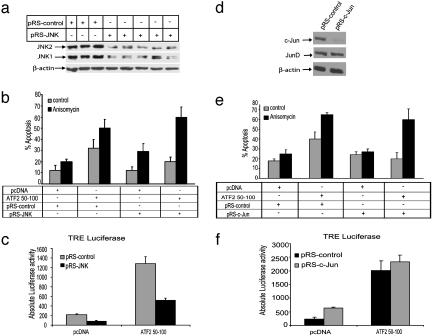
JNK Is Important for the Sensitization of Melanoma Cells to Apoptosis by ATF250–100. RNAi against the two major forms of JNK, JNK1/2 (termed pRS-JNK) were used to assess the role of JNK in mediating the activities of ATF2 peptide. Expression of pRS-JNK reduced the levels of JNK by 50% (Fig. 3a), and the degree of basal (spontaneous) apoptosis induced by the ATF250–100 peptide from 30% to 20%) (Fig. 3b; compare second and fourth gray bars). Surprisingly, suppression of JNK by pRS-JNK did not affect the degree of apoptosis in SW1-ATF250–100 cells treated with anisomycin. The latter data suggest that the degree of apoptosis seen after anisomycin treatment (50%) can be attributed to changes elicited by a kinase(s) other than JNK or to the incomplete inhibition of JNK expression by pRS-JNK. Inhibition of JNK caused ≈50% inhibition of TRE-dependent luciferase activity (Fig. 3c), suggesting that a JNK substrate is in part responsible for regulating TRE-containing promoters.
Fig. 3.
JNK and c-Jun are required for sensitization of melanoma cells to basal apoptosis by ATF250–100. (a) Infection with pRS-JNK virus of SW1 cells reduces JNK expression. SW1 cells were infected with pRS control or pRS-JNK viruses. Proteins were prepared 48 h after infection and subjected to Western blot analysis with antibodies to JNK. (Lower) Depiction of the same immunoblot but with β-actin antibodies to assure equal loading. The multiple lanes truly represent the degree of inhibition of JNK expression achieved by this RNAi. (b) Inhibition of JNK expression inhibits basal but not inducible apoptosis. SW1 cells were infected with pRS-JNK and control virus followed (24 h later) by transfection of the ATF2 peptide and (20 h later) treatment with anisomycin. Twenty-four hours after treatment, the degree of apoptosis was monitored by fluorescence-activated cell sorter analysis. (c) RNAi by pRS-JNK inhibits TRE-dependent transcription. SW1 cells were infected with pRS control or pRS JNK viruses, and 48 h later the cells were transfected with the corresponding control or ATF250–100 constructs and TRE-Luc. Proteins were assayed for luciferase activity after an additional 20 h. Values depict absolute luciferase activity. (d) Infection of SW1 cells with pRS-c-Jun inhibits c-Jun expression. SW1 cells were infected with control construct; the pRS-c-Jun construct and proteins were prepared 48 h after infection and subjected to Western blot analysis with antibodies to c-Jun. (Middle) Control for lack of pRS-Jun effect on JunD expression. β-Actin was used as a loading control. (e) Suppression of c-Jun expression affects the degree of basal apoptosis induced in ATF250–100-expressing cells. SW1 cells were infected with pRS-c-Jun or control pRS followed by transfection of ATF250–100 or control vectors 48 h later. Twenty hours later, cells were subjected to anisomycin treatment, and the degree of apoptosis was monitored by fluorescence-activated cell sorter analysis after an additional 24 h. (f) pRS-c-Jun does not affect TRE-dependent transcription in ATF250–100-expressing cells. SW1 cells were infected with the pRS control pRS-c-Jun, followed by cotransfection of ATF250–100 or control vectors with TRE-Luc 48 h later. Twenty hours later, proteins were prepared and assayed for luciferase activity. Values depict absolute luciferase activity.
c-Jun Is Important for the Sensitization of Melanoma Cells to Basal Apoptosis. To directly assess the role of c-Jun in the sensitization of melanoma expressing ATF250–100 to apoptosis, we generated c-Jun RNAi constructs (termed pRS-Jun) that efficiently decrease the expression of endogenous c-Jun expression (Fig. 3d) and inhibited cell growth (data not shown). Expression of pRS-Jun decreased the degree of basal apoptosis from 40% to ≈20% (Fig. 3e; compare second and fourth gray bars), suggesting that c-Jun is required for sensitization of melanoma cells to basal apoptosis during ATF250–100 expression. However, like pRS-JNK, inhibition of c-Jun expression had no effect on anisomycin-induced apoptosis (Fig. 3e). Surprisingly, inhibition of c-Jun expression did not affect the degree of TRE-dependent transcription (Fig. 3f), indicating that c-Jun affects transcriptional targets that are not regulated by TRE.
JunD Is Required for TRE-Dependent Transcription and Sensitization of Melanoma Cells Expressing ATF250–100 to Apoptosis. Although inhibition of JNK or c-Jun appears to play a key role in sensitizing SW1-ATF250–100 to anisomycin-induced apoptosis, the expression of TAM67, a dominant negative form of the Jun family of transcription factors, was efficient in mediating such inhibition (27). Because TAM67 is equally potent in inhibiting JunD (35, 36), we tested the role of JunD by using RNAi that efficiently inhibits JunD (termed pRS-JunD) (Fig. 4a) but not c-Jun expression (data not shown). Infection of SW1-ATF250–100 cells with pRS-JunD resulted in increased Jun2-mediated transcription to levels higher than seen in control cells (Fig. 4b), suggesting that JunD limited the degree of Jun2 transcription. In contrast, inhibition of JunD expression decreased TRE-dependent activities ≈40% (Fig. 4c). These findings suggest that JunD mediates, in part, the increase in TRE-dependent transcriptional activities seen in melanoma cells that express ATF250–100. Partial inhibition is also seen after inhibition of JNK expression (data not shown), suggesting that these two factors may complement each other in mediating TRE-dependent transcription. Indeed, analysis of TRE-Luc activity in melanoma cells that express both pRS-JNK and pRS-JunD resulted in complete inhibition of TRE-Luc (Fig. 4d), suggesting that JunD and JNK complement each other's activities.
Inhibition of JunD transcription also caused the degree of ATF250–100-induced apoptosis to decrease from 33% to 19% (Fig. 4e; compare second and fourth black bars) and the degree of anisomycin-induced apoptosis to decrease from 65% to 35% (Fig. 4e, compare second and fourth gray bars). These data identify JunD as the primary transcription factor that elicits sensitization of SW1-ATF250–100 melanoma cells to apoptosis after anisomycin treatment. Overexpression of the wild-type but not the transcriptionally inactive mutant form of ATF2 (mutated on amino acid 69,71) in JunD RNAi-expressing cells reduced the sensitization of these cells to apoptosis (Fig. 8, which is published as supporting information on the PNAS web site), further substantiating the role of JunD in mediating ATF250–100 effect.
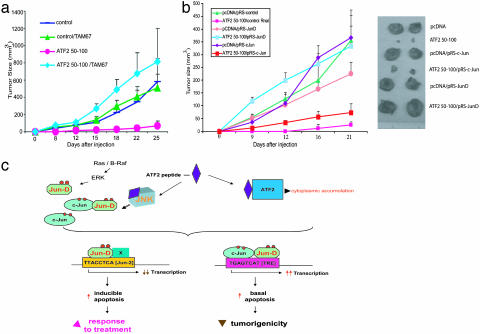
JunD Inhibits Tumorigenicity of SW1 Melanomas in C3H Mice. Further assessment of the role of JunD in mediating the effect of ATF250–100 was carried out in vivo. First, we evaluated the effect of TAM67 expression on tumor growth of SW1 cells implanted in C3H mice. As we described before, expression of ATF250–100 in SW1-melanoma cells suppressed almost completely their ability to produce tumors, compared with SW1 control vectors, which produced tumors of ≈500 mm3 within 25 d after injection (Fig. 5a). However, expression of TAM67 in SW1-ATF250–100 melanoma cells abolished the peptide's suppressive effect on tumorigenesis, increasing tumor size from 15 mm3 to >750 mm3 (Fig. 5a).
Fig. 5.
JunD is the primary factor mediating inhibition of melanoma tumorigenicity during expression of ATF250–100.(a) Expression of TAM67 attenuates the inhibition of melanoma tumorigenicity after expression of ATF250–100. SW1 cells that stably express control or ATF250–100 were infected with TAM67, and clones expressing TAM67 clones were selected on the basis of resistance to puromycin. Cells expressing TAM67 in combination with control or ATF250–100 were injected (104) s.c. into C3H mice, and tumor size was monitored for 25 d. Data represent mean values (P < 0.003; t test) based on analysis of eight mice per experimental group. (b) Expression of pRS-JunD but not pRS-c-Jun abolishes ATF250–100 ability to inhibit tumorigenicity of SW1 melanoma. The experiment was carried out as indicated in a, except that SW1 cells were infected with indicated pRS-constructs in vitro followed by their s.c. injection into C3H mice. The analysis was performed in groups of eight mice per experimental condition (P < 0.001; t test). (Right) Pictures provide representative data on tumor size at the end of the experiment. (c) Proposed model. The expression of the ATF250–100 results in cytoplasmic accumulation of ATF2 with concomitant inhibition of ATF2 transcription. Consequently, the Jun2 promoter sequences are occupied by c-Jun/JunD, which sensitizes melanoma to apoptosis after treatment (Left). Consistent with this model is the finding that expression of a 10-aa ATF2 peptide, which no longer inhibits ATF2 or Jun2-dependent transcription, suffices to sensitize melanoma cells to basal apoptosis by up-regulating c-Jun/JunD (unpublished data). ATF250–100 association with JNK results in increased activity of c-Jun, which sensitizes melanoma cells to spontaneous (basal) apoptosis, and suffices to reduce tumorigenicity of this otherwise aggressive tumor (Right). ERK, extracellular response kinase.
Because TAM67 equally affects c-Jun and JunD, we next assessed the possible role of each transcription factor on tumorigenesis by employing SW1-ATF250–100 melanoma cells stably expressing RNAi to the respective transcription factors. Stable expression of pRS-JunD, but not pRS-c-Jun, attenuated the ATF250–100-mediated inhibition of SW1 tumorigenicity (Fig. 5b). Furthermore, inhibition of JunD caused a >16-fold increase in the tumor size of SW1-ATF250–100 cells (from 18.7 to 315 mm3), similar to the changes seen in response to TAM67 expression. These results directly support the role of JunD in mediating the ATF250–100 inhibition of melanoma tumorigenicity and confirm that the effects elicited by TAM67 are primarily mediated through JunD.
Discussion
This work provides insight into mechanisms underlying the ability of ATF250–100 to sensitize melanoma to apoptosis and inhibit its tumorigenic potential. At least three distinct changes are elicited in advanced melanoma cells that express the ATF2 peptide (model outlined in Fig. 5c). First, ATF250–100 increases ATF2 localization within the cytoplasm. The mechanism of this effect may involve the peptide's affect on a protein kinase(s) that could regulate ATF2 translocation. Consistent with these findings is the observation that melanoma patients that exhibit cytoplasmic localization of ATF2 also had better prognoses (28).
Second, ATF250–100 associates with JNK and increases its basal kinase activity, resulting in higher levels of c-Jun expression and phosphorylation. Yet, neither TRE-Luc activity nor the induction of apoptosis after anisomycin treatment, depended on c-Jun, suggesting that c-Jun regulates a yet-unidentified subset of genes that contribute to the sensitization of ATF250–100-expressing melanomas to apoptosis. Consistent with our findings, c-Jun was previously shown to be capable of inducing apoptosis in nontransformed cultures (37, 38), conditions that might have been generated during expression of ATF250–100. It is of interest to note that inhibition of ATF2 expression per se (by RNAi) suffices to increase c-Jun levels (27), thereby pointing to alternate mechanisms that may underlie regulation of c-Jun by ATF2 (or its absence).
Increased JNK basal activities are likely to be a key factor in the phosphorylation and stabilization of c-Jun, whose stability is inversely correlated with its activities (33). Equally plausible is that increase in c-Jun expression is due to reduced availability of active ATF2, which would have otherwise formed an unstable complex with c-Jun (39). The association of ATF250–100 with JNK may also relieve histone deacetylase 3-dependent suppression of c-Jun transcription, which depends on JNK phosphorylation (40). The increased basal activities of JNK could be attributed to its binding to ATF250–100, explaining the increase in Jun phosphorylation and stability. Such binding may also out-compete JNK phosphorylation of other substrates and/or protect JNK from protein phosphatases that would otherwise limit its activities.
Third is the activity of JunD, which was found to play a central role in the sensitization of melanoma cells to apoptosis. Inhibition of JunD expression by RNAi reduced the transcriptional activities mediated by TRE and the degree of anisomycin-induced apoptosis seen in ATF250–100-expressing cells. Further, expression of TAM67, which effectively inhibits JunD (35, 36), also abolished the sensitization of ATF250–100-expressing melanomas to apoptosis (27). Both TAM67 and pRS-JunD, but not pRS-c-Jun, attenuated ATF250–100 ability to inhibit tumorigenicity of SW1 melanoma. Because JunD expression in primary and nontransformed cells coincides with protection from apoptosis (36, 41), it is likely that JunD displaces ATF2 on the corresponding promoter sequences, which would impair its antiapoptotic capacity. Given that >70% of melanomas harbor activating mutations in either N-Ras or a B-RAF that stimulate MAPK/extracellular response kinase (9, 10), which phosphorylates JunD (42), it is likely that JunD is constitutively active in these tumors. Because ATF250–100 elicits an increase in TRE-Luc and sensitization to apoptosis only in advanced tumors (unpublished observations), it is possible that active JunD is required for ATF2 peptide's activities.
Association of ATF250–100 with p38 and its effect on p38 activation after stress were dispensable for the inhibition of melanoma tumorigenicity by ATF250–100 peptide. Nevertheless, such association is expected to play an important role in the activities elicited by the peptide, albeit, in response to drug treatment. Thus, the responsiveness of melanomas expressing ATF250–100 to treatment in vivo is expected to depend on its ability to reduce p38 kinase activity.
Collectively, our observations provide insight into mechanisms underlying the sensitization of highly resistant melanoma cells to apoptosis and decreased tumorigenicity by ATF250–100. Similar changes were observed during expression of ATF250–100 in human tumors tested in nude mouse models (unpublished observations). The observations highlight the ability to elicit a transcriptional switch after inhibition of a single transcription factor, ATF2, pointing to its role in melanoma progression and to its possible use as a target for novel drug design.
Supplementary Material
Acknowledgments
We thank Reuven Agami for providing us with the pRS construct, Michael Karin and Roger Davis for JNK and p38 constructs, Adrian Ting for retroviral packaging constructs, Laurie Owen-Schaub for SW1 melanoma cells, Pablo-Lopez Bergami and Koh Nayakama for discussions, and Ruth Halaban for helpful comments. This work was supported by National Cancer Institute Grants CA99961 and CA51995 and by a Sharp Foundation grant to Z.R.
Abbreviations: ATF2, activating transcriptional factor 2; HA, hemagglutinin; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; pRS, pRETRO-SUPER; RNAi, RNA interference; TRE, 12-o-tetradecanoylphorbol 13-acetate response element.
References
- 1.Pawlik, T. M. & Sondak, V. K. (2003) Crit. Rev. Oncol. Hematol. 45, 245-264. [DOI] [PubMed] [Google Scholar]
- 2.Soengas, M. S. & Lowe, S. W. (2003) Oncogene 22, 3138-3151. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov, V. N., Bhoumik, A. & Ronai, Z. (2003) Oncogene. 22, 3152-3161. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov, V. N., Bhoumik, A., Krasilnikov, M., Ras, R., Owen-Schaub, L. B., Levy, D., Horvath, C. M. & Ronai, Z. (2001) Mol. Cell 7, 517-528. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov, V. N., Lopez, B. P., Maulit, G., Sato, T. A., Sassoon, D. & Ronai, Z. (2003) Mol. Cell. Biol. 23, 3623-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soengas, M. S., Capodieci, P., Polsky, D., Mora, J., Esteller, M., Opitz-Araya, X., McCombie, R., Herman, J. G., Gerald, W. L., Lazebnik, Y. A., et al. (2001) Nature 409, 207-211. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, X. D., Franco, A. V., Nguyen, T., Gray, C. P. & Hersey, P. (2000) J. Immunol. 164, 3961-3970. [DOI] [PubMed] [Google Scholar]
- 8.Ivanov, V. N., Kehrl, J. H. & Ronai, Z. (2000) Oncogene 19, 933-942. [DOI] [PubMed] [Google Scholar]
- 9.Davies, H., Bignell, G.R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M.J., Bottomley, W., et al. (2002) Nature 417, 949-954. [DOI] [PubMed] [Google Scholar]
- 10.Smalley, K. S. M. (2003) Int. J. Cancer. 104, 527-532. [DOI] [PubMed] [Google Scholar]
- 11.Bohm, M., Moellmann, G., Cheng, E., Alvarez-Franco, M., Wagner, S., Sassone-Corsi, P. & Halaban, R. (1995) Cell Growth Differ. 6, 291-302. [PubMed] [Google Scholar]
- 12.De Cesare, D., Jacquot, S., Hanauer, A. & Sassone-Corsi, P. (1998) Proc. Natl. Acad. Sci. USA 95, 12202-12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGill, G. G., Horstmann, M., Widlund, H. R., Du. J., Motyckova, G., Nishimura, E. K., Lin, Y. L., Ramaswamy, S., Avery, W., Ding, H. F., et al (2002) Cell 109, 707-718. [DOI] [PubMed] [Google Scholar]
- 14.Wilund, H. R., Hortsmann, M. A., Price, E. R., Cui, J., Lessnick, S. L., Wu, M., He, X. & Fisher, D. E. (2002) J. Cell. Biol. 158, 1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCawley, L. J., LI, S., Wattenberg, E. V. & Hudson, L. G. (1999) J. Biol. Chem. 274, 4347-4353. [DOI] [PubMed] [Google Scholar]
- 16.Woods, D., Cherwinski, H., Venetsanakos, E., Bhat, A., Gysin, S., Humbert, M., Bray, P. F., Saylor, V. L. & McMahon, M. (2001) Mol. Cell. Biol. 21, 3192-31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasilnikov, M., Ivanov, V. N., Dong, J. & Ronai, Z. (2003) Oncogene, 22, 4092-4101. [DOI] [PubMed] [Google Scholar]
- 18.Ouwens, D. M., de Ruiter, N. D., van der Zon, G. C., Carter, A. P., Schouten, J., van der Burgt, C., Kooistra, K., Bos, J. L., Maassen, J. A. & van Dam, H. (2002) EMBO J. 21, 3782-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutpa, S., Campbell, D., Derijard, B. & Davis, R. J. (1995) Science 267, 389-393. [DOI] [PubMed] [Google Scholar]
- 20.Kerppola, T. K. & Curran, T. (1993) Mol. Cell. Biol. 13, 5479-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falvo, J. V., Parekh, B. S., Lin, C. H., Fraenkel, E. & Maniatis, T. (2000) Mol. Cell. Biol. 20, 4814-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaszubska, W., Hooft van Huijsduijnen, R., Ghersa, P., DeRAemy-Schenk, A. M., Chen, B. P., Hai, T., DeLamarter, J. F. & Whelan, J. (1993) Mol. Cell. Biol. 13, 7180-7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, S. J., Wagner, S., Liu, F., O'Reilly, M. A., Robbins, P. D. & Green, M. R. (1992) Nature 358, 331-334. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, V. N. & Ronai, Z. (1999) J. Biol. Chem. 274, 14079-14089. [DOI] [PubMed] [Google Scholar]
- 25.Ronai, Z., Yang., Y. M., Fuchs, S. Y., Adler, V., Sardana, M. & Herlyn, M. (1998) Oncogene 16, 523-531. [DOI] [PubMed] [Google Scholar]
- 26.Bhoumik, A., Ivanov, V. & Ronai, Z. (2001) Clin. Cancer Res. 2, 331-342. [PubMed] [Google Scholar]
- 27.Bhoumik, A., Huang, T. G., Ivanov, V., Gangi, L., Qiao, R. F., Woo, S. L., Chen, S. H. & Ronai, Z. (2002) J. Clin. Invest. 110, 643-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger, A., Harriet, K., Ning, L., Eric, K., Halaban, R., Ronai, Z. & Rimm, D. L. (2003) Cancer Res., 63, 8103-8107. [PubMed] [Google Scholar]
- 29.Habelhah, H., Shah, K., Huang, L., Burlingame, A. L., Shokat, K. M. & Ronai, Z. (2001) J. Biol. Chem. 276, 18090-18095. [DOI] [PubMed] [Google Scholar]
- 30.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Cancer Cell. 2, 243-247. [DOI] [PubMed] [Google Scholar]
- 31.He, K.L. & Ting, A.T. (2002) Mol. Cell. Biol. 17, 6034-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Didier, C., Broday, L., Bhoumik, A., Israeli, S., Takahashi, S., Nakayama, K., Thomas, S.M., Turner, C.E., Henderson, S., Sabe, H. & Ronai, Z. (2003) Mol. Cell. Biol. 15, 5331-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchs, S. Y., Dolan, L., Davis, R. J. & Ronai, Z. (1996) Oncogene 13, 1531-1535. [PubMed] [Google Scholar]
- 34.Owen-Schaub, L. B., van Golen, K. L., Hill, L. L. & Price, J. E. (1998) J. Exp. Med. 188, 1717-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmani, M., Peron, P., Weitzman, J., Bakiri, L., Lardeux, B. & Bernuau, D. (2001) Oncogene 20, 5132-5142. [DOI] [PubMed] [Google Scholar]
- 36.Lamb, J.A., Ventura, J.J., Hess, P., Flavell, R.A. & Davis, R.J. (2003) Mol. Cell. 11, 1479-1489. [DOI] [PubMed] [Google Scholar]
- 37.Bossy-wetzel, E., Bakiri, L. & Yaniv, M. (1997) EMBO J. 16, 1695-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrens, A., Sibilia, M. & Wagner, E.F. (1999) Nat. Genet. 3, 326-329. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs, S. Y. & Ronai, Z. (1999) Mol. Cell. Biol. 5, 3289-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, C., Schneider, S., Wagner, E.F., Zhang, X., Seto, E. & Bohmann, D. (2003) EMBO J. 22, 3686-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weitzman, J. B., Fiette, L., Matsuo, K. & Yaniv, M. (2000) Mol. Cell. 6, 1109-1119. [DOI] [PubMed] [Google Scholar]
- 42.Gallo, A., Cuozzo, C., Esposito, I., Maggiolini, M., Bonofiglio, D., Vivacqua, A., Garramone, M., Weiss, C., Bohmann, D. & Musti, A.M. (2002) Oncogene 21, 6434-6445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.