Abstract
Hepatitis C virus (HCV) is a nonretroviral oncogenic RNA virus, which is frequently associated with hepatocellular carcinoma (HCC) and B cell lymphoma. We demonstrated here that acute and chronic HCV infection caused a 5- to 10-fold increase in mutation frequency in Ig heavy chain, BCL-6, p53, and β-catenin genes of in vitro HCV-infected B cell lines and HCV-associated peripheral blood mononuclear cells, lymphomas, and HCCs. The nucleotide-substitution pattern of p53 and β-catenin was different from that of Ig heavy chain in HCV-infected cells, suggesting two different mechanisms of mutation. In addition, the mutated protooncogenes were amplified in HCV-associated lymphomas and HCCs, but not in lymphomas of nonviral origin or HBV-associated HCC. HCV induced error-prone DNA polymerase ζ, polymerase ι, and activation-induced cytidine deaminase, which together, contributed to the enhancement of mutation frequency, as demonstrated by the RNA interference experiments. These results indicate that HCV induces a mutator phenotype and may transform cells by a hit-and-run mechanism. This finding provides a mechanism of oncogenesis for an RNA virus.
Hepatitis C is an emerging infectious disease, affecting 200 million people worldwide. Hepatitis C virus (HCV) causes chronic hepatitis, liver cirrhosis, hepatocellular carcinoma (HCC), and occasionally, non-Hodgkin's B cell lymphoma (1). HCV contains an RNA genome, which replicates in the cytoplasm, does not contain an obvious oncogene, and does not integrate into host genomes; the mechanism of its oncogenesis remains unclear. Some HCV-associated HCCs have mutations in the tumor suppressor p53, the protooncogene β-catenin (2, 3) and several other genes. However, the long latency period of HCV infection makes it difficult to demonstrate the causal association between protooncogene mutations and HCV infection. Furthermore, many HCV-associated HCCs do not have detectable HCV RNA (1, 4), suggesting that HCV-induced tumorigenesis may employ a hit-and-run mechanism. HCV infects not only hepatocytes, but also B cells in vitro and in vivo (5, 6). Significantly, the HCV envelope protein E2 can bind CD81 in the CD21/CD19/CD81 costimulatory complex (7), suggesting the ability of HCV to alter the intracellular signaling of B cells. In HCV-infected individuals, oligoclonal lymphoproliferative disorders (8) and chromosomal translocation (9) have frequently been observed in B lymphocytes, suggesting that HCV may cause chromosomal instability. The hypervariable region of V(D)J in heavy chain of Ig (VH) in HCV-associated lymphoma has frequent somatic mutations and intraclonal diversity (10), suggesting the strong association between HCV infection and activation of somatic hypermutation. To understand the molecular mechanism of these changes, we set out to investigate whether HCV infection enhances the mutation frequencies of cellular genes.
Materials and Methods
Cloning and Sequencing of Peripheral Blood Mononuclear Cell (PBMC), Cellular, and Tumor DNA. Genomic DNA was extracted from PBMCs, cell lines, or tumor tissues. Cloning was performed by using Pfu Turbo DNA polymerase (Stratagene), and the reported primers as described in Supporting Text, which is published as supporting information on the PNAS web site.
Cell Culture. Raji cells and Ramos cells were obtained from the American Type Culture Collection and were grown in RPMI medium 1640 (Invitrogen) containing 20% and 10% FBS, respectively. Raji and JT cells (5) were further cloned by singl-cell dilution and were used for HCV infection by using the culture supernatant of non-Hodgkin's lymphoma cell culture (SB cell, ref. 5). A control infection using UV-irradiated SB culture supernatant was included in all of the experiments.
Plasmid Constructs. To generate the plasmid pI-Enh, the Ig intron enhancer was cloned into AflII site of pEGFPN1 (Clontech; ref. 11). A stop codon TAG that contains an RGYW motif was introduced as reported (11). To generate the plasmid pI-TAA, a stop codon mutation TAA at amino acid position 107 was introduced into the GFP gene. A similar construct without the premature stop codon in GFP was also made. Plasmids were transfected into cells by electroporation in a Bio-Rad electroporator. Flow cytometric analysis for detection of GFP was performed 5 days after transfection on a FACSCalibur cell sorter (Becton Dickinson; ref. 11).
Ligation-Mediated PCR (LM-PCR) Analysis of Double-Stranded DNA Breaks (DSBs). DSBs were identified by using a modified procedure (12). For descriptions of methods used in detail, see Supporting Text.
Gene Knock-Down by Using Small Interfering RNA (siRNA). More details concerning the siRNA sequences can be found in Supporting Text.
Statistical Analysis. Statistical analysis of the data in Tables 1, 2, 3 was performed by using a χ2 test. Statistical analysis of the data for Fig. 1 was performed by the independent-sample t test. Values of P < 0.05 were considered to be statistically significant.
Table 1. Mutation frequencies of cellular genes in HCV-infected cells.
| HCV(-)*
|
HCV(+)
|
|||
|---|---|---|---|---|
| Locus | Clones mutated† | Mutation frequency‡ × 10-4 | Clones mutated† | Mutation frequency‡ × 10-4 |
| VH | ||||
| Raji | 2/20 | 2.5 | 11/20 | 17.3 |
| JT | 1/20 | 1.2 | 6/20 | 9.9 |
| BCL-6 (area B) | ||||
| Raji | 0/20 | 0 | 8/20 | 8.3 |
| JT | 1/20 | 0.7 | 7/19 | 7.3 |
| PBMC | 2/54 | 0.8 | 27/80 | 6.4 |
| PBMC (area A) | — | — | 9/72 | 3.7 |
| P53 | ||||
| Raji | 0/30 | 0§ | 4/30 | 6.7 |
| JT | 0/33 | 0 | 6/26 | 11.5 |
| PBMC | 1/172 | 0.3 | 36/400 | 4.6 |
| β-catenin | ||||
| Raji | 0/20 | 0 | 5/20 | 5.6 |
| JT | 0/19 | 0 | 6/18 | 7.4 |
| PBMC | 2/60 | 0.7 | 20/64 | 7.8 |
| β-globin | ||||
| Raji | 0/21 | 0 | 4/21 | 3.6 |
| JT | 0/24 | 0 | 6/24 | 4.7 |
| PBMC | 1/56 | 0.6 | 8/58 | 4.2 |
HCV(-) represents PBMCs from healthy individuals or B cell lines inoculated with UV-inactivated HCV and are negative for HCV RNA. HCV(+) were from the infected counterparts
The numbers of PCR clones containing one or more mutations vs. the total numbers of clones sequenced are given
Mutation frequencies (mutation per base pair) are calculated as the total numbers of single-nucleotide mutations in all clones vs. the total number of nucleotides sequenced
The heterozygous mutations in the p53 gene (codon 213 CGA→CAA and codon 234 TAC→CAC) of Raji cells were excluded from the calculation
Table 2. Features of mutations in cellular genes in HCV-infected cells.
| Locus | Ratio of dG·dC/dA·dT* | R/S† | Ratio of Trs/Trv‡ | RGYW/WRCY bias per mutation,§ % |
|---|---|---|---|---|
| VH | 4.3 | 4.2 | 1.5 | 45.2 (P < 0.0001) |
| BCL-6 | ||||
| Area B | 2.5 | — | 2.3 | 32.7 (P < 0.0005) |
| Area A | 0.13 | — | 3.0 | 11.1 (NS) |
| p53 | 0.41 | 1.9 | 7.0 | 11.4 (NS) |
| β-catenin | 0.18 | 2.0 | 4.5 | 10.8 (NS) |
| β-globin | 0.26 | 2.3 | 5.8 | 12.3 (NS) |
Data were derived from Tables 1 and 5.
The ratios of dG·dC over dA·dT in the normal genomic sequence of VH areas B and A of BCL-6, p53, β-catenin, and β-globin are 1.05, 1.03, 0.60, 1.37, 0.72 and 1.14, respectively
Ratio of replacement/silent mutations
Ratio of transition overtransversion
Mutations at dG or dC nucleotides within RGWY or WRCY are defined as hot-spot mutations. The statistical significance for the excessive frequency of hot-spot mutations in each gene, as calculated by the χ2 test, is given in parentheses. NS, not significant
Table 3. Mutation frequencies of cellular genes in HCV-infected cells with siRNA and antisense strategy.
| HCV(-)*
|
HCV(+)
|
||||
|---|---|---|---|---|---|
| Gene | Scilenced target | Clones mutated | Mutation frequency × 10-4 | Clones mutated | Mutation frequency × 10-4 |
| VH | None | 2/21 | 2.4 | 13/20 | 18.3 |
| AID | 1/23 | 1.0 | 3/20 | 4.2† | |
| Polζ | 2/20 | 2.5 | 8/21 | 13.3 | |
| Pol ι | 1/20 | 1.2 | 4/24 | 6.3‡ | |
| p53 | None | 0/21 | 0* | 7/30 | 11.7 |
| AID | 0/20 | 0* | 6/28 | 10.7 | |
| Pol ζ | 0/23 | 0* | 1/31 | 1.6‡ | |
| Polι | 0/22 | 0* | 1/35 | 1.4‡ | |
, P<0.001;
, P<0.05. The statistical significance for the excessive frequency of mutations in each gene of siRNA/antisense-treated cells vs. control cells was calculated by the χ2 test
Methods are as described in Table 1
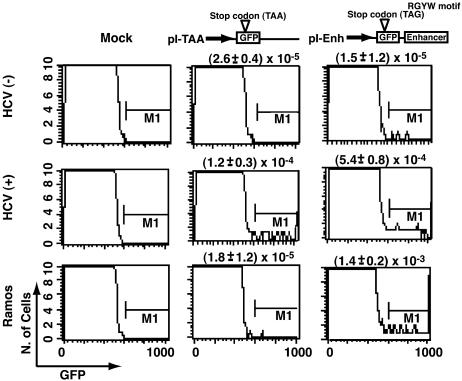
Fig. 1.
Determination of mutation frequencies by using reporter GFP genes in B cells. Cloned Raji cells infected with HCV or UV-irradiated HCV [HCV(-)] were transfected with the indicated reporter plasmid (pI-TAA or pI-Enh) on day 8 after infection (11). pI-Enh reporter plasmids without the enhancer element were used as the control. The cells were analyzed for expression of GFP by fluorescence-activated cell sorting analysis. Frequencies of mutations are shown.
Results
HCV Infection Induces Hypermutation of VH and BCL-6 in B Cell Lines. To examine the possible association between HCV infection and somatic mutations of cellular genes, we took advantage of the recently established systems of in vitro and in vivo HCV infection of B cells (5). Two B cell lines (Raji and JT) that had been either acutely or chronically infected with HCV in vitro were used (5). We first examined the effects of HCV infection on somatic hypermutation of the VH gene. The VH gene of the HCV-infected cells was PCR-amplified by using a proofreading polymerase, and multiple PCR clones were sequenced. A control Raji cell line, which had been infected with UV-irradiated HCV and is negative for HCV RNA, showed a point mutation frequency of 2.5 × 10-4 mutations per base pair (Table 1). By comparison, the mutation frequency of the VH gene in the HCV-infected Raji cell was >6-fold higher, at 17.3 × 10-4 mutations per base pair (Table 1). Similarly, the HCV-infected JT cells showed an 8-fold higher mutation frequency over the uninfected cells. These data indicated that HCV infection enhanced the somatic hypermutation rate of the VH gene in B cells. To confirm this conclusion, we determined the mutation frequency of the intronic residues +683 through +1408 of the BCL-6 gene (area B, Fig. 4, which is published as supporting information on the PNAS web site), which also hypermutates in normal germinal-center B lymphocytes (13) as well as diffuse large-cell lymphoma (14), by the same somatic hypermutation mechanism as for the VH gene. Both the HCV-infected Raji and JT cells showed nearly a 10-fold higher frequency of mutations than the Raji or JT cells infected with UV-inactivated HCV. Some of the HCV-associated mutations of the BCL-6 gene were located in the silencer element, which regulates the expression of BCL-6. Thus, HCV infection enhanced somatic hypermutation of both VH and BCL-6 genes in B cells.
To rule out the possible artifacts associated with HCV infection in vitro, we performed a similar analysis on the BCL-6 gene of PBMCs of HCV-infected individuals. We did not analyze the VH gene because it is likely to be heterogeneous in the unselected PBMC population. Among the 10 HCV patients studied, HCV RNA was detected in the PBMC of seven individuals, whereas it could not be detected in any of the six healthy individuals (data not shown). Sequence analysis of multiple PCR-amplified BCL-6 clones (area B) from PBMC of the HCV patients showed a >8-fold higher mutation frequency than that of the healthy individuals (Table 1). Most of the PCR-amplified clones contained unique mutations that were rarely detected in healthy individuals, indicating that they were not the results of selection and amplification of a particular clone; nor do they represent genetic polymorphism. Thus, they reflect the true mutation frequency. These results indicate that HCV infection enhances somatic hypermutations of VH and BCL-6 both in cell culture and in PBMCs.
HCV Infection Induces Mutations of Tumor Suppressor and Protooncogenes in B Cells. To examine whether HCV infection induces mutations in other somatic genes, we next examined cellular genes that normally are not mutated by the somatic hypermutation mechanism of the Ig gene. We chose p53 and β-catenin for analysis because they have been reported to be frequently mutated in HCV-associated HCCs (2, 3). The mutation frequencies of p53 and β-catenin genes were significantly higher in both PBMC from HCV-infected individuals, and in vitro HCV-infected B cell lines than those in the uninfected counterparts (Table 1). In Raji and JT cells infected with UV-inactivated HCV, no mutation was found in p53 or β-catenin (Table 1). The sites of mutation in HCV-infected cells were nearly randomly distributed (Fig. 4). An enhanced mutation frequency was also observed for a second region of the BCL-6 gene (area A) and the β-globin gene (Table 1). These results indicate that HCV infection directly or indirectly induces mutations in somatic genes other than the Ig gene.
We next examined p53 and VH in Raji cells at several time points after HCV infection in vitro. We observed the enhanced mutation frequency in both genes as early as 8 days after infection, and for as long as 40 days after infection (Tables 5 and 6, which are published as supporting information on the PNAS web site). These results suggest that HCV infection induces and stably maintains mutations in p53 and VH.
HCV Infection Induces Two Types of Nucleotide Substitution Patterns. Analysis of the nature of the mutations in VH genes from HCV-infected cells (combining the data from both HCV-infected B cell lines and PBMC) showed that these mutations exhibited most of the features characteristic of the somatic hypermutation of the Ig gene in normal B cells (ref. 15 and Table 2). These features include the preferential dG·dC mutations over dA·dT mutations, preferential mutations in RGYW and WRCY [R, purine (A/G); Y, pyrimidine (C/T); W, A/T] (16) motifs and a high replacement/silent mutation ratio. Thus, the enhanced mutation frequency of the VH gene probably represents the enhancement of the normal somatic hypermutation mechanism of the VH gene, which typically affects genomic sequences within ≈2 kbp downstream from the transcription initiation sites of the Ig gene (17). Area B of BCL-6 had dG·dC-biased mutations, which preferentially targeted the RGYW motif, similar to the pattern seen for VH (Table 2). In contrast, the nucleotide substitution pattern of p53, β-catenin, and area A of BCL-6 showed that mutations preferentially occurred on dA·dT over dG·dC (most commonly, from dA to dG or dT to dC) and that there was no RGYW preference (Table 2). Furthermore, the ratio of replacement/silent mutation was substantially lower for p53 and β-catenin than for VH and the ratio of transition/transversion mutations in the former was relatively higher. Thus, the mechanism of the enhanced mutations within p53, β-catenin, and area A of the BCL-6 gene sequence is likely to be different from that of VH and area B of BCL-6. These results suggest that HCV infection activates both the normal hypermutation mechanism of the VH gene and the general mutation mechanism of other somatic genes.
HCV Infection Activates the Machinery of Somatic Hypermutation in B Cells. We next used reporter plasmids to score mutation frequencies in HCV-infected cells. We engineered a mutation substrate (named pI-Enh) that expressed GFP, which is located upstream of an Igκ enhancer, because somatic hypermutation requires the presence of the Ig enhancer (18); we inserted a stop codon (TAG) that consists of the mutational hot spot RGYW motif (16) in the GFP gene (Fig. 1). Mutations occurring at the somatic hypermutation hotspot of the Ig gene could revert the stop codon, allowing the expression of GFP and fluorescence-based detection. Raji cells were infected with HCV- or UV-irradiated HCV and were then transfected with pI-Enh plasmids on day 8 after infection. Flow cytometry analysis detected significantly higher numbers of GFP+ cells in HCV(+) Raji cells than in the mock-transfected cells or in the cells infected with UV-inactivated HCV (35-fold difference, P = 0.0046; Fig. 1). The mutation frequency in HCV(+) cells was in the same order of magnitude as in Ramos cells, which are a B cell line undergoing a high frequency of Ig mutation (19). To determine the mutation frequency of other cellular genes, we used another reporter plasmid (named pI-TAA), in which GFP is terminated by a stop codon (TAA) without the RGYW motif or the Igκ enhancer. This plasmid also yielded significantly higher numbers of GFP+ cells in HCV-infected Raji cells than in the control cells (5-fold difference, P = 0.0397; Fig. 1). Significantly, the mutation frequency of pI-TAA in Ramos cells was similar to that in the HCV(-) Raji cells, but was substantially lower than that in the HCV(+) Raji cells. Thus, the enhanced mutations observed with both pI-Enh and pI-TAA plasmids in HCV(+) cells were clearly related to HCV infection. These results indicate that HCV infection causes enhanced hypermutation in both Ig and general somatic genes of B cells by at least two different mechanisms.
HCV-Infection-Induced DSBs. To examine the molecular mechanism of enhanced hypermutation in HCV-infected cells, we studied several reported mechanisms associated with Ig hypermutation. First, we examined the possible occurrence of DSBs, which contribute to somatic hypermutation of the Ig V(D)J region (20, 21) in HCV-infected and control Raji cells. We used LM-PCR (12) which can detect both general and locus-specific DNA breaks. Bluntended DSBs were readily detectable over the entire genome or specifically in the VH or p53 region of HCV-infected Raji cells as early as 2 ays after HCV infection (Fig. 2 A–C, and Fig. 5, which is published as supporting information on the PNAS web site). The extent of DSBs increased with time. Correspondingly, HCV RNA could be detected only in HCV-infected cells (Fig. 2E). In contrast, only a very small degree of DSBs was detectable in Raji cells infected with UV-inactivated HCV. Similarly, HCV-infected JT cells also had a higher degree of DSBs than did the HCV(-) cells (Fig. 2F). To rule out the possibility that the high mutation frequency was due to growth stimulation by HCV, we determined the growth rate of HCV-infected cells. HCV infection was found to retard the cell growth in comparison with the uninfected counterparts, indicating that HCV-infected cells undergo a comparably lower number of divisions (Fig. 2G). Therefore, cell growth rate could not explain the higher frequency of mutations in HCV-infected cells. These results indicate that HCV infection induces DSBs, the repair process of which may be associated with the introduction of mutations. Furthermore, these DSBs could not be examined by apoptosis alone (Fig. 5).
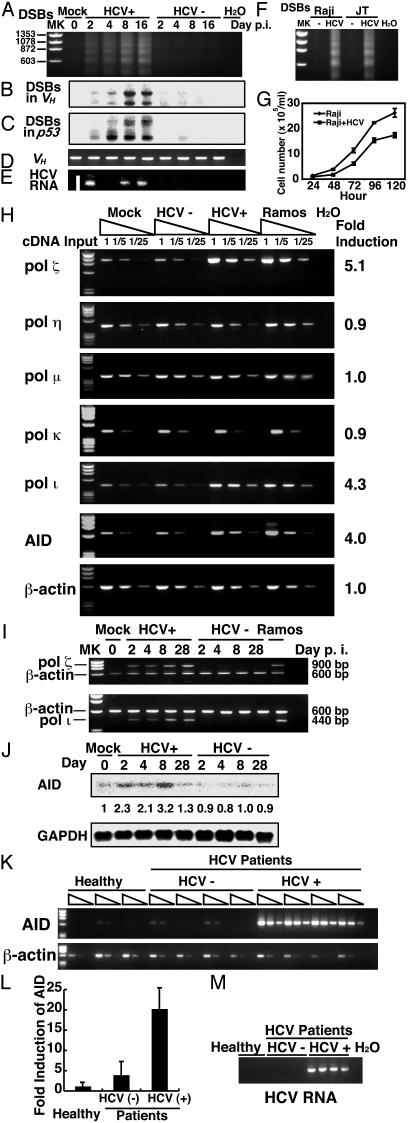
Fig. 2.
LM-PCR amplification of DSBs in HCV-infected and control Raji cells. (A–C) Blunt-ended DSBs were detected over the entire genome (A) or specifically in the Ig (B) or p53 (C). Raji cells were infected with HCV or UV-irradiated HCV [HCV(-)] and were examined at different days after infection. HCV(-) cells were infected with UV-irradiated HCV and are negative for HCV RNA. MK, DNA size marker; H2O, water control. (D) PCR amplification of the VH region was used as a loading control. (E) (M) HCV-RNA was detected by RT-PCR. (F) DSBs in HCV-infected JT cells were compared with those in the infected Raji cells. (G) Cell growth curve of HCV-infected Raji cells as determined by Trypan blue exclusion assay. (H) Semiquantitative RT-PCR detection of error-prone DNA polymerases and AID in Raji cells. RNA transcripts of various polymerases and the AID gene were reverse-transcribed in Raji cells on day 8 after HCV infection. cDNA of different dilutions (no dilution, 1:5, and 1:25) were then used for PCR amplification. Ramos cells were included as a control. (I) RNA samples from Raji cells on different days after HCV infection were used for amplification of polymerase ζ or polymerase ι, and β-actin in the same reaction. (J) Northern blots for AID and GAPDH in Raji cells on different days after infection. The relative amounts of the AID transcript on different days after infection were quantified by an image analyzer and are indicated below. (K) RT-PCR of AID in HCV-infected and non-infected PBMC from 3–4 individual HCV patients and healthy individuals. Three different concentrations of cDNA from each PBMC sample were used. RT-PCR of β-actin served as internal control. MK, DNA size marker, H2O, water control. (L) Fold induction of AID was normalized with the expression of β-actin.
HCV Infection also Induces Error-Prone DNA Polymerases and Activation-Induced Cytidine Deaminase (AID). Mutations occur while homologous recombination repairs DSBs (20, 21) or single-strand DNA breaks (22), possibly mediated by error-prone DNA polymerases (23–26). In particular, error-prone polymerases ζ, η, ι, and μ have been postulated to be the mutagenic polymerases for somatic hypermutation (23–26). Therefore, we determined the expression levels of various error-prone DNA polymerases by semiquantitative RT-PCR of their transcripts. Polymerases η, κ, and μ were expressed at similar levels in the HCV-infected Raji cells and the control cells at day 8 after infection (Fig. 2H). In contrast, the expression levels of polymerases ζ and ι were approximately five times higher in HCV-infected cells than in HCV(-) cells. The mRNA amounts of these two polymerases in HCV-infected cells were almost the same as those in Ramos cells. Kinetic analysis of the polymerases ζ and ι transcripts at various times after HCV infection showed that the expression of polymerases ζ and ι in HCV-infected Raji cells started to increase at day 2 and remained at high level, even at day 28 (Fig. 2I). These data together indicated that HCV infection activated error-prone DNA polymerases ζ and ι.
We have also examined the expression of AID, which plays a role in the hypermutation of Ig, probably by deaminating dC (27). AID induces hypermutation in actively transcribed genes with a strong bias toward dG·dC base pairs (28), which resembles the pattern of mutations seen in the Ig gene of HCV-infected cells. Semiquantitative RT-PCR analysis showed that HCV infection activated the expression of AID in Raji cells (Fig. 2H). Northern blot analysis confirmed that HCV infection enhanced the expression of AID in Raji cells at various time points (from day 2 to day 8) after infection (Fig. 2 J). AID expression level was also higher in PBMCs from HCV-infected patients than uninfected individuals (Fig. 2 K and L). This finding was also observed in normal PBMC infected with HCV in vitro. Eight days after infection, the mRNA level of AID increased, as determined by semiquantitative RT-PCR (Fig. 6, which is published as supporting information on the PNAS web site), although PBMC eventually died at 2 weeks after infection. These results suggest that AID may also contribute to dG·dC-biased mutations observed in VH and area B of BCL-6 in HCV-infected Raji cells.
In addition, we used semiquantitative RT-PCR to examine whether HCV infection alters the expression of mismatch repair enzymes (pms1, pms2, mlh1, msh2, msh3, and msh6) because these enzymes may play a role in the error-prone repair process (29). The expression level of these enzymes in the HCV-infected cells was not different from the uninfected cells (Fig. 7, which is published as supporting information on the PNAS web site).
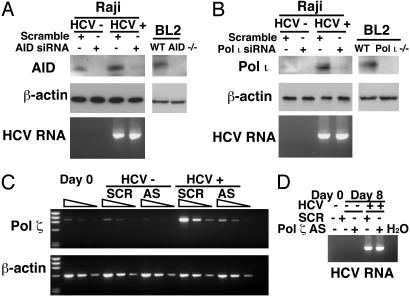
siRNA and Antisense Oligodeoxynucleotide Targeting AID or Error-Prone Polymerases Reduced the Mutations of Cellular Genes. We used RNA interference and antisense strategy to test the hypothesis that activated AID and error-prone DNA polymerases are the enzyme responsible for enhanced mutations of cellular genes in HCV-infected cells. Toward that end, we first evaluated the activity of an siRNA targeting to AID as well as siRNA targeting error-prone polymerase ι and antisense oligodeoxynucleotides for polymerase ζ. As shown in Fig. 3, the introduction of AID and polymerase ι siRNA and polymerase ζ antisense oligodeoxynucleotides resulted in the 6-fold reduction of the expression of AID polymerase ι and ζ, respectively, without significantly affecting the level of β-actin protein.
Fig. 3.
(A and B) The effect of siRNA on the levels of AID and DNA polymerase ι during HCV infection. Western blot analysis shows AID and error-prone polymerases expression at 8-days after transfection in HCV-infected cells in the presence of the specific or scramble siRNA. Western blot of β-actin was used as loading control. AID-/- or polymeraseι-/- BL2 cells served as negative control. HCV-RNA was detected with RT-PCR. (C) Antisense oligodeoxynucleotides against polymerase ζ were used as reported (23). SCR, scramble oligonucleotide; AS, antisense oligonucleotide against polymerase ζ.(D) HCV RNA was detected with RT-PCR.
We next determined whether the introduction of these siRNAs affected the mutation frequency of cellular genes. Introduction of the AID siRNA caused a 4.5-fold reduction of mutation frequency of the VH gene (Table 3). Polymerase ζ-knock-down did not have significant effects on VH hypermutations, but polymerase ι-knockdown significantly reduced the mutation frequencies of VH genes. Thus, AID and polymerase ι are responsible for most of the mutations that occur in the VH region. The polymerase ζ and ι-knock-down also reduced significantly mutation frequencies of p53 (Table 3). In contrast, p53 mutation was not affected by AID knock-down. Thus, the HCV-induced polymerase ι affected the mutations of both the VH and p53 genes. These results together established that activation of error-prone polymerases and AID is responsible for HCV-induced mutagenesis of cellular genes.
HCV-Associated Neoplasias Showed Amplified Mutations of Cellular Genes. To determine whether the HCV-induced mutations in protooncogenes may lead to oncogenic transformation of the infected cells and are thereby amplified in the HCV-associated tumors, we examined the mutation frequency and pattern of cellular genes in HCV-associated neoplasias. For this purpose, we performed direct sequencing of p53, β-catenin, and BCL-6 (area B) genes to identify those mutations that have been amplified and become predominant in the tumors. The HCV-associated B cell lymphomas showed several predominant mutations in p53, β-catenin, and BCL-6 (Fig. 4), with a mutation frequency of 3.0 × 10-3, 3.3 × 10-3,4.1 × 10-3 mutations per base pair, respectively (Table 4). By contrast, the number of the amplified mutations in B cell lymphomas not associated with HCV was at least seven times fewer (Table 4). Significantly, the nucleotide substitution pattern of p53 and β-catenin in the HCV-associated tumors exhibited a low dG·dC/dA·dT substitution ratio and a high transition/transversion ratio very similar to those seen in HCV-infected Raji cells and PBMCs (Table 2). However, HCV-associated lymphoma has a very high replacement/silent mutation ratio, which is unlike that in the HCV-infected PBMC (Table 2) and is significantly higher than the expected ratio of 2.9 in the absence of selection pressure (30). We further sequenced multiple individual PCR plasmid clones to determine the mutation frequencies within the p53 and β-catenin genes in these tumors. After excluding those predominant mutations previously detected by direct sequencing of PCR products, we determined the mutation frequency of p53 and β-catenin to be 6.5 × 10-4 and 8.2 × 10-4 mutations per base pair, respectively (data not shown). These frequencies were similar to those in the HCV-infected Raji cells (Table 1). These findings together are consistent with the interpretation that some of the HCV-induced somatic mutations are amplified as a result of selection during tumorigenesis.
Table 4. Features of the predominant (heterozygous) mutations in HCV-associated neoplasias as determined by direct sequencing.
| Mutation frequency (mutation per base pair) × 10-4
|
|||||||
|---|---|---|---|---|---|---|---|
| Type | Tumor/nontumor | p53 | β-cat | BCL-6 (Area B) | Ratio of dG·dC/dA·dT* | R/S*† | Ratio of Trs/Trv*‡ |
| B cell | HCV T | 30 | 33 | 41 | 0.6 | 6.0 | 7.0 |
| Nonviral | 3.9 | 0 | 4.6 | — | — | — | |
| HCC | HCV T | 31 | 30 | ND | 0.64 | 5.7 | 4.8 |
| HCV NT | 5.9 | 11 | ND | 0.5 | 2.0 | 5.0 | |
| HBV T | 3.9 | 7.4 | ND | — | — | — | |
| HBV NT | 0 | 7.4 | ND | — | — | — | |
| Nonviral | 2.5 | 4.4 | ND | — | — | — | |
ND, not determined. —, not calculated due to the small numbers of mutations in these tumors. T, tumor; NT, nontumor.
Only p53 and β-catenin (β-cat) were included for these calculations
The expected R/S for a random sequence in the absence of selection pressure is 2.9 (30)
Ratio of transition over transversion
We further determined whether these findings could be extended to HCV-associated HCC. We examined p53 and β-catenin mutations in tumors and their neighboring nontumor tissues. Very similar mutation frequency and nucleotide substitution patterns to those of HCV-associated B cell lymphomas were observed in HCV-associated HCC. Interestingly, the number of mutations in the tumors was 3–5 times higher than that in the nontumor parts (Table 4). Nevertheless, the patterns of nucleotide substitutions were similar between the tumors and the neighboring nontumor tissues, except that the ratio of replacement/silent mutations in HCV-associated HCC was higher in tumors than in the nontumor tissues. These results further confirm the conclusions reached in the HCV-associated B cell lymphoma, suggesting that some of the HCV-induced mutations were amplified and selected in the HCV-associated HCC. In contrast, HBV-associated HCC and HCC of nonviral origin had fewer mutations than HCV-associated HCC. These results suggest that HCV-induced mutations contribute to the development of B-cell lymphoma and HCC.
Discussion
In this study, we demonstrated that HCV infection induced a mutator phenotype, which involves enhanced mutations of many somatic genes. We propose that these mutations contributed to the eventual selection and amplification of certain deleterious mutations of protooncogenes or tumor-suppressor genes in tumors. However, the long latency period of HCV-associated malignancies suggests that multiple hits are required for HCV-associated oncogenesis. This interpretation is supported by the fact that the mutations observed in HCV-associated lymphoma had similar nucleotide substitution patterns to those of HCV-infected Raji or PBMC (Tables 2 and 4), and yet, only the lymphomas contained amplified mutations. Furthermore, the mutations detected in the HCV-associated lymphoma, but not in the unselected HCV-infected cells, had a replacement/silent mutation ratio higher than the expected ratio in the absence of selection. Similar findings were obtained for HCV-associated HCC. It is striking that other types of tumors, including lymphomas not associated with HCV, HBV-associated HCCs, and HCCs of nonviral origin, did not show such an amplification of protooncogene mutations. Thus, the ability of HCV to induce the mutator phenotype, with subsequently amplified mutations, is unique among oncogenic viruses. The heterogeneity of the Ig gene may perturb the immune response to HCV infection and contribute to the occurrence of B cell lymphomas.
The HCV-induced mutator phenotype can be explained by the abilities of HCV to cause DSBs and activate error-prone DNA polymerase ζ, polymerase ι and AID. Repair of DSBs by homologous recombination has been reported to result in an ≈100-fold increase in the rate of point mutations in the vicinity of the breaks in Saccharomyces cerevisiae (31). These mutations depend on the error-prone polymerase ζ, which has the DNA-damage bypass activity (32). Polymerase ι behaves as a dA·dT mutator in the middle of DNA templates but as a dG·dC mutator at their ends, when acting on a primer terminus with a long template overhang, with extraordinarily low fidelity (33). Polymerase ι also induces somatic hypermutation in Ig genes in the BL2 cell line (26). Both error-prone polymerases ζ and ι could be activated as a result of B cell receptor stimulation (23, 24). It is interesting that error catastrophe and lethal mutagenesis reported in several RNA viruses may extend to DNA error catastrophe, which has been described as “melting” of genetic information (34). Significantly, the HCV envelope protein E2 has been shown to bind to CD81, which is expressed on both B cells and hepatocytes (7), and to the B cell receptor of HCV-associated lymphoma (35), thus activating the intracellular signal transduction pathway. On the other hand, HCV-induced DNA breaks might be the result of reactive oxygen species; it has been reported that core, NS3 and NS5A proteins can induce reactive oxygen species (36). The enhanced DSBs may also explain the occurrence of apoptosis associated with hepatitis and chromosomal instability found in the B cells and hepatocytes of HCV-infected individuals. Furthermore, the ability of HCV to induce high mutation frequency of cellular genes suggests that HCV may cause tumor formation by a hit-and-run mechanism.
Supplementary Material
Acknowledgments
We thank Dr. M. Lieber and Dr. M. F. Goodman (University of Southern California, Los Angeles) for critical reading of our manuscript and Dr. C.-A. Reynaud (Institut National de la Santé et de la Recherche Médicale U373) for AID-/- and Pol ι-/- BL2 cells. This work was supported by National Institutes of Health Grant AI 40038.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HCV, hepatitis C virus; PBMC, peripheral blood mononuclear cell; VH, heavy chain of Ig; DSB, double-stranded DNA break; AID, activation-induced cytidine deaminase; HCC, hepatocellular carcinoma; LM-PCR, ligation-mediated PCR; siRNA; small interfering RNA.
References
- 1.Ferri, C., Caracciolo, F., Zignego, A. L., La Civita, L., Monti, M., Longombardo, G., Lombardini, F., Greco, F., Capochiani, E., Mazzoni, A., et al. (1994) Br. J. Haematol. 88, 392-394. [DOI] [PubMed] [Google Scholar]
- 2.Teramoto, T., Satonaka, K., Kitazawa, S., Fujimori, T., Hayashi, K. & Maeda, S. (1994) Cancer Res. 54, 231-235. [PubMed] [Google Scholar]
- 3.Huang, H., Fujii, H., Sankila, A., Mahler-Araujo, B. M., Matsuda, M., Cathomas, G. & Ohgaki, H. (1999) Am. J. Pathol. 155, 1795-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda, S., Shibata, M., Morishima, T., Harada, A., Nakao, A., Takagi, H. & Nagai, Y. (1992) Cancer 70, 2255-2259. [DOI] [PubMed] [Google Scholar]
- 5.Sung, V. M., Shimodaira, S., Doughty, A. L., Picchio, G. R., Can, H., Yen, T. S., Lindsay, K. L., Levine, A. M. & Lai, M. M. (2003) J. Virol. 77, 2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zignego, A. L., De Carli, M., Monti, M., Careccia, G., La Villa, G., Giannini, C., D'Elios, M. M., Del Prete, G. & Gentilini, P. (1995) J. Med. Virol. 47, 58-64. [DOI] [PubMed] [Google Scholar]
- 7.Pileri, P., Uematsu, Y., Campagnoli, S., Galli, G., Falugi, F., Petracca, R., Weiner, A. J., Houghton, M., Rosa, D., Grandi, G. & Abrignani, S. (1998) Science 282, 938-941. [DOI] [PubMed] [Google Scholar]
- 8.Silvestri, F., Pipan, C., Barillari, G., Zaja, F., Fanin, R., Infanti, L., Russo, D., Falasca, E., Botta, G. A. & Baccarani, M. (1996) Blood 87, 4296-4301. [PubMed] [Google Scholar]
- 9.Kitay-Cohen, Y., Amiel, A., Hilzenrat, N., Buskila, D., Ashur, Y., Fejgin, M., Gaber, E., Safadi, R., Tur-Kaspa, R. & Lishner, M. (2000) Blood 96, 2910-2912. [PubMed] [Google Scholar]
- 10.Ivanovski, M., Silvestri, F., Pozzato, G., Anand, S., Mazzaro, C., Burrone, O. R. & Efremov, D. G. (1998) Blood 91, 2433-2442. [PubMed] [Google Scholar]
- 11.Bachl, J., Carlson, C., Gray-Schopfer, V., Dessing, M. & Olsson, C. (2001) J. Immunol. 166, 5051-5057. [DOI] [PubMed] [Google Scholar]
- 12.Schlissel, M., Constantinescu, A., Morrow, T., Baxter, M. & Peng, A. (1993) Genes Dev. 7, 2520-2532. [DOI] [PubMed] [Google Scholar]
- 13.Pasqualucci, L., Migliazza, A., Fracchiolla, N., William, C., Neri, A., Baldini, L., Chaganti, R. S., Klein, U., Kuppers, R., Rajewsky, K. & Dalla-Favera, R. (1998) Proc. Natl. Acad. Sci. USA 95, 11816-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Migliazza, A., Martinotti, S., Chen, W., Fusco, C., Ye, B. H., Knowles, D. M., Offit, K., Chaganti, R. S. & Dalla-Favera, R. (1995) Proc. Natl. Acad. Sci. USA 92, 12520-12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorner, T., Brezinschek, H. P., Brezinschek, R. I., Foster, S. J., Domiati-Saad, R. & Lipsky, P. E. (1997) J. Immunol. 158, 2779-2789. [PubMed] [Google Scholar]
- 16.Rogozin, I. B. & Kolchanov, N. A. (1992) Biochim. Biophys. Acta 1171, 11-18. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualucci, L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature 412, 341-346. [DOI] [PubMed] [Google Scholar]
- 18.Goyenechea, B., Klix, N., Yelamos, J., Williams, G. T., Riddell, A., Neuberger, M. S. & Milstein, C. (1997) EMBO J. 16, 3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, W., Bardwell, P. D., Woo, C. J., Poltoratsky, V., Scharff, M. D. & Martin, A. (2001) Int. Immunol. 13, 1175-1184. [DOI] [PubMed] [Google Scholar]
- 20.Bross, L., Fukita, Y., McBlane, F., Demolliere, C., Rajewsky, K. & Jacobs, H. (2000) Immunity 13, 589-597. [DOI] [PubMed] [Google Scholar]
- 21.Papavasiliou, F. N. & Schatz, D. G. (2000) Nature 408, 216-221. [DOI] [PubMed] [Google Scholar]
- 22.Kong, Q. & Maizels, N. (2001) Genetics 158, 369-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zan, H., Komori, A., Li, Z., Cerutti, A., Schaffer, A., Flajnik, M. F., Diaz, M. & Casali, P. (2001) Immunity 14, 643-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poltoratsky, V., Woo, C. J., Tippin, B., Martin, A., Goodman, M. F. & Scharff, M. D. (2001) Proc. Natl. Acad. Sci. USA 98, 7976-7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gearhart, P. J. & Wood, R. D. (2001) Nat. Rev. Immunol. 1, 187-192. [DOI] [PubMed] [Google Scholar]
- 26.Faili, A., Aoufouchi, S., Flatter, E., Gueranger, Q., Reynaud, C. A. & Weill, J. C. (2002) Nature 419, 944-947. [DOI] [PubMed] [Google Scholar]
- 27.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 28.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296, 2033-2036. [DOI] [PubMed] [Google Scholar]
- 29.Buermeyer, A. B., Deschenes, S. M., Baker, S. M. & Liskay, R. M. (1999) Annu. Rev. Genet. 33, 533-564. [DOI] [PubMed] [Google Scholar]
- 30.Chang, B. & Casali, P. (1994) Immunol. Today 15, 367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strathern, J. N., Shafer, B. K. & McGill, C. B. (1995) Genetics 140, 965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holbeck, S. L. & Strathern, J. N. (1997) Genetics 147, 1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank, E. G., Tissier, A., McDonald, J. P., Rapic-Otrin, V., Zeng, X., Gearhart, P. J. & Woodgate, R. (2001) EMBO J. 20, 2914-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domingo, E. (2000) Virology 270, 251-253. [DOI] [PubMed] [Google Scholar]
- 35.Quinn, E. R., Chan, C. H., Hadlock, K. G., Foung, S. K., Flint, M. & Levy, S. (2001) Blood 98, 3745-3749. [DOI] [PubMed] [Google Scholar]
- 36.Lai, M. M. (2002) Gastroenterology 122, 568-571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.