Abstract
We report studies of a Croatian boy, a proven case of human S-adenosylhomocysteine (AdoHcy) hydrolase deficiency. Psychomotor development was slow until his fifth month; thereafter, virtually absent until treatment was started. He had marked hypotonia with elevated serum creatine kinase and transaminases, prolonged prothrombin time and low albumin. Electron microscopy of muscle showed numerous abnormal myelin figures; liver biopsy showed mild hepatitis with sparse rough endoplasmic reticulum. Brain MRI at 12.7 months revealed white matter atrophy and abnormally slow myelination. Hypermethioninemia was present in the initial metabolic study at age 8 months, and persisted (up to 784 μM) without tyrosine elevation. Plasma total homocysteine was very slightly elevated for an infant to 14.5–15.9 μM. In plasma, S-adenosylmethionine was 30-fold and AdoHcy 150-fold elevated. Activity of AdoHcy hydrolase was ≈3% of control in liver and was 5–10% of the control values in red blood cells and cultured fibroblasts. We found no evidence of a soluble inhibitor of the enzyme in extracts of the patient's cultured fibroblasts. Additional pretreatment abnormalities in plasma included low concentrations of phosphatidylcholine and choline, with elevations of guanidinoacetate, betaine, dimethylglycine, and cystathionine. Leukocyte DNA was hypermethylated. Gene analysis revealed two mutations in exon 4: a maternally derived stop codon, and a paternally derived missense mutation. We discuss reasons for biochemical abnormalities and pathophysiological aspects of AdoHcy hydrolase deficiency.
This paper reports a proven human case of inherited deficiency of S-adenosylhomocysteine (AdoHcy) hydrolase (E.C.3.3.1.1) activity. AdoHcy hydrolase catalyzes the hydrolysis of AdoHcy to adenosine and homocysteine (1). In eukaryotes, this is the major route for disposal of the AdoHcy formed as a common product of each of many S-adenosylmethionine (AdoMet)-dependent methyltransferases (Fig. 1). The reaction is reversible, but under normal conditions the removal of both adenosine and homocysteine is sufficiently rapid to maintain the flux in the direction of hydrolysis (2). AdoHcy is an inhibitor of many AdoMet-dependent methyltransferases. Because the inhibition is generally competitive, for any given methyltransferase the effect on activity will depend on the Km for AdoMet, the Ki for AdoHcy, and the concentrations of these two metabolites (3). Thus, AdoHcy hydrolysis serves physiologically not only to sustain the flux of methionine sulfur toward cysteine, but is believed also to play a critical role in the regulation of biological methylations (2, 4). Here we report both the clinical findings as well as the metabolic, enzymatic and gene studies that established a Croatian infant to be severely AdoHcy hydrolase deficient.
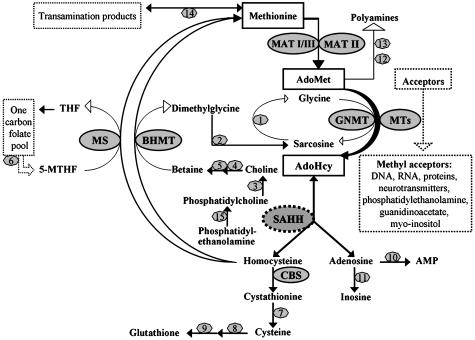
Fig. 1.
Methionine, AdoMet, and AdoHcy metabolism. AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; THF, tetrahydrofolate; 5-MTHF, 5-methyltetrahydrofolate; AMP, adenosine monophosphate; MAT, methionine adenosyltransferase (E.C.2.5.1.6); GNMT, glycine N-methyltransferase (E.C.2.1.1.20); MTs, a variety of AdoMet-dependent methyltransferases; SAHH, AdoHcy hydrolase (E.C.3.3.1.1); CBS, cystathionine β-synthase (E.C.4.2.1.22); MS, methionine synthase (5-MTHF-homocysteine methyltransferase) (E.C.2.1.1.13); BHMT, betaine-homocysteine methyltransferase (E.C.2.1.1.5). The numbers represent the following: 1, sarcosine dehydrogenase (E.C.1.5.99.1); 2, N,N-dimethylglycine dehydrogenase (E.C.1.5.99.2); 3, phospholipase D (E.C.3.1.4.4); 4, choline dehydrogenase (E.C.1.1.99.1); 5, betaine aldehyde dehydrogenase (E.C.1.2.1.8); 6, methylenetetrahydrofolate reductase (MTHFR) (E.C.1.5.1.20); 7, γ-cystathionase (E.C.4.4.1.1); 8, glutamate–cysteine ligase (E.C.6.3.2.2); 9, glutathione synthase (E.C.6.3.2.3); 10, adenosine kinase (E.C.2.7.1.20); 11, adenosine deaminase (E.C.3.5.4.4); 12, AdoMet decarboxylase (E.C.4.1.1.50); 13, spermidine (spermine) synthase (E.C.2.5.1.16 and E.C.2.5.1.22); 14, methionine transamination pathway; and 15, phosphatidylethanolamine methyltransferase (PEMT) (E.C.2.1.1.17).
Methods
Metabolite Assays. Amino acids were measured by ion exchange chromatography using a Biochrom 20 plus analyzer (Amersham Pharmacia). Plasma total homocysteine (tHcy) was assayed by using an IMX, Abbott analyzer. Plasma methionine and tHcy were measured also by capillary gas chromatography-mass spectrometry, as were cystathionine, total cysteine, dimethylglycine, and sarcosine (N-methylglycine) (5–7). Plasma and cerebrospinal fluid (CSF) AdoMet and AdoHcy (8), and plasma phosphatidylcholine, free choline, and betaine (9) were assayed as described. Guanidino compounds were measured by cation-exchange chromatography with postcolumn derivatization (10, 11). Leucocyte DNA methylation was quantitated in packed blood cells (12). Purines and pyrimidines were analyzed by using the method of Morris and Simmonds (13).
Assays of AdoHcy Hydrolase Activity. These were carried out by a modification of the method of Hershfield and coworkers (14). Enzyme- and homocysteine-dependent conversion of radiolabeled adenosine to radiolabeled S-adenosylhomocysteine was measured. Details are described in Supporting Text, which is published as supporting information on the PNAS web site.
Gene Analysis. RNA isolation from blood samples was carried out by the RNAzol method (Biogenesis, U.K.). DNA extraction from blood samples was performed by using standard procedures (15).
A PCR-based approach was used to amplify the coding regions of the AdoHcy hydrolase gene. PCR templates were either cDNA or genomic DNA. Primers used for PCR amplification are given in Supporting Text. cDNA was synthesized from 200 ng of whole RNA and a poly(T)-20mer oligonucleotide by using Superscript II Reverse Transcriptase (Invitrogen). Amplified genomic DNA or cDNA fragments were either subcloned as described (16) or submitted directly to the sequencing service (MWG Biotech, Ebersberg, Germany). Sequencing chromatograms were analyzed by using the chromas software (Conor McCarthy, Southport, Australia).
Amplified fragment-length polymorphism analysis was used to screen for an A-to-G point mutation in codon 143 of the AdoHcy hydrolase gene of a total of 65 random individuals from the Croatian population. DNA was amplified by using primers specific for the flanking region of exon 4. The obtained 240-bp PCR product was incubated with restriction enzyme Bsp1286 and analyzed by polyacrylamide gel electrophoresis for a restriction pattern different from that expected for the wild-type sequence (130 and 110 bp).
MR Imaging and [1H]MRS Studies. Details are published as supporting information on the PNAS web site.
Informed consent was obtained from the parents for all patient studies.
Results
Case Report. The patient is a boy, the first child of healthy nonconsanguineous parents, born at term by normal delivery after a normal pregnancy. The father's psychomotor development was somewhat slow in early infancy. Otherwise, the family history was unremarkable. Birth length was 50 cm, weight was 3,900 g, and head circumference was 34.5 cm. From the beginning, psychomotor development was slow, but there was a continuous progress until about his 5th month. Subsequently, stagnation and then regression were noted. Hypotonia, sluggishness, lack of interest, and very poor head control (especially elevation) were the main problems. Physical therapy resulted in only slight improvement.
At age 8 months, an initial neuropediatric evaluation showed marked muscular hypotonia, more in the legs than the arms. When supine, he was able to elevate his arms only for a brief period. In the prone position, head elevation and elbow support were also sustained only briefly. While being held in a standing position, his extremities dangled limply. He could not maintain the sitting position. When the trunk of the body was pulled from a lying to a sitting position, head control was incomplete. Tendon reflexes could not be elicited. Concomitant, convergent strabismus was noted at age 11 months. Subsequently, the boy made little further progress until the metabolic defect was defined when he was 12.8 months old. At that age, he could sit holding his legs and stand only very briefly with support. Subcutaneous fat was abundant, and his muscles were soft. His feet were in planovalgus position. Because of his brief attention span and quick tiring, cognitive functions could not be evaluated precisely. His length was 77 cm (25th to 50th percentile); weight was 11 kg (nearly 75th percentile); head circumference was 45 cm (≈3rd percentile).
Normal laboratory diagnostic tests included complete blood count; basic urine screening tests and sediment; acid-base balance; urinary organic acids, purines and pyrimidines; deletion screening by multiplex-PCR for Duchenne muscular dystrophy; electroencephalogram; ECG; and heart ultrasound. The following tests in plasma or serum were also normal: sodium, potassium, glucose, urea, creatinine, uric acid, γ-glutamyltranspeptidase, alkaline phosphatase, 5-nucleotidase, triglycerides, ammonia, lactate, pyruvate, total and free carnitine, acylcarnitines, very long chain fatty acids, vitamin B12, folate, viral hepatitis markers, thyroid hormones, thyroid-stimulating hormone, copper, ceruloplasmin. CSF contained normal cells, proteins, lactate, and glucose.
Several major biochemical abnormalities were detected. Plasma alanine aminotransferase, aspartate aminotransferase, and creatine kinase (almost solely the muscle isoenyzme) were elevated. Plasma albumin was low and the prothrombin time was prolonged (Table 1). Fibrinogen was also low, from 1.2 to 1.6 g/liter. Electromyography at age 8 months showed myopathic potentials. Histological examination of right quadriceps muscle at age of 13.1 months revealed moderate variability in fiber size, with a few necrotizing fibers undergoing phagocytosis, and occasional basophilic regenerating fibers. Histochemistry demonstrated no specific pathological changes, whereas electron microscopy (Fig. 4, which is published as supporting information on the PNAS web site) showed numerous myelin figures of different sizes and shapes; both in many muscle fibers as well as extracellularly. Larger myelin figures were mainly located in the subsarcolemma or in areas of myofibrilar disorganization, whereas smaller figures were predominantly intermyofibrillary. Numerous enlarged and abnormally shaped mitochondria were found within a few muscle fibers.
Table 1. Plasma or serum concentrations of methionine, related metabolites, guanidinoacetate, creatine, and other biochemical parameters in the patient and in his parents.
| Concentrations
|
||||
|---|---|---|---|---|
| Metabolite, units | Patient | Mother | Father | Reference |
| Methionine, μM | 477-784 | 24 | 21 | 13-45 |
| AdoMet, nM | 2,971 | 81 | 83 | 93 ± 16 |
| AdoHcy, nM | 5,044 | 21 | 22 | 15-45 |
| tHcy, μM | 14.5-15.9 | 6.0 | 7.2 | 5.4-13.9* |
| Cystathionine, nM | 552 | 163 | 133 | 44-342 |
| tCys, μM | 244 | 208 | 244 | 203-369 |
| PtdCho, μM | 780 | - | - | 1,500-2,559 |
| Free choline, μM | 7.8 | - | - | 10.7-19.7 |
| Betaine, μM | 211 | - | - | 26-67 |
| Dimethylglycine, μM | 11.8 | 3.7 | 3.9 | 1.4-5.3 |
| Sarcosine, μM | 7.5 | 1.2 | 1.4 | 0.6-2.7 |
| Guanidinoacetate, μM | 6.1, 2.1 | - | - | 0.83 ± 0.32 |
| Creatine, μM | 528, 42 | - | - | 87 ± 19 |
| Albumin, g/liter | 33.7 | - | - | 35.7-51.3 |
| ALT, units/liter | 252-1304 | - | - | 11-40 |
| AST, units/liter | 146-716 | - | - | 14-55 |
| Creatine kinase, units/liter | 2,000-4,360 | - | - | <228 |
| PT, INR† | 1.31-2.21 | - | - | 0.83-1.30 |
tCys, total cysteine; ALT, alanine aminotransferase; AST, aspartate amino transferase; PtdCho, phosphatidylcholine; -, not determined.
This reference range applies to subjects of ages 18-65 years. Representative published upper normal limits for young children range from 8.3 to 10.7 μM (17-19)
Prothrombin time as international normalized ratio
Magnetic resonance (MR) examination of the brain, performed at age of 12.7 months (Fig. 2), showed diffuse white matter hyperintensity on FSE T2 images as a consequence of T2 prolongation, most prominent around the frontal horns and peritrigonal white matter bilaterally. The ventricles were moderately enlarged secondary to white matter atrophy. The subarachnoid space was enlarged, predominantly in the frontal and temporal cortical areas. There was partial myelinization in the posterior part of the internal capsule and brainstem.
Fig. 2.

MR studies of the brain at age 12.7 months, before therapy. Myelination is present only in the posterior part of the internal capsule (arrowhead). Note clear delineation of globi pallidi because of unmyelinated lateral and medial lamina (double black arrow). Pattern of myelination corresponds to an age of 2–3 months.
MR spectroscopy of cerebral white matter (Fig. 5, which is published as supporting information on the PNAS web site) showed definite peaks of creatine and choline with a low choline/creatine ratio and an unidentified atypical, inverted peak near 3.78 ppm. Gray matter was normal.
The liver was diffusely hypoechogenic on ultrasound examination. Histologically there was normal architecture with signs of mildly active chronic hepatitis with only moderate portal fibrosis. Electron microscopy (Fig. 6, which is published as supporting information on the PNAS web site) showed hyperplasia of the smooth endoplasmic reticulum in the form of numerous small vesicles and a decreased number of rough endoplasmic reticulum cisterns. Many hepatocytes seemed to lack any rough endoplasmic reticulum. Mitochondria were numerous and mainly small with sparse and short cristae.
Metabolite and Enzyme Activity Assays Establishing Deficient Activity of AdoHcy Hydrolase. Initial biochemical studies (Table 1) revealed plasma methionine concentrations elevated to 477–784 μmol/liter (normal, 13–45 μmol/liter), but tHcy level was only slightly above normal for an infant at 14.5–15.9 μM (normal up to 10.7 μM). Plasma tyrosine was normal in several assays. Striking elevations of both plasma AdoHcy (>150-fold above the mid-point of the reference range) and AdoMet (≈30-fold) were discovered. Plasma cystathionine was slightly elevated, as was sarcosine. In CSF, methionine was elevated at 118 μmol/liter (normal 2.7–5.7 μmol/liter). CSF AdoHcy was 141 nM, highly elevated compared to a mean of 13.2 ± 10.6 nM (SD; n = 11). CSF AdoMet was, at most, moderately elevated at 548 nM (normal 386 ± 123 nM). To our knowledge, a similar pattern of abnormalities had never before been reported. The most plausible interpretation was that the patient had a severe deficiency of AdoHcy hydrolase activity. Subsequently, assays of the enzyme (Table 2) established that the patient has abnormally low activities of AdoHcy hydrolase, with perhaps 3% of control activity in liver extracts and 5–10% in fibroblast extracts and erythrocyte hemolysates. Only minor inconstant inhibition was detected when patient's fibroblast extract was mixed with extract of control cells. Frozen fibroblasts were homogenized and analyzed on the same day. 20 μl of patient's extract resulted in 64 × 103 dpm of the AdoHcy product, 10 μl of control extract in 170 × 103 dpm, and a mixture of the extracts (20 μl + 10 μl) in 196 × 103 dpm. The extracts were frozen, and the experiment repeated the next day. Twenty microliters of patient's extract resulted in 39 × 103 dpm, 10 μl of control extract in 121 × 103 dpm, the mixture extracts in 157 × 103 dpm.
Table 2. AdoHcy hydrolase activities in extracts of tissues from the patient, his parents, and control subjects.
| AdoHcy hydrolase activities, nmol/mg protein per h
|
||||
|---|---|---|---|---|
| Tissue | Patient | Mother | Father | Controls |
| Red blood cells | ||||
| Thawed × 1 | 0.7, 0.7* | ND | ND | 3.0-5.8 (n = 7) |
| Thawed × 2 | 0.4, 0.5* | 4.9 | 4.8 | 6.1 (n = 1) |
| Fibroblasts | 0.17 | ND | ND | 1.5-2.1 (n = 4) |
| Liver | 0.54 | ND | ND | 15.4, 17.8 |
Erythrocyte hemolysates were prepared from cells that had been stored frozen for various periods. For the parents only samples thawed twice were available for this assay. Extracts were prepared from fresh cultured skin fibroblasts or from liver samples that had been stored frozen. ND, not determined.
Repeat assays
Studies of Methylated Compounds Before Treatment. To assess whether compounds formed by AdoMet-dependent reactions might be depleted because of inhibition of the relevant methyltransferases by the accumulated AdoHcy, we assayed phosphatidylcholine and related metabolites (Table 1). Both phosphatidylcholine (normally formed in part by AdoMet-dependent methylation of phosphatidylethanolamine) (Fig. 1) and free choline (normally formed in part from phosphatidylcholine) (Fig. 1) were below their reference ranges in the patient's plasma (Table 1).
Creatine synthesis via AdoMet-dependent methylation of guanidinoacetate, catalyzed by guanidinoacetate methyltransferase (E.C.2.1.1.2), consumes substantial quantities of AdoMet. Guanidinoacetate was elevated in two independent pretreatment plasma samples from the patient, suggesting that some inhibition of this reaction was occurring. In one sample, creatine was abnormally low, but in the other, for reasons that are not clear, it was high (Table 1). In CSF, guanidinoacetate was 0.150 μmol/liter (normal, 0.015–0.1 μmol/liter) and creatine was 31 μmol/liter (normal 35–90 μmol/liter).
In an assay of the extent of genome-wide DNA methylation in leucocytes by [3H]dCTP incorporation (12), the values for repeat samples from the patient (age 11 months) were 1,174 and 1,250 dpm/μg DNA. The mean value for control children was 4,075 dpm/μg DNA (range 3,318 and 5,307 dpm/μg DNA; n = 4). In the assay used, incorporation of radioactivity is directly proportional to the number of unmethylated sites in the DNA being studied. Thus, the decreased radiolabel incorporation into the patient's DNA indicates that, at least in his leucocytes, DNA was hypermethylated relative to DNA from control children.
Analysis of the AdoHcy Hydrolase Gene. Sequencing of the AdoHcy hydrolase genes of the patient and his parents revealed that our patient has two point mutations in exon 4: a maternally derived nonsense mutation that introduces a stop codon at amino acid 112 (TGG to TGA), and a paternally derived mutation that changes tyrosine 143 to cysteine (codon TAC to TGC) and introduces a new restriction site for enzyme Bsp1286 (recognition sequence GTGCCC) (Fig. 3). So, the amplified fragment-length polymorphism analysis of the patient's exon 4 revealed a restriction pattern of 130, 59, and 51 bp, differing from the wild-type pattern of 130 and 110 bp found in 65 random Croatian DNA samples.
Fig. 3.
Partial nucleotide sequences of directly sequenced PCR products of patient's exon 4. Each allele bears one mutation as indicated by arrows. M, maternally derived allele, the TGG → TGA nonsense mutation in codon 112 causes replacement of a tryptophan by a premature stop codon. F, paternally derived allele, the TAC → TGC missense mutation in codon 143 causes replacement of a tyrosine by a cysteine. WT, related wild-type sequences of each mutated region are presented (Left).
Initial Therapeutic Approach. To diminish accumulation of AdoHcy, considered as the major pathogenetic factor, a methionine-restricted diet was started at age 12.8 months, providing methionine at 15 mg/kg per day. In parallel, to compensate for inhibition of methyltransferases, supplemental phosphatidylcholine was provided mainly as two half egg yolks per day. Supplemental creatine monohydrate, 5 g daily, was added at age 13.3 months. The diet resulted in marked decreases of AdoHcy, AdoMet, and methionine. Early clinical results were encouraging, with gradual gains in muscle strength and mental responsiveness. A longer interval will be required to permit complete evaluation of these interventions.
Discussion
Diagnosis and Mode of Inheritance. The initial finding was a very elevated plasma methionine that persisted until intervention, when the patient was >1 year old. The absence of elevations of plasma tyrosine and urinary succinylacetone together with the clinical presentation ruled out tyrosinemia type 1 as a cause of the hypermethioninemia. The slight to moderate elevations of serum transaminases made it unlikely that hepatocellular disease caused hypermethioninemia of this magnitude (20). The absence of markedly elevated tHcy as well as an elevated plasma cystathionine ruled out cystathionine β-synthase deficiency (21, 22). The patient thus fell within a group that has been termed “isolated persistent hypermethioninemia” (23). The majority of such patients have been found to have deficient methionine adenosyltransferase I/III (MAT I/III) activity. Elevated plasma AdoMet in the present patient demonstrated conclusively that he did not have MAT I/III deficiency (24). Isolated persistent hypermethioninemia associated with elevated AdoMet may result from a deficiency of glycine N-methyltransferase (GNMT), a defect that has been reported in three children (20, 25, 26). In these patients, a diagnostically important finding was the absence of N-methylglycine elevation in the face of elevated AdoMet (20). Elevation of both plasma N-methylglycine (sarcosine) and plasma AdoHcy in the present patient provided strong evidence against GNMT deficiency.
Deficient activity of AdoHcy hydrolase has now been demonstrated in extracts of liver and cultured fibroblasts and red blood cell hemolysates from the patient (Table 2). He has received no medication known to suppress AdoHcy hydrolase activity. Mixing experiments did not indicate significant inhibition of control activity by the patient's fibroblast extract.
Gene analysis revealed two mutations, one inherited from each parent. The maternal mutation generates a significantly truncated protein. The paternal mutation, a missense mutation, could not be found in 130 control alleles, indicating that it is not a common polymorphism.
Both heterozygous parents had normal plasma concentrations of AdoHcy, AdoMet, and methionine (Table 1). AdoHcy hydrolase activities in hemolysates of their red blood cells were only slightly below the activity in a hemolysate from control cells that had been treated similarly (Table 2). Thus, the biochemical studies, and the presence of two mutations in the patient's AdoHcy hydrolase gene, offer proof that he suffers from primary AdoHcy hydrolase deficiency inherited as an autosomal recessive trait.
Causes of the Pretreatment Metabolite Abnormalities. AdoHcy hydrolase deficiency readily explains the extreme elevations of the patient's pretreatment AdoHcy in plasma and CSF. The abnormal elevation of AdoMet is plausibly secondary to generalized inhibition by AdoHcy of multiple AdoMet-dependent methyltransferases. The elevations of methionine are explained by down-regulation of the flux from methionine into AdoMet when AdoMet is greatly increased, as was found in the children with deficient activity of GNMT (20, 25). The abnormally low plasma phosphatidylcholine and free choline presumably resulted from inhibition by AdoHcy of phosphatidylethanolamine methyltransferase. The pretreatment elevations of plasma guanidinoacetate suggest inhibition of guanidinoacetate methyltransferase. That the latter was incomplete is indicated by the presence of a readily detected creatine peak in MR spectroscopy of the brain (Fig. 5). In patients with genetically determined deficient guanidinoacetate methyltransferase, a creatine peak has been absent in MR studies (27–29). The elevation of sarcosine is attributed to an increased rate of AdoMet-dependent methylation of glycine. Several kinetic properties of GNMT especially suit this enzyme to synthesize sarcosine more rapidly as AdoMet concentrations rise, even in the face of elevated AdoHcy (3, 20, 26).
The reasons for elevated plasma concentrations of tHcy, betaine, dimethylglycine, and cystathionine remain uncertain. Also unexplained is the increased extent of DNA methylation. Discussion of the effects that might explain these changes is deferred until it becomes clear whether these changes occur in further cases of AdoHcy hydrolase deficiency.
Previous Cases for Which AdoHcy Hydrolase Deficiency Might Be Considered. To delineate better this enzyme defect, we reviewed previous reports of unexplained hypermethioninemia. Two of them deserved special attention. Labrune and coworkers (30) described three siblings with isolated hypermethioninemia for whom they raised the question of AdoHcy hydrolase deficiency. MAT activity in liver ruled out MAT I/III deficiency. In comparison to our patient, clinical presentation was quite different. Especially, there was no myopathy, and muscle enzymes and histology were normal. Plasma AdoMet and AdoHcy were not measured. In urine, AdoMet was high, whereas AdoHcy could not be detected. AdoHcy hydrolase activity in liver was decreased by ≈80%, but normal in fibroblasts and inconstant in red blood cells (from 10% to 100%). Primary generalized AdoHcy hydrolase deficiency seemed unlikely, and the authors concluded that the decreased hepatic AdoHcy activity might be secondary to liver disease. Gaull et al. (31) described a girl with isolated hypermethioninemia and clinical findings and histopathology very similar to our patient: myopathy, retarded psychomotor development, mild hepatic dysfunction, and severely elevated creatine kinase. Hepatic MAT activity was not low, ruling out MAT I/III deficiency. Neither AdoMet nor AdoHcy were assayed in plasma, and Gaull et al. did not posit a specific enzyme deficiency in their patient. However the clinical and biochemical similarities between this patient and ours, especially the myopathy that has been absent in other cases of persistent isolated hypermethioninemia, suggests that she may have had AdoHcy hydrolase deficiency.
Distribution of Pathologic Process and Pathophysiological Considerations. In our patient, the disease affected predominantly muscle, liver, and brain. In muscle, the process can be characterized as slowly progressive, destructive myopathy. In liver, the histological changes resembled mildly active chronic hepatitis with hyperplasia of smooth endoplasmic reticulum and decreased rough endoplasmic reticulum. Lack of rough endoplasmic reticulum, the major site of synthetic activity of liver cells, could explain the low albumin and prolonged prothrombin time. In brain, there were diffuse changes, with white matter chiefly affected.
Clarke and Banfield (3) list 39 AdoMet-dependent methyltransferases known in mammals as of 2001 (and the list has almost surely been extended since that time). Virtually all these enzymes are inhibited to a greater or lesser extent by AdoHcy (3). With the many potential metabolic derangements, it seems likely that the pathophysiology of AdoHcy hydrolase deficiency will be extremely complex. Among the variety of potentially inhibited methyltransferase reactions, we have obtained evidence about three: conversion of phosphatidylethanolamine to phosphatidylcholine, conversion of guanidinoacetate to creatine, and methylation of DNA. The first two were selected because they consume significant amounts of AdoMet (32–34) and produce reaction products that are both known to be physiologically important and readily replaced by dietary supplementation, thus providing the possibility of clinical benefit.
Choline is essential for normal hepatic function in both experimental animals and human subjects. Thus, knock-out of phosphatidylethanolamine methyltransferase in mice leads to severe, but reversible, liver pathology if dietary choline intake is not maintained (35, 36). Normal humans fed a choline-deficient diet for 6 weeks had 12- to 60-fold elevations of creatine kinase, associated with rises in serum alanine aminotransferase,q changes similar to those observed in our patient. Similarly, there is evidence for the essentiality of creatine, either synthesized or ingested, for normal neuromuscular function in children. Individuals with guanidinoacetate methyltransferase deficiency experience early developmental delay or arrest, heterogeneous neurological symptoms, muscular hypotonia, and are benefited clinically by dietary creatine administration (27–29). We assessed DNA methylation because changes in DNA methylation patterns are heritable and could negatively affect tissue-specific gene expression during embryogenesis and after birth (37, 38). Without further studies, the finding that leucocyte DNA was hypermethylated cannot be extrapolated to other tissues or other similar patients. However, because the silencing of genes by inappropriate methylation is the functional equivalent of somatic mutations (39), the inheritability of DNA methylation patterns (40) raises concern for the future that restoration of “normal” genomic methylation patterns may not occur.
Of course, other abnormalities could contribute to the pathophysiology of AdoHcy hydrolase deficiency. These might include (i) inhibitions of additional methyltransferases; (ii) adverse effects of abnormally high concentrations of AdoMet and AdoHcy and/or the abnormally low ratio of AdoMet/AdoHcy on vascular endothelium and other cells (41–45); (iii) adenosine depletion due to inadequate cleavage of AdoHcy, which might result in renal dysfunction (46), although glomerular and tubular function were normal in our patient; (iv) disturbed copper metabolism based on the finding that AdoHcy hydrolase is an important copper binding protein (47, 48), although the normal serum copper and ceruloplasmin levels in our patient and his clinical presentation do not suggest disturbed copper metabolism; and (v) glutathione depletion consequent to impaired conversion of methionine to cysteine, the metabolic precursor of glutathione. In turn, glutathione depletion may result in vulnerability to oxidative stress and xenobiotic toxicities, which could lead to liver dysfunction (20).
Definition of the pathophysiology of AdoHcy hydrolase deficiency may have significance beyond the apparently rare aptient with the genetic disorder. Several groups have examined inhibition of this enzyme in attempts to define new therapies for viral and neoplastic diseases (49–51). Animal models of AdoHcy deficiency would be invaluable in this research. However previous studies with knock-out mice must be analyzed cautiously because additional genes were removed by the deletion studied (52). Thus, the observed embryonic fatality cannot be attributed with certainty to the complete AdoHcy deficiency. Our patient maintained 3–10% residual activity, a finding that might better simulate the effects of exogenous inhibitors. In addition, AdoHcy hydrolase activity is readily reversible, and when homocysteine accumulates abnormally, so does AdoHcy. Therefore, insight into the effects of AdoHcy elevation may provide clues as to the adverse clinical effects of hyperhomocysteinemia, a field now under broad and intensive study.
Supplementary Material
Acknowledgments
This work was supported by Ministry of Science and Technology Republic of Croatia Grant 0108016 and National Institutes of Health Grants DK55865, DK15289, and DK54859.
Abbreviations: AdoHcy, S-adenosylhomocysteine; AdoMet, S-adenosylmethionine; tHcy, total homocysteine; CSF, cerebrospinal fluid; MAT, methionine adenosyltransferase; GNMT, glycine N-methyltransferase.
Footnotes
da Costa, K., Kwock, L., Hooker, J. & Zeisel, S. H. (2002) FASEB J. 16, A1023 (abstr.).
References
- 1.De La Haba, G. & Cantoni, G. L. (1959) J. Biol. Chem. 234, 603-608. [PubMed] [Google Scholar]
- 2.Cantoni, G. L. & Chiang, P. K. (1980) in Novel Biochemical and Structural Aspects, eds. Cavallini, D., Gaull, G. E. & Zappia, V. (Plenum, New York), pp. 67-80.
- 3.Clarke, S. & Banfield, K. (2001) in Homocysteine in Health and Disease, eds. Carmel, R. & Jacobsen, D. W. (Cambridge Univ. Press, Cambridge, U.K.), pp. 63-78.
- 4.Hoffman, D. R., Cornatzer, W. E. & Duerre, J. A. (1979) Can. J. Biochem. 57, 56-65. [DOI] [PubMed] [Google Scholar]
- 5.Allen, R. H., Stabler, S. P. & Lindenbaum, J. (1993) Metabolism 42, 1448-1460. [DOI] [PubMed] [Google Scholar]
- 6.Stabler, S. P., Marcell, P. D., Podell, E. R. & Allen, R. H. (1987) Anal. Biochem. 162, 185-196. [DOI] [PubMed] [Google Scholar]
- 7.Stabler, S. P., Lindenbaum, J., Savage, D. G. & Allen, R. H. (1993) Blood 81, 3404-3413. [PubMed] [Google Scholar]
- 8.Capdevila, A. & Wagner, C. (1998) Anal. Biochem. 264, 180-184. [DOI] [PubMed] [Google Scholar]
- 9.Koc, H., Mar, M.-H., Ranasinghe, A., Swenberg, J. A. & Zeisel, S. H. (2002) Anal. Chem. 74, 4734-4740. [DOI] [PubMed] [Google Scholar]
- 10.Marescau, B., Deshmukh, D. R., Kockx, M., Possemiers, I., Qureshi, I. A., Wiechert, P. & De Peyn, P. P. (1992) Metabolism 41, 526-532. [DOI] [PubMed] [Google Scholar]
- 11.Schulze, A., Ebinger, F., Rating, D. & Mayatepek, E. (2001) Mol. Genet. Metab. 74, 413-419. [DOI] [PubMed] [Google Scholar]
- 12.Pogribny, I., Yi, P. & James, S. J. (1999) Biochem. Biophys. Res. Commun. 262, 624-628. [DOI] [PubMed] [Google Scholar]
- 13.Morris, G. S. & Simmonds, H. A. (1985) J. Chromatogr. 344, 101-113. [DOI] [PubMed] [Google Scholar]
- 14.Hershfield, M. S., Kredich, N. M., Ownby, D. R., Ownby, H. & Buckley, R. (1979) J. Clin. Invest. 63, 807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 16.Marchuk, D., Drumm, M., Saulino, A. & Collins, F. (1991) Nucleic Acids. Res. 19, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilaseca, M. A., Moyano, D., Ferrer, I. & Artuch, R. (1997) Clin. Chem. 43, 690-692. [PubMed] [Google Scholar]
- 18.Delvin, E. E., Rozen, R., Merouani, A., Genest, J., Jr., & Lambert, M. (2000) Am. J. Clin. Nutr. 72, 1469-1473. [DOI] [PubMed] [Google Scholar]
- 19.Reddy, M. N. (1997) Clin. Chim. Acta. 262, 153-155. [DOI] [PubMed] [Google Scholar]
- 20.Mudd, S. H., Cerone, R., Schiaffino, M. C., Fantasia, A. R., Minniti, G., Caruso, U., Lorini, R., Watkins, D., Matiaszuk, N., Rosenblatt, D., et al. (2001) J. Inherit. Metab. Dis. 24, 448-464. [DOI] [PubMed] [Google Scholar]
- 21.Mudd, S. H., Levy, H. L. & Kraus, J. P. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), 8th Ed., pp. 2007-2056.
- 22.Stabler, S. P., Steegborn, C., Wahl, M. C., Allen, R. H., Wagner, C. & Mudd, S. H. (2002) Metabolism 51, 981-988. [DOI] [PubMed] [Google Scholar]
- 23.Mudd, S. H., Levy, H. L., Tangerman, A., Boujet, C., Buist, N., Davidson-Mundt, A., Hudgins, L., Oyanagi, K., Nagao, M. & Wilson, W. G. (1995) Am. J. Hum. Genet. 57, 882-892. [PMC free article] [PubMed] [Google Scholar]
- 24.Mudd, S. H., Jenden, D. J., Capdevila, A., Roch, M., Levy, H. L. & Wagner, C. (2000) Metabolism 49, 1542-1547. [DOI] [PubMed] [Google Scholar]
- 25.Augoustides-Savvopoulou, P., Luka, Z., Karyda, S., Stabler, S. P., Allen, R. H., Patsiaoura, K., Wagner, C. & Mudd, S. H. (2003) J. Inherit. Metab. Dis. 26, 1-15. [DOI] [PubMed] [Google Scholar]
- 26.Luka, Z., Cerone, R., Phillips, J. A., III, Mudd, S. H. & Wagner, C. (2002) Hum. Genet. 110, 68-72. [DOI] [PubMed] [Google Scholar]
- 27.Schulze, A. (2003) Mol. Cell. Biochem. 244, 143-150. [PubMed] [Google Scholar]
- 28.Stöckler, S., Holzbach, U., Hanefeld, F., Marquardt, I., Helms, G., Requart, M., Hänicke, W. & Frahm, J. (1994) Pediatr. Res. 36, 409-413. [DOI] [PubMed] [Google Scholar]
- 29.von Figura, K., Hanefeld, F., Isbrandt, D. & Stöckler-Ipsiroglu, S. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), 8th Ed., pp. 1897-1908.
- 30.Labrune, P., Perignon, J. L., Rault, M., Brunet, C., Lutun, H., Charpentier, C., Saudubray, J. M. & Odievre, M. (1990) J. Pediatr. 117, 220-226. [DOI] [PubMed] [Google Scholar]
- 31.Gaull, G. E., Bender, A. N., Vulovic, D., Tallan, H. H. & Schaffner, F. (1981) Ann. Neurol. 9, 423-432. [DOI] [PubMed] [Google Scholar]
- 32.Mudd, S. H., Ebert, M. H. & Scriver, C. R. (1980) Metabolism 29, 707-720. [DOI] [PubMed] [Google Scholar]
- 33.Mudd, S. H. & Poole, J. R. (1975) Metabolism 24, 721-735. [DOI] [PubMed] [Google Scholar]
- 34.Noga, A. A., Stead, L. M., Zhao, Y., Brosnan, M. E., Brosnan, J. T. & Vance, D. E. (2003) J. Biol. Chem. 279, 5952-5955. [DOI] [PubMed] [Google Scholar]
- 35.Waite, K. A., Cabillo, N. R. & Vance, D. E. (2002) J. Nutr. 132, 68-71. [DOI] [PubMed] [Google Scholar]
- 36.Walkey, C. J., Yu, L., Agellon, L. B. & Vance, D. E. (1998) J. Biol. Chem. 273, 27043-27046. [DOI] [PubMed] [Google Scholar]
- 37.Razin, A. & Shemer, R. (1995) Hum. Mol. Genet. 4, 1751-1755. [DOI] [PubMed] [Google Scholar]
- 38.Turker, M. S. & Bestor, T. H. (1997) Mutat. Res. 386, 119-130. [DOI] [PubMed] [Google Scholar]
- 39.Baylin, S. B., Herman, J. G., Graff, J. R., Vertino, P. & Issa, J.-P. (1998) Adv. Cancer. Res. 72, 141-196. [PubMed] [Google Scholar]
- 40.Robertson, K. D. & Jones, P. A. (2000) Carcinogenesis 21, 461-467. [DOI] [PubMed] [Google Scholar]
- 41.Fu, W., Dudman, N. P. B., Perry, M. A., Young, K. & Wang, X. L. (2000) Biochem. Biophys. Res. Commun. 271, 47-53. [DOI] [PubMed] [Google Scholar]
- 42.Dayal, S., Bottiglieri, T., Arning, E., Maeda, N., Malinow, M. R., Sigmund, C. D., Heistad, D. D., Faraci, F. M. & Lentz, S. R. (2001) Circ. Res. 88, 1203-1209. [DOI] [PubMed] [Google Scholar]
- 43.Kerins, D. M., Koury, M. J., Capdevila, A., Rana, S. & Wagner, C. (2001) Am. J. Clin. Nutr. 74, 723-729. [DOI] [PubMed] [Google Scholar]
- 44.Loehrer, F. M. T., Tschöpl, M., Angst, C. P., Litynski, P., Jäger, K., Fowler, B. & Haefeli, W. E. (2001) Atherosclerosis 154, 147-154. [DOI] [PubMed] [Google Scholar]
- 45.James, S. J., Melnyk, S., Pogribna, M., Pogribny, I. P. & Caudill, M. A. (2002) J. Nutr. 132, 2362S-2366S. [DOI] [PubMed] [Google Scholar]
- 46.Chen, Y.-F., Li, P.-L. & Zou, A.-P. (2002) Circulation 106, 1275-1281. [DOI] [PubMed] [Google Scholar]
- 47.Bethin, K. E., Cimato, T. R. & Ettinger, M. J. (1995) J. Biol. Chem. 270, 20703-20711. [DOI] [PubMed] [Google Scholar]
- 48.Bethin, K. E., Petrovic, N. & Ettinger, M. J. (1995) J. Biol. Chem. 270, 20698-20702. [DOI] [PubMed] [Google Scholar]
- 49.Chiang, P. K. (1998) Pharmacol. Ther. 77, 115-134. [DOI] [PubMed] [Google Scholar]
- 50.Turner, M. A., Yang, X., Yin, D., Kuczera, K., Borchardt, R. T. & Howell, P. I. (2000) Cell Biochem. Biophys. 33, 101-125. [DOI] [PubMed] [Google Scholar]
- 51.De Clercq, E. (2002) Mini. Rev. Med. Chem. 2, 163-175. [DOI] [PubMed] [Google Scholar]
- 52.Miller, M. W., Duhl, D. M. J., Winkes, B. M., Arredondo-Vega, F., Saxon, P. J., Wolff, G. L., Epstein, C. J., Hershfield, M. S. & Barsh, G. S. (1994) EMBO J. 13, 1806-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.