Abstract
Background
Lutzomyia umbratilis (a probable species complex) is the main vector of Leishmania guyanensis in the northern region of Brazil. Lutzomyia anduzei has been implicated as a secondary vector of this parasite. These species are closely related and exhibit high morphological similarity in the adult stage; therefore, they have been wrongly identified, both in the past and in the present. This shows the need for employing integrated taxonomy.
Methods
With the aim of gathering information on the molecular taxonomy and evolutionary relationships of these two vectors, 118 sequences of 663 base pairs (barcode region of the mitochondrial DNA cytochrome oxidase I – COI) were generated from 72 L. umbratilis and 46 L. anduzei individuals captured, respectively, in six and five localities of the Brazilian Amazon. The efficiency of the barcode region to differentiate the L. umbratilis lineages I and II was also evaluated. The data were analyzed using the pairwise genetic distances matrix and the Neighbor-Joining (NJ) tree, both based on the Kimura Two Parameter (K2P) evolutionary model.
Results
The analyses resulted in 67 haplotypes: 32 for L. umbratilis and 35 for L. anduzei. The mean intra-specific genetic distance was 0.008 (0.002 to 0.010 for L. umbratilis; 0.008 to 0.014 for L. anduzei), whereas the mean interspecific genetic distance was 0.044 (0.041 to 0.046), supporting the barcoding gap. Between the L. umbratilis lineages I and II, it was 0.009 to 0.010. The NJ tree analysis strongly supported monophyletic clades for both L. umbratilis and L. anduzei, whereas the L. umbratilis lineages I and II formed two poorly supported monophyletic subclades.
Conclusions
The barcode region clearly separated the two species and may therefore constitute a valuable tool in the identification of the sand fly vectors of Leishmania in endemic leishmaniasis areas. However, the barcode region had not enough power to separate the two lineages of L. umbratilis, likely reflecting incipient species that have not yet reached the status of distinct species.
Keywords: Leishmania vectors, Barcode region, Molecular entomology, Speciation
Background
Phlebotomine sand flies (Diptera: Psychodidae) are vectors of human leishmaniasis, a disease caused by trypanosomatids of the genus Leishmania. Leishmania infection is characterized by a species-specific pathology, varying from cutaneous lesions to the potentially fatal visceral form [1,2]. This disease occurs in the tropical, subtropical and Mediterranean regions of the world and its global burden has been estimated to be ~500,000 cases of visceral leishmaniasis (VL) and ~1.1-1.5 million cases of cutaneous leishmaniasis (CL) per year [2,3]. Out of the six genera belonging to the subfamily Phlebotominae, only Lutzomyia and Phlebotomus include the vectors of human leishmaniasis. The former is restricted to the New World, where approximately 32 out of more than 500 species described [4] are implicated as vectors, whereas the latter is found in the Old World [1,5]. In the New World, Lutzomyia (Lutzomyia) longipalpis (likely a species complex) is recognized as the main vector of visceral leishmaniasis in the Neotropics [6,7], whereas Lutzomyia (Nyssomyia) umbratilis, Lutzomyia (Nyssomyia) flaviscutellata, Lutzomyia (Nyssomyia) whitmani sensu lato, Lutzomyia (Nyssomyia) intermedia sensu lato, Lutzomyia (Nyssomyia) neivai, Lutzomyia (Nyssomyia) olmeca, Lutzomyia (Lutzomyia) gomezi, Lutzomyia (Psychodopygus) wellcomei, Lutzomyia (Viannamyia) furcata, among others, are important vectors of cutaneous leishmaniasis [7].
Lutzomyia umbratilis is a highly anthropophilic sand fly that has been appointed as the main vector of Leishmania guyanensis, the etiological agent of cutaneous leishmaniasis (CL) in northern Brazil [8-14] and probably in other countries of northern South America [15-18]. Lutzomyia anduzei, its probable sister taxon, is also an anthropophilic species [9,19]. Arias and Freitas [11] isolated Leishmania spp. (likely Le. guyanensis) from wild-caught L. anduzei females collected near the city of Manaus, in the State of Amazonas, Brazil. Similar findings were also reported in the State of Pará, Brazil [9]. Hence, the sporadic records of infections by L. anduzei suggest that it could be a secondary vector in the Brazilian Amazon [7,9,19]. There are actually only a few studies on L. anduzei, and the available data are limited to species diversity and abundance in areas of leishmaniasis transmission. Consequently, only little is known about the accurate geographic distribution, ecology and genetics of this species and its efficiency as vector of this parasite. Additionally, the data published regarding L. anduzei before L. umbratilis had been described may in fact refer to the latter [8,20].
Lutzomyia anduzei and L. umbratilis are two closely related species exhibiting high morphological similarity in the adult stage [19,21]. Both species are geographically distributed in northern South America, with extensive overlapping areas [19]. Lutzomyia anduzei was described by Rozeboom [22], who used specimens (females) from Gran Sabana in Venezuela. Lutzomyia umbratilis was described by Ward and Fraiha [23], based on specimens captured in the Jari River region, State of Pará, Brazil. Because of the high morphological similarity between them, L. umbratilis has been wrongly identified as L. anduzei in the past [8,20]. This misidentification may still occur today, compromising the accurate identification of the vectors involved. Along with the difficulty of separating these species, recent studies have demonstrated that L. umbratilis may represent a cryptic species complex of at least two distinct or incipient species which are separated across opposite banks of the largest rivers in the central region of the Brazilian Amazon [24,25] and are probably different in vector competence [11]. Therefore, the use of molecular markers combined with morphology (integrated taxonomy) could make the identification of L. umbratilis and L. anduzei more accurate and differentiate the two lineages within L. umbratilis, which in turn is relevant for understanding the epidemiology and the distinct patterns of transmission of Le. guyanensis, thus facilitating the vector control efforts in this region.
Main morphological and chromatic differences between L. umbratilis and L. anduzei
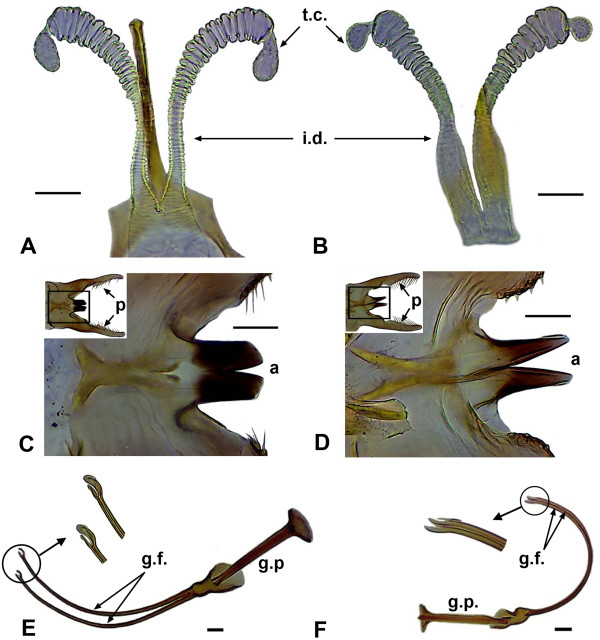
The morphological differences between L. umbratilis and L. anduzei are subtle [19, R. A. Freitas, personal communication] (Figure 1). The adults of these species can show slight differences in color, with L. umbratilis exhibiting a light brown body and L. anduzei an almost pale coloration. The females of L. umbratilis have a well developed spermatheca compared to those of L. anduzei. In the latter, the terminal tubercle of this organ (spermatheca head) is thinner than the terminal ring. In L. umbratilis, the common and individual ducts show sharper transverse striations (Figure 1A), whereas in L. anduzei these striations are weak and fade toward the common duct (Figure 1B). Furthermore, in L. anduzei there is an evident narrowing of individual ducts at the junction with the body. As for the males, in L. umbratilis, the aedeagus apex is truncated (Figure 1C), whereas in L. anduzei it is slender (Figure 1D). The apex of the genital filament is slightly bifid in L. umbratilis males (Figure 1E) but bezel-shaped in L. anduzei males (Figure 1F).
Figure 1.
Female and male internal and external genitals of Lutzomyia umbratilis and Lutzomyia anduzei. A and B: Spermathecae of L. umbratilis and L. anduzei females, respectively; C and D: Aedeagi of L. umbratilis and L. anduzei males, respectively (upper left corners, showing the position of aedeagi regarding the parameres); E and F: Genital filaments apex of L. umbratilis and L. anduzei males, respectively. t.t.: terminal tubercles; i.d.: individual ducts; p: parameres; a: aedeagus; g.f.: genital filaments; g.p.: genital pumps. The arrows indicate in highlight the apex of the genital filaments. Bar = 20 μm.
The barcode region comprising 648 base pairs (bp) at the 5′ end of the mitochondrial DNA (mtDNA) cytochrome oxidase I - COI has emerged as the standard region for the identification of a wide variety of invertebrate and vertebrate species [26], although failures have been reported due to non-monophyly [27]. This region has shown to be exceptionally promising for species-level identification in insects [28-30], including Anopheles[31,32] and other mosquitoes [33]. However, the barcode region has so far been little investigated in sand flies in general [34] and not at all in sand flies from Brazil.
In the Brazilian Amazon region, although there are areas of high transmission of leishmaniasis [35] and of sand fly vectors presenting morphological variations as well as isomorphic taxa [7], studies on molecular taxonomy, phylogenetic relationships, molecular evolution and population genetics are scarce [5,25,36,37], and nothing is known about the genetic diversity of these sand flies, which could contribute to their management. Hence, the aim of this study was to seek information on the molecular taxonomy and to preliminarily clarify the evolutionary relationships of L. umbratilis and L. anduzei using the DNA barcode region to help in their accurate identification and, thereby better determine the role of each one in the leishmaniasis foci. Additionally, this study assessed the efficiency of this fragment in differentiating the two lineages or incipient L. umbratilis species, previously described by Scarpassa and Alencar [25].
Methods
Samples and collection sites
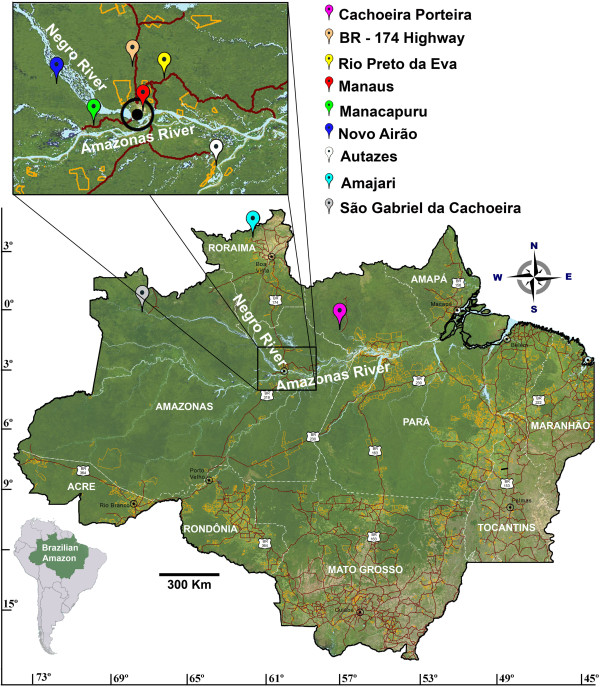
Lumbratilis umbratilis adults were collected in six localities of the Brazilian Amazon region (Table 1, Figure 2), including Cachoeira Porteira in the State of Pará; km 43 of the BR-174 Highway; km 65 of the AM-010 Highway in the municipality of Rio Preto da Eva; a fragment of urban forest in Manaus; km 60 of the AM-070 Highway in the municipality of Manacapuru; and km 60 and km 70 of the AM-352 Highway in the municipality of Novo Airão, in the State of Amazonas, as described in Scarpassa and Alencar [25]. The collection sites of Cachoeira Porteira, BR-174 Highway, Rio Preto da Eva and Manaus are situated on the left bank of the Negro River and north of the Amazonas River, and the samples from these sites were named lineage I. The localities of Manacapuru and Novo Airão are situated on the right bank of the Negro River and the samples from these sites were named lineage II [25]. Lutzomyia anduzei adults were captured in five locations of the Brazilian Amazon, comprising four sites in the State of Amazonas (urban forest fragment in Manaus; municipality of Autazes; km 60 and km 70 of the AM-352 Highway in the municipality of Novo Airão; municipality of São Gabriel da Cachoeira), and one in the State of Roraima (Amajari) (Table 1, Figure 2). The two species were captured sympatrically in Manaus and Novo Airão. All information regarding collection data, coordinates and sample size of the species is displayed in Table 1.
Table 1.
Localities data and haplotype frequency of the Lutzomyia umbratilis and Lutzomyia anduzei samples from the Brazilian Amazon
| Species | Localities, State | Co-ordinates (Lat., Long.) | N | Haplotype frequency |
|---|---|---|---|---|
|
Lutzomyia umbratilis |
Cachoeira Porteira, Oriximiná, Pará |
1° 28′ S; 56° 22′ W |
12 |
H1(1), H2(2), H3(1), H4(1), H5(1), H6(1), H7(1), H8(1), H9(1), H10(2) |
| |
Km 43 of BR-174 Highway, Amazonas |
2° 36′ S; 60° 02′ W |
12 |
H11(1), H12(1), H13(6), H14(1), H15(1), H16(1), H17(1) |
| |
Rio Preto da Eva, Amazonas |
2° 43′ S; 59° 47′ W |
13 |
H12(5), H13(5), 18(1), H19(1), H20(1) |
| |
Manaus, Amazonas |
3° 04′ S; 59° 57′ W |
12 |
H13(3), H15(1), H19(4), H21(1), H22(1), H23(1), H24(1) |
| |
Manacapuru, Amazonas |
3° 14′ S; 60° 31′ W |
13 |
H25(2), H26(8), H27(1), H28(1), H29(1) |
| |
Novo Airão, Amazonas |
2° 47′ S; 60° 55′ W |
10 |
H26(5), H29(2), H30(1), H31(1), H32(1) |
|
Subtotal |
|
|
72 |
|
|
Lutzomyia anduzei |
Manaus, Amazonas |
3° 04′ S; 59° 57′ W |
10 |
H33(1), H34(2), H35(1), H36(1), H37(1), H38(1), H39(1), H40(1), H41(1) |
| |
Autazes, Amazonas |
3º 42′ S; 59º 07′ W |
2 |
H42(1), H43(1) |
| |
Novo Airão, Amazonas |
2° 47′ S; 60° 55′ W |
13 |
H42(2), H44(6), H45(2), H46(1), H47(1), H48(1) |
| |
Amajari, Roraima |
3º 46′ N; 61º 44′ W |
8 |
H49(1), H50(1), H51(2), H52(1), H53(1), H54(1), H55(1) |
| |
São Gabriel da Cachoeira, Amazonas |
0º 03′ S; 66º 59′ W |
13 |
H41(1), H56(1), H57(1), H58(1), H59(1), H60(1), H61(1), H62(1), H63(1), H64(1), H65(1), H66(1), H67(1) |
|
Subtotal |
|
|
46 |
|
| TOTAL | 118 |
N sample size, H1 to H32 Haplotypes observed in the samples of Lutzomyia umbratilis, H33 to H67 Haplotypes observed in the samples of Lutzomyia anduzei. Inside the parentheses is the number of individuals observed for each haplotype. The underlined haplotypes are shared among localities.
Figure 2.
Collection sites of Lutzomyia umbratilis and Lutzomyia anduzei samples from the Brazilian Amazon.Lutzomyia umbratilis: Cachoeira Porteira, BR-174 Highway, Rio Preto da Eva, Manaus, Manacapuru and Novo Airão. Lutzomyia anduzei: Manaus, Autazes, Novo Airão, Amajari and São Gabriel da Cachoeira.
A 663 base pairs (bp) fragment (DNA barcode) was generated from 72 L. umbratilis and 46 L. anduzei specimens captured in the Brazilian Amazon region (Table 1). Adults of both species were collected with CDC (Centers for Disease Control) miniature light traps and with aspirators placed on the bases of tree trunks, as described in Scarpassa and Alencar [25]. This study and catch protocol was reviewed and approved by the Institutional Review Board of the National Institute of Amazonian Research (INPA), of the Brazilian Ministry of Science, Technology and Innovation (MCTI). The sample collections were authorized by the Brazilian Institute for the Environment and Renewable Natural Resources (IBAMA) and by the System of Authorization and Information in Biodiversity (SISBIO), license number 12733–1 for the collection of L. umbratilis and L. anduzei from Amazonas and Roraima States, Brazil, and license number 14054–5 for the collection of L. umbratilis from the Cachoeira Porteira, State of Pará, Brazil. After collection, the sand flies were preserved in 95% ethanol and stored at −20°C until processing for DNA extraction. Morphological identification of the species was based on the internal and external genitalia of males and females [19], using an optical microscope at a magnification of 10x, 40x and 100x (Carl Zeiss, Primo Star, 3119000947, Germany). The male and female genitalia of L. umbratilis and L. anduzei shown in Figure 1 are from specimens collected in Cachoeira Porteira (State of Pará) and Autazes (State of Amazonas), respectively (see Table 1).
DNA extraction, PCR and sequencing
Total genomic DNA was extracted individually from whole sand flies using the phenol and chloroform method [38], resuspended in 20 μL of 1x TE buffer (10 mM Tris-Cl pH 8.0, 1 mM EDTA pH 8.0) or sterile water, and then stored at −80°C until amplification by polymerase chain reaction (PCR). The primers (10 μM) used in the amplification reaction were LCO 1490 and HCO 2198 [39]. All PCR reactions included negative controls. The PCR products were visualized on 1% agarose gels under UV light, purified with PEG and both DNA strands were sequenced in an ABI 3130 XL Automated DNA Sequencer (Applied Biosystems), available at INPA (Manaus, Brazil).
Data analysis
All sequences were automatically aligned with the Clustal W and then compiled and edited in BIOEDIT v. 7.0.8.0 [40]. Following, sequence identity searches were performed using BLAST (Basic Local Alignment Search Tool), available at http://www.ncbi.nlm.nih.gov/BLAST/. The haplotype numbers for each species, number of polymorphic sites and other measures of genetic diversity were estimated using the DNASP v. 5.10 [41] and TCS v. 1.21 [42] softwares. The nucleotide frequencies, numbers of transitions and transversions, the number of variable sites among haplotypes, and intra and interspecific genetic distances (sequence divergence), based on the Kimura Two Parameter (K2P) evolutionary model, were calculated in MEGA v. 5.1 [43]. The phylogenetic relationship of the haplotypes was inferred using a Neighbor Joining (NJ) tree, with 2,000 replicates, and constructed based on the K2P distances. The tree was rooted using sequences of Anopheles triannulatus sensu lato (Diptera: Culicidae: Anophelinae) (V. M. Scarpassa, unpublished data) as outgroup.
The 663 bp barcode region analyzed in this study overlapped between sites 283 and 663 with those of L. umbratilis analyzed by Scarpassa and Alencar [25] who studied a 1181 bp fragment of the COI gene. Haplotypes sequences are deposited in GenBank under the accession numbers KF467531 to KF467597.
Results
The DNA barcode region analyzed in this study comprised 663 bp. All alignments were unambiguous and no insertions or deletions were detected in the dataset. The amino acid translations showed no stop codons, ensuring that the dataset did not constitute nuclear mitochondrial DNA sequences (NUMTs). The amino acid reading frame starts at the first base of the primer-edited sequences. All sequences, including those from L. umbratilis and L. anduzei (n=118), yielded 75 (11.31%) variable sites and 53 (8%) of these were parsimoniously informative. Transitions were more common than transversions (Table 2). The average nucleotide composition was similar in the two species, with an overall average of 38.2% for Thymine (T), 30.3% for Adenine (A), 15.4% for Cytosine (C) and 16.2% for Guanine (G). The A+T content were rich (68.5%), as observed in other insects.
Table 2.
Mean of nucleotide frequencies, and transitions and transversions rate of the Lutzomyia umbratilis, Lutzomyia anduzei and total
| Identical pairs | TS (%) | TV (%) | T | C | A | G | Total bases | |
|---|---|---|---|---|---|---|---|---|
|
L. umbratilis |
|
|
|
|
|
|
|
|
| Average |
659 |
4 |
0 |
38.2 |
15.2 |
30.5 |
16.1 |
663 |
| 1st |
221 |
0 |
0 |
28 |
14.9 |
28 |
29.4 |
221 |
| 2nd |
221 |
0 |
0 |
43 |
26.2 |
13.6 |
17.6 |
221 |
| 3rd |
217 |
4 |
0 |
44 |
4.6 |
50 |
1.1 |
221 |
|
L. anduzei |
|
|
|
|
|
|
|
|
| Average |
656 |
6 |
1 |
38.2 |
15.6 |
29.9 |
16.3 |
663 |
| 1st |
220 |
1 |
0 |
28 |
14.5 |
28 |
29.4 |
221 |
| 2nd |
221 |
0 |
0 |
43 |
26.2 |
13.6 |
17.6 |
221 |
| 3rd |
215 |
5 |
1 |
44 |
6 |
48.2 |
1.8 |
221 |
|
Total general |
|
|
|
|
|
|
|
|
| Average |
647 |
12 |
4 |
38.2 |
15.4 |
30.3 |
16.2 |
663 |
| 1st |
220 |
1 |
0 |
28 |
14.8 |
28 |
29.4 |
221 |
| 2nd |
221 |
0 |
0 |
43 |
26.2 |
13.6 |
17.6 |
221 |
| 3rd | 206 | 11 | 4 | 44 | 5.1 | 49.3 | 1.4 | 221 |
TS number of transitions, TV number of transversions.
One hundred and eighteen sequences resulted in a total of 67 haplotypes (Table 1), reflecting a high haplotype diversity for the two species. Out of 32 haplotypes (H1-H32) observed for 72 L. umbratilis specimens, six (H12, H13, H15, H19, H26, H29) were common to the analyzed localities. The sample of Cachoeira Porteira, situated at the north of the Amazonas River (lineage I), however, did not share any haplotype with other localities, likely due to the geographic distance. The samples from the three other locations (BR-174 Highway, Rio Preto da Eva and Manaus), situated on the left bank of the Negro River (also lineage I), shared the haplotypes H12, H13, H15 and H19, suggesting gene flow. The samples from Manacapuru and Novo Airão, situated on the right bank of the Negro River (lineage II), shared the haplotypes H26 and H29. H13 was the most frequent in lineage I, whereas the H26 was the most frequent in lineage II. No haplotypes were shared between samples from opposite river banks (lineages I and II), as previously detected by Scarpassa and Alencar [25].
The analysis of 46 L. anduzei specimens yielded 35 haplotypes (H33-H67) (Table 1). Of these, only two (H41 and H42) were shared among samples. The samples from Manaus and São Gabriel da Cachoeira, geographically distant, shared H41, whereas the samples from Autazes and Novo Airão shared H42. The sample from Amajari did not share any haplotype with another locality. Additional file 1 shows the nucleotide substitutions among 67 haplotypes, where ten fixed mutations (four transitions, at positions 366, 369, 538 and 660; six transversions, at positions 45, 102, 222, 459, 528 and 558) between L. umbratilis and L. anduzei were observed. Lutzomyia umbratilis lineages I and II, however, had one fixed mutation (a T↔C transition at position 21) and three almost fixed mutations (all A↔G transitions, at positions 540, 567 and 624). Table 3 portrays the summary of genetic diversity measures for L. umbratilis and L. anduzei, which were higher in the latter.
Table 3.
Summary of genetic diversity measures of Lutzomyia umbratilis and Lutzomyia anduzei
| Summary statistics | L. umbratilis | L. anduzei |
|---|---|---|
| Nº of sequences (N) |
72 |
46 |
| Nº of polymorphic sites (S) |
28 |
48 |
| Total Nº of mutations |
29 |
51 |
| Nº of haplotypes |
32 |
35 |
| Haplotype diversity (h) ± SD |
0.921 ± 0.019 |
0.979 ± 0.012 |
| Nucleotide diversity (π) ± SD |
0.00602 ± 0.00025 |
0.01070 ± 0.00096 |
| Average nucleotide differences (k) | 3.993 | 7.095 |
SD Standard deviation.
Table 4 shows the pairwise intra and interspecific genetic distances (K2P) of the two species. The average intraspecific genetic distance was 0.008, ranging from 0.002 to 0.010 in L. umbratilis and from 0.008 to 0.014 in L. anduzei. The mean interspecific genetic distance between L. umbratilis and L. anduzei was ~six-fold higher (0.044 ± 0.007; varying from 0.041 to 0.046), supporting the barcoding gap. Within species L. umbratilis, the genetic distance between lineages I and II varied from 0.009 to 0.010, corroborating the previous observations of Scarpassa and Alencar [25].
Table 4.
Mean intra and interspecific genetic distances, based on the K2P, of the Lutzomyia umbratilis and Lutzomyia anduzei samples
| Samples | N | CPumbr | BRumbr | RPumbr | MNumbr | MCumbr | NAumbr | MNandu | AUandu | NAandu | AJandu | SGandu |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPumbr |
12 |
0.005 |
|
|
|
|
|
|
|
|
|
|
| BRumbr |
12 |
0.005 |
0.003 |
|
|
|
|
|
|
|
|
|
| RPumbr |
13 |
0.005 |
0.002 |
0.002 |
|
|
|
|
|
|
|
|
| MNumbr |
12 |
0.005 |
0.004 |
0.004 |
0.003 |
|
|
|
|
|
|
|
| MCumbr |
13 |
0.009 |
0.010 |
0.009 |
0.009 |
0.002 |
|
|
|
|
|
|
| NAumbr |
10 |
0.009 |
0.010 |
0.009 |
0.009 |
0.002 |
0.001 |
|
|
|
|
|
| MNandu |
10 |
0.042 |
0.043 |
0.043 |
0.043 |
0.044 |
0.044 |
0.007 |
|
|
|
|
| AUandu |
2 |
0.041 |
0.041 |
0.041 |
0.042 |
0.042 |
0.042 |
0.008 |
0.011 |
|
|
|
| NAandu |
13 |
0.045 |
0.045 |
0.045 |
0.045 |
0.046 |
0.046 |
0.012 |
0.011 |
0.014 |
|
|
| AJandu |
8 |
0.043 |
0.044 |
0.044 |
0.044 |
0.044 |
0.044 |
0.009 |
0.010 |
0.014 |
0.010 |
|
| SGandu | 13 | 0.042 | 0.044 | 0.043 | 0.043 | 0.044 | 0.044 | 0.008 | 0.011 | 0.014 | 0.010 | 0.007 |
CP Cachoeira Porteira, BR BR-174 Highway, RP Rio Preto da Eva, MN Manaus, MC Manacapuru, NA Novo Airão, AU Autazes, AJ Amajari, SG São Gabriel da Cachoeira, umbrLutzomyia umbratilis, anduLutzomyia anduzei, N sample sizes. Values in bold indicate genetic distances between haplotypes intra-sample. The underlined values are related to comparisons between samples of the lineages I and II of Lutzomyia umbratilis.
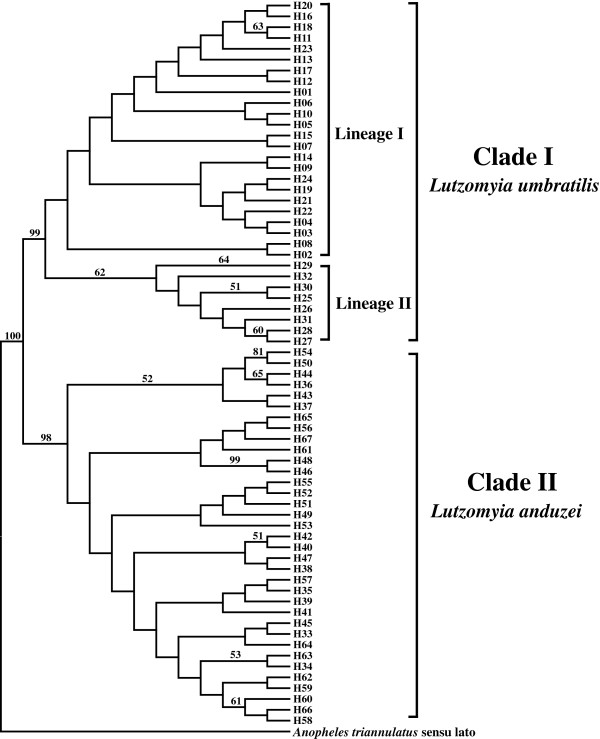
The phylogenetic relationships of haplotypes visualized in the NJ tree (Figure 3), using K2P, supported monophyly for L. umbratilis and L. anduzei, with bootstrap values of 99% and 98%, respectively, indicating that these species can be recognized by their barcode region. Clade I, comprising L. umbratilis lineages I and II, formed two monophyletic subclades, but had low bootstrap support. Lineage I haplotypes (H1 to H24) were clustered in several subdivisions, whereas all haplotypes of lineage II (H25 to H32) were grouped in another subclade, with bootstrap support of 62%. Clade II, comprising L. anduzei haplotypes (H33 to H67), consisted of smaller subdivisions, regardless of the geographic origin of the haplotypes (Table 1), but was also poorly supported.
Figure 3.
Neighbor Joining (NJ) tree of the 67 haplotypes observed in Lutzomyia umbratilis and Lutzomyia anduzei, inferred under the Kimura 2 Parameter (K2P) model. Numbers above branch represent bootstrap supports. Values lower than 50 are not represented on branch. Sequences of Anopheles triannulatus sensu lato were used as outgroup.
Discussion
The phylogenetic relationships of haplotypes suggest that L. umbratilis and L. anduzei are monophyletic, with strongly supported clades; therefore, the barcode region can be used for differentiating these two species. The genetic distance between them was rather small (4.4%) and similar to the values found between members of species complexes in mosquitoes [31,32,44]. Barcode region analysis provided resolution for 13 sand fly species of genus Lutzomyia, with the probable existence of cryptic species in two of them [34]; the authors, however, did not mention the genetic distances among the species studied. The genetic distance observed in this study is supported by the great morphological similarity between L. umbratilis and L. anduzei[19]. Scarpassa and Alencar [25] observed a higher value (5.8%) for the genetic distance between L. umbratilis and L. anduzei. These differences may be explained by the fact that the COI fragment at the 3′ end analyzed by these authors was more variable, resulting in higher differentiation. Additionally, in this study the most of the transitions and transversions observed between L. umbratilis and L. anduzei occurred in the third codon position, except for one transition, located in the first codon position (Table 2). Taken together, these findings may suggest that L. umbratilis and L. anduzei are either closely related species or of a recent common origin. However, our data brought no evidence of genetic introgression between them, at least at the mitochondrial DNA level.
The phylogenetic relationships observed in this study for the L. umbratilis lineages I and II indicated two monophyletic subclades, but were poorly supported statistically. The fragment of end 3′ analyzed by Scarpassa and Alencar [25] generated trees with better resolution and two moderately supported monophyletic clades, likely due to its higher variation level. On the other hand, biological and morphological differentiation [24,45] and probable differences in vector competence [11] have been described for lineages I and II. In contrast, preliminary data obtained with nuclear markers are indicating few differences between the two lineages (V. M. Scarpassa and R. B. Alencar, unpublished data), which could be the result of recent speciation, where the differences are visible only in mitochondrial genes.
In the present study, the value of the genetic distance between L. umbratilis lineages I and II was low (0.9-1.0%), identical to the one found previously by Scarpassa and Alencar [25]. This distance falls within the range of intraspecific nucleotide differences, which has been less than 2% among Diptera species for the barcode region [31], but there is an exception [46]. However, the absence of shared haplotypes detected in the present and in previous studies [25] may indicate lack of gene flow between lineages and may be an indicator of genetic discontinuity. Species that have diverged very recently are expected to share ancestral variation in high proportions, a situation that may confound their phylogenetic reconstruction; hence, the fixed differences between these species may be evidenced only in genes involved in the speciation process [47,48]. Taken together, the low genetic distance and the poorly supported monophyletic clades observed in the present study, combined with the previous results, may suggest that two lineages of L. umbratilis are incipient species [49]. Whether or not these lineages consist of cryptic species within L. umbratilis requires further inquiry. Moreover, this differentiation between two L. umbratilis lineages, although low, could probably affect the genes that control vector competence and consequently lead to distinct patterns of Le. guyanensis transmission in the central region of the Brazilian Amazon, a hypothesis initially raised by Arias and Freitas [11]. Altogether, these data allows us to propose that the populations belonging to lineage I (left bank of the Negro River and north of the Amazonas River) could be susceptible to the development (i. e., could be vectors) of Le. guyanensis, whereas the populations belonging to lineage II (right bank of the Negro River and south of the Amazonas River) are likely not to be involved in the transmission (not vectors) or could be less susceptible to the development of Le. guyanensis. However, further studies, including entomological parameters and transmission dynamics (anthropophily and natural infection rate of samples from the field) and experimental infections of these populations are needed to either support or refute this hypothesis.
The findings of this study (Tables 1 and 3) indicated that L. anduzei has a greater genetic diversity than its closely related species, L. umbratilis, at least with regard to the barcode region. Out of the 35 haplotypes observed in L. anduzei, only two were shared among the samples analyzed, suggesting restricted gene flow between them. Curiously, the genetic distance observed in L. anduzei were slightly larger than those detected between lineages I and II of L. umbratilis, probably indicating that the L. anduzei populations consist of a significant genetic structure or display a large effective population size (Ne) [50] or highly divergent haplotypes [51]. In fact, highly divergent haplotypes were detected in the sample from Novo Airão, consequently the comparisons involving this sample yielded the highest genetic distance (0.012, 0.014) and intra-sample (0.014) values (Table 4).
Similar to what was found for L. umbratilis in the present and a previous study [25], no haplotype was shared among the L. anduzei samples from opposite banks of the Negro and Amazonas Rivers. This raises the hypothesis that the large Rivers of the Amazon region may act as possible barriers to the sand fly species, as already discussed by Scarpassa and Alencar [25]. All these findings, including the high haplotype diversity with a great number of unique haplotypes and the strong evidence of genetic structure, demonstrate the need of population genetics studies in L. anduzei across its range.
Conclusions
This is the first study of molecular taxonomy in two Leishmania vectors, L. umbratilis and L. anduzei, from the Brazilian Amazon region. We consider the COI barcode region to be a robust marker for differentiating sand fly species, even those closely related, and it may constitute a valuable tool for epidemiologic studies and for leishmaniasis control programs throughout South America. The barcode region, however, had not enough power to separate the two lineages of L. umbratilis, which may indicate that they are incipient species that have not yet reached the status of distinct species [49].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VMS: Designed the experiment, generated the results, analyzed the data, and wrote the manuscript. RBA: Captured the sand flies in the field, identified the samples, generated and processed morphological analyses and revised the manuscript. All authors read and approved the final version of the manuscript.
Supplementary Material
Variable sites of the 67 haplotypes observed for Lutzomyia umbratilis and Lutzomyia anduzei.
Contributor Information
Vera Margarete Scarpassa, Email: vera@inpa.gov.br.
Ronildo Baiatone Alencar, Email: ronildo@inpa.gov.br.
Acknowledgments
This study was funded by MCTI/INPA and by grants from the National Council of Scientific and Technological Development (CNPq) to Toby Vincent Barrett (process number 409058/2006-8) and to Vera Margarete Scarpassa (process number 575482/2008-7), from the Amazonas Research Foundation (FAPEAM) to Vera Margarete Scarpassa (process numbers 1501/2008 and 3111/2012), from the INCT-CENBAM/CNPq/FAPEAM/INPA to William Ernest Magnusson, and from PRO-EQUIPAMENTO/CAPES, Brazil. The authors thank the team of the Thematic Laboratory of Molecular Biology at INPA for their help in injecting samples into the DNA Sequencer.
References
- Lane RP. In: Medical insects and arachnids. Lane RP, Crosskey RW, editor. London: Chapman and Hall; 1993. Sandflies; pp. 78–119. [Google Scholar]
- WHO. First WHO report on neglected tropical diseases: working to overcome the global impact of neglected tropical diseases. Geneva: World Health Organization; 2010. p. 172. Available at: http://www.who.int/neglected_diseases. [Google Scholar]
- Reithnger R. Leishmaniases’ burden of disease: ways forward for getting from speculation to reality. PLoS Negl Trop Dis. 2008;2:e285. doi: 10.1371/journal.pntd.0000285. doi:10.1371/journal.pntd.0000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro PHF, Galati EAB. Checklist dos Phlebotominae (Diptera, Psychodidae) do Estado de São Paulo, Brasil, com comentários sobre sua distribuição geográfica. Biota Neotrop. 2011;11 http://www.biotaneotropica.org.br/v11n1a/pt/abstract?inventory +bn0361101a2011. [Google Scholar]
- Mazzoni CJ, Gomes CA, Souza NA, Queiroz RG, Justiniano SCB, Ward RD, Kyriacou CP, Peixoto AA. Molecular evolution of the period gene in sandflies. J Mol Evol. 2002;55:553–562. doi: 10.1007/s00239-002-2351-z. [DOI] [PubMed] [Google Scholar]
- Lainson R, Ward RD, Shaw JJ. Experimental transmission of Leishmania chagasi causative agent of neotropical visceral leishmaniasis by the sandfly Lutzomyia longipalpis. Nature. 1977;266:628–630. doi: 10.1038/266628a0. [DOI] [PubMed] [Google Scholar]
- Lainson R. Espécies neotropicais de Leishmania: uma breve revisão histórica sobre sua descoberta, ecologia e taxonomia. Rev Pan-Amaz Saude. 2010;1(2):13–32. [Google Scholar]
- Lainson R, Ward D, Shaw JJ. Cutaneous leishmaniasis in North Brazil: Lutzomyia anduzei as a major vector. Trans R Soc Trop Med Hyg. 1976;70:171–172. doi: 10.1016/0035-9203(76)90202-9. [DOI] [PubMed] [Google Scholar]
- Lainon R, Shaw JJ, Ward RD, Ready PD, Naiff RD. Leishmaniasis in Brazil: XIII. Isolation of Leishmania from armadillos (Dasypus novemcinctus), and observation on the epidemiology of cutaneous leishmaniasis in north Pará State. Trans R Soc Trop Med Hyg. 1979;73:239–242. doi: 10.1016/0035-9203(79)90225-6. [DOI] [PubMed] [Google Scholar]
- Lainson R, Shaw JJ, Ward RD, Ready PD, Miles M, Póvoa MM. Leishmaniasis in Brazil: XVI. Isolation and identification of Leishmania species from sandflies, wild mammals and man in north Pará State, with particular reference to L. braziliensis guyanensis causative agent of “pian-bois”. Trans R Soc Trop Med Hyg. 1981;75:530–536. doi: 10.1016/0035-9203(81)90192-9. [DOI] [PubMed] [Google Scholar]
- Arias JR, Freitas RA. Sobre os vetores da leishmaniose cutânea na Amazônia central do Brasil. 2. Incidência de flagelados em flebotomíneos selváticos. Acta Amaz. 1978;8:387–396. [Google Scholar]
- Pinheiro FG, Luz SLB, Franco AMR. Infecção natural por tripanosomatídeos (Kinetoplastida: Trypanosomatidae) em Lutzomyia umbratilis (Diptera: Psychodidae) em áreas de leishmaniose tegumentar americana no Amazonas, Brasil. Acta Amaz. 2008;38:165–172. doi: 10.1590/S0044-59672008000100019. [DOI] [Google Scholar]
- Azevedo ACR, Costa SM, Pinto MCG, Souza JL, Cruz HC, Vidal J, Rangel EF. Studies on the sand fly fauna (Diptera: Psychodidae: Phlebotominae) from transmission areas of American Cutaneous Leishmaniasis in state of Acre, Brazil. Mem Inst Oswaldo Cruz. 2008;103:760–767. doi: 10.1590/S0074-02762008000800003. [DOI] [PubMed] [Google Scholar]
- Gil LHS, Araújo MS, Villalobos JM, Camargo LMA, Ozaki LS, Fontes CJF, Ribolla PEM, Katsuragawa TH, Cruz RM, Silva AA, Silva LHP. Species structure of sand fly (Diptera: Psychodidae) fauna in the Brazilian western Amazon. Mem Inst Oswaldo Cruz. 2009;104:955–959. doi: 10.1590/S0074-02762009000700002. [DOI] [PubMed] [Google Scholar]
- Le Pont F, Pajot FX. La leishmaniose em Guyane Française. 1. Étude de l´écologie et du taux d’infection naturelle de Lutzomyia (Nyssomyia) umbratilis Ward et Fraiha, 1977 em saison seche. Considérations épidémiologiques. Cahiers ORSTOM Série Ent Méd et Parasitologie. 1980;18:359–382. [Google Scholar]
- Gentile B, Le Pont F, Pajot FX, Besnard R. Dermal leishmaniasis in French Guiana: the sloth (Choloepus didactylus) as reservoir host. Trans R Soc Trop Med Hyg. 1981;75:612–613. doi: 10.1016/0035-9203(81)90223-6. [DOI] [PubMed] [Google Scholar]
- Pajot FX, Le Pont F, Gentile B, Besnard R. Epidemiology of leishmaniasis in French Guiana. Trans R Soc Trop Med Hyg. 1982;76:112–113. doi: 10.1016/0035-9203(82)90033-5. [DOI] [PubMed] [Google Scholar]
- Feliciangeli MD, Pérez JR, Ramirez A. First Venezuelan record of Lutzomyia umbratilis Ward & Fraiha 1977 (Diptera: Psychodidae), a proven vector of Leishmania braziliensis guyanensis. Trans R Soc Trop Med Hyg. 1985;79:878. doi: 10.1016/0035-9203(85)90150-6. [DOI] [PubMed] [Google Scholar]
- Young DG, Duncan NA. Guide to the identification and geographic distribution of Lutzomyia sandflyies in Mexico, the West Indies, Central and South America (Diptera: Psychodidade) Mem Am Entomol Inst. 1994;54:1–881. [Google Scholar]
- Almeida FB. Flebótomos da Amazônia. I – Sobre a presença de Lutzomyia anduzei (Rozeboom, 1942) no Brasil (Diptera: Psychodidade) Boletim do Instituto Nacional de Pesquisas da Amazônia: Série Patologia Tropical. 1970;3:1–16. [Google Scholar]
- Azevedo ACR, Lainson R, Souza AA, Fé NF, Feliciangeli DM, Menezes CRV, Rangel EF. Comparative studies of populations of Lutzomyia umbratilis (Diptera: Psychodidae) in Brazil and Venezuela. J Med Entomol. 2002;39:587–600. doi: 10.1603/0022-2585-39.4.587. [DOI] [PubMed] [Google Scholar]
- Rozeboom LE. Phlebotomus anduzei, a new phlebotomus from Venezuela. Bol Ent Venez. 1942;1:91–94. In: Floch H, Abonnenc E: Phlébotomos de La Guyane Française. – (X) Sur lês famelles a 5° segment dês palpes court. Description du male de P. anduzei. Inst Pasteur Guyane Territ Inini Publ 1944, 88: 22 pp. [Google Scholar]
- Ward RD, Fraiha H. Lutzomyia umbratilis, a new species of sandfly from Brazil (Diptera: Psychodidae) J Med Entomol. 1977;14:313–317. [Google Scholar]
- Justiniano SCB, Chagas AC, Pessoa FAC, Queiroz RG. Comparative biology of two populations of Lutzomyia umbratilis (Diptera: Psychodidae) of central Amazonia, Brazil, under laboratory conditions. Braz J Biol. 2004;64:227–235. doi: 10.1590/S1519-69842004000200007. [DOI] [PubMed] [Google Scholar]
- Scarpassa VM, Alencar RB. Lutzomyia umbratilis, the Main Vector of Leishmania guyanensis, Represents a Novel Species Complex? PLoS ONE. 2012;7(5):e37341. doi: 10.1371/journal.pone.0037341. doi:10.1371/journal.pone.0037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, de Waard JR. Biological identification through DNA barcode. Proc R Soc Lond B. 2003;270:313–322. doi: 10.1098/rspb.2002.2218. [DOI] [Google Scholar]
- Cognato AI. Standard percent DNA sequence difference for insects does not predict species boundaries. J Econ Entomol. 2006;99:1037–1045. doi: 10.1603/0022-0493-99.4.1037. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PD. DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci USA. 2006;103:968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Wood DM, Jansen DH, Hallwachs W, Hebert PDN. DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera: Tachnidae) are not all generalists. Proc Natl Acad Sci USA. 2007;104:4967–4972. doi: 10.1073/pnas.0700050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio M, Backeljau T, Nevado B, De Meyer M. Comparative performances of DNA barcoding across insets orders. BMC Bioinforma. 2010;11:206. doi: 10.1186/1471-2105-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon SN, Lehr MA, Wilkerson RC, Ruiz JF, Sallum MA, Lima JB, Póvoa MM, Conn JE. Lineage divergence detected in the malaria vector Anopheles marajoara (Diptera: Culicidae) in Amazonian Brazil. Malar J. 2010;9:271. doi: 10.1186/1475-2875-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopes F, Wilkerson RC, Conn JE, Mckeon SN, Levin DM, Quiñones ML, Póvoa MM, Linton YM. DNA barcoding reveals both known and novel taxa in the Albitarsis group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasit Vectors. 2012;5:44. doi: 10.1186/1756-3305-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PD. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Azpurua J, de la Cruz D, Valderama A, Windsor D. Lutzomyia sandfly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis. 2010;4(3):e627. doi: 10.1371/journal.pntd.0000627. doi:10.1371/journal.pntd.0000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra JAO, Talhari S, Paes MG, Garrido M, Talhari JM. Aspectos clínicos e diagnósticos da leishmaniose tegumentar americana em militares simultaneamente expostos à infecção na Amazônia. Rev Soc Bras Med Trop. 2003;36:587–590. doi: 10.1590/S0037-86822003000500008. [DOI] [PubMed] [Google Scholar]
- Ready PD, Day JC, de Souza AA, Rangel EF, Davies CR. Mitochondrial DNA characterization of populations of Lutzomyia whitmani (Diptera: Psychodidae) incriminated in the peri-domestic and silvatic transmission of Leishmania species in Brazil. Bull Entom Res. 1997;87:187–195. doi: 10.1017/S0007485300027346. [DOI] [Google Scholar]
- Lins RMMA, Oliveira SG, Souza NA, Queiroz RG, Justiniano SCB, Ward RD, Kyriacou CP, Peixoto AA. Molecular evolution of the cacophony IVS6 region in sandflies. Insect Mol Biol. 2002;11(2):117–122. doi: 10.1046/j.1365-2583.2002.00315.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial Cytochrome C Oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Librado P, Rozas J. DnaSP v. 5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpassa VM, Conn JE. Molecular differentiation in natural populations of Anopheles oswaldoi sensu lato (Diptera: Culicidae) from the Brazilian Amazon, using sequences of the COI gene from mitochondrial DNA. Genetics Mol Res. 2006;5:493–502. [PubMed] [Google Scholar]
- Justiniano SCB. Biologia comparada de populações de Lutzomyia umbratilis (Diptera: Psychodidae) da Amazônia Central Brasileira. Manaus, Amazonas, Brazil: Doctoral Thesis. Instituto Nacional de Pesquisas da Amazônia; 2004. p. 151. [Google Scholar]
- Moreno M, Bickersmith S, Harlow W, Hildebrandt J, McKeon SN, Silva-do- Nascimento TF, Loaiza JR, Ruiz F, Lourenço-de-Oliveira R, Sallum MAM, Bergo ES, Fritz GN, Wilkerson RC, Linton YM, Juri MJD, Rangel Y, Póvoa MM, Gutiérrez-Builes LA, Correa MM, Conn JE. Phylogeography of the neotropical Anopheles triannulatus complex (Diptera: Culicidae) supports deep structure and complex patterns. Parasit Vectors. 2013;6:47. doi: 10.1186/1756-3305-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular Systematic of Anopheles: From Subgenera to Subpopulations. Annu Rev Entomol. 2003;48:111–139. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malaria J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Velzen RV, Weitschek E, Felici G, Bakker FT. DNA barcoding fo recently diverged species: relative performance of matching methods. PLoS ONE. 2012;7(1):e30490. doi: 10.1371/journal.pone.0030490. doi:10.1371/journal.pone.0030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpassa VM, Conn JE. Mitochondrial DNA detects a complex evolutionary history with Pleistocene Epoch divergence for the Neotropical Malaria Vector Anopheles nuneztovari Sensu Lato. Am J Trop Med Hyg. 2011;85:857–867. doi: 10.4269/ajtmh.2011.11-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variable sites of the 67 haplotypes observed for Lutzomyia umbratilis and Lutzomyia anduzei.